LIFE.PTML Model Development Targeting Calmodulin Pathway Proteins †
Abstract
1. Introduction
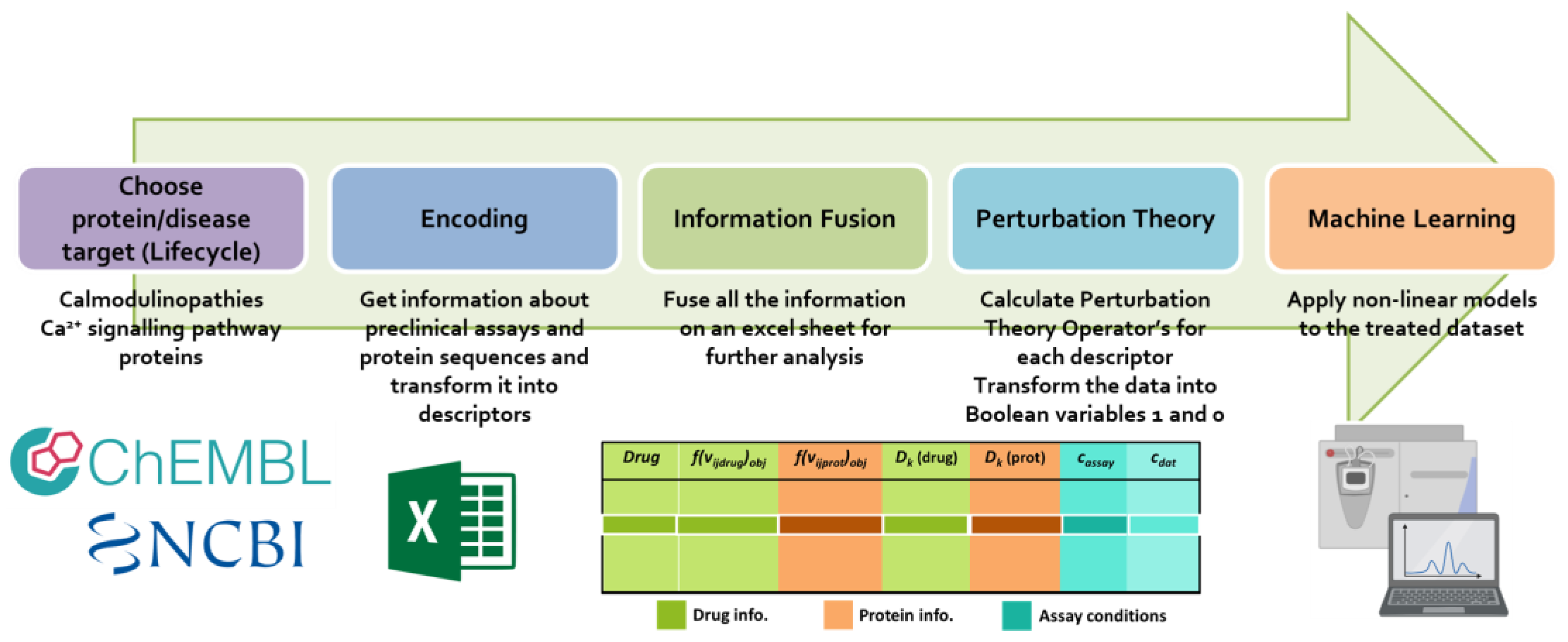
2. Materials and Methods
2.1. Information Fusion
2.2. Perturbation Theory Operator’s Calculation
2.3. Objective and Reference Function Calculation
2.4. Machine Learning Model Development
3. Results and Discussion
3.1. Dataset Construction
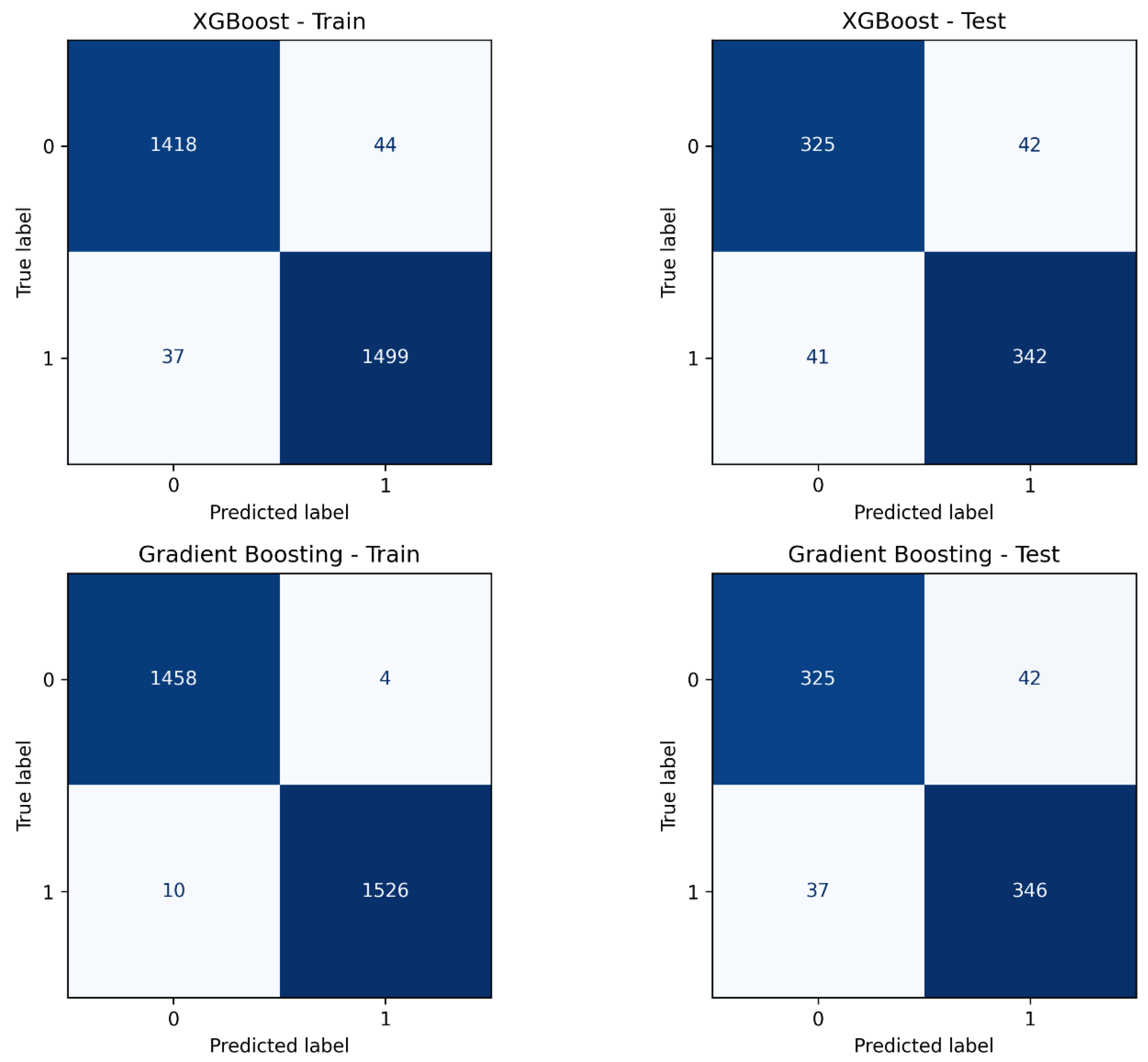
3.2. LIFE.PTML Non-Linear Models
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CaM | Calmodulin |
| E | Encoding Processes |
| EGFR | Epidermal Growth Factor Receptor |
| f(vij)obj | Objective Function: binary label indicating compound activity under specific assay conditions (1 = active, 0 = inactive) |
| f(vij)ref | Reference Function: probability of compound being active under specific boundary conditions (baseline for predictions) |
| IC50 | Half Maximal Inhibitory Concentration |
| Ki | Inhibition Constant |
| KNN | K-Nearest Neighbors |
| LIFEPTML | Life cycle (L) + Information Fusion (IF) + Encoding processes (E) + Perturbation Theory (PT) + Machine Learning (ML) |
| IF | Information Fusion |
| ML | Machine Learning |
| MA | Moving Average |
| PT | Perturbation Theory |
| PTO | Perturbation Theory Operator |
| ROC AUC | Area Under the Receiver Operating Characteristic curve |
| SVM | Support Vector Machine |
References
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the Pharmaceutical Industry: New Estimates of R&D Costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Sadybekov, A.V.; Katritch, V. Computational Approaches Streamlining Drug Discovery. Nature 2023, 616, 673–685. [Google Scholar] [CrossRef] [PubMed]
- García, I.; Fall, Y.; Gómez, G.; González-Díaz, H. First Computational Chemistry Multi-Target Model for Anti-Alzheimer, Anti-Parasitic, Anti-Fungi, and Anti-Bacterial Activity of GSK-3 Inhibitors In Vitro, In Vivo, and in Different Cellular Lines. Mol. Divers. 2011, 15, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Ni, D.; Liu, D.; Zhang, J.; Lu, S. Computational Insights into the Interactions between Calmodulin and the c/nSH2 Domains of P85α Regulatory Subunit of PI3Kα: Implication for PI3Kα Activation by Calmodulin. Int. J. Mol. Sci. 2018, 19, 151. [Google Scholar] [CrossRef]
- Ferreira da Costa, J.; Silva, D.; Caamaño, O.; Brea, J.M.; Loza, M.I.; Munteanu, C.R.; Pazos, A.; García-Mera, X.; González-Díaz, H. Perturbation Theory/Machine Learning Model of ChEMBL Data for Dopamine Targets: Docking, Synthesis, and Assay of New l-Prolyl-l-Leucyl-Glycinamide Peptidomimetics. ACS Chem. Neurosci. J. 2018, 9, 2572–2587. [Google Scholar] [CrossRef]
- He, S.; Nader, K.; Abarrategi, J.S.; Bediaga, H.; Nocedo-Mena, D.; Ascencio, E.; Casanola-Martin, G.M.; Castellanos-Rubio, I.; Insausti, M.; Rasulev, B.; et al. NANO.PTML Model for Read-across Prediction of Nanosystems in Neurosciences. Computational Model and Experimental Case of Study. J. Nanobiotechnol. 2024, 22, 435. [Google Scholar] [CrossRef]
- Baltasar-Marchueta, M.; Llona, L.; M-Alicante, S.; Barbolla, I.; Ibarluzea, M.G.; Ramis, R.; Salomon, A.M.; Fundora, B.; Araujo, A.; Muguruza-Montero, A.; et al. Identification of Riluzole Derivatives as Novel Calmodulin Inhibitors with Neuroprotective Activity by a Joint Synthesis, Biosensor, and Computational Guided Strategy. Biomed. Pharmacother. 2024, 174, 116602. [Google Scholar] [CrossRef]
- Martínez-Arzate, S.G.; Tenorio-Borroto, E.; Barbabosa Pliego, A.; Díaz-Albiter, H.M.; Vázquez-Chagoyán, J.C.; González-Díaz, H. PTML Model for Proteome Mining of B-Cell Epitopes and Theoretical–Experimental Study of Bm86 Protein Sequences from Colima, Mexico. J. Proteome Res. 2017, 16, 4093–4103. [Google Scholar] [CrossRef]
- Zhang, M.; Abrams, C.; Wang, L.; Gizzi, A.; He, L.; Lin, R.; Chen, Y.; Loll, P.J.; Pascal, J.M.; Zhang, J. Structural Basis for Calmodulin as a Dynamic Calcium Sensor. Structure 2012, 20, 911–923. [Google Scholar] [CrossRef]
- Kotta, M.-C.; Sala, L.; Ghidoni, A.; Badone, B.; Ronchi, C.; Parati, G.; Zaza, A.; Crotti, L. Calmodulinopathy: A Novel, Life-Threatening Clinical Entity Affecting the Young. Front. Cardiovasc. Med. 2018, 5, 175. [Google Scholar] [CrossRef]
- O’Day, D.H. Calmodulin Binding Proteins and Alzheimer’s Disease: Biomarkers, Regulatory Enzymes and Receptors That Are Regulated by Calmodulin. Int. J. Mol. Sci. 2020, 21, 7344. [Google Scholar] [CrossRef] [PubMed]
- Mustaly-Kalimi, S.; Gallegos, W.; Marr, R.A.; Gilman-Sachs, A.; Peterson, D.A.; Sekler, I.; Stutzmann, G.E. Protein Mishandling and Impaired Lysosomal Proteolysis Generated through Calcium Dysregulation in Alzheimer’s Disease. Proc. Natl. Acad. Sci. USA 2022, 119, e2211999119. [Google Scholar] [CrossRef] [PubMed]
- Blay, V.; Yokoi, T.; González-Díaz, H. Perturbation Theory–Machine Learning Study of Zeolite Materials Desilication. J. Chem. Inf. Model. 2018, 58, 2414–2419. [Google Scholar] [CrossRef] [PubMed]
- Nocedo-Mena, D.; Cornelio, C.; Camacho-Corona, M.d.R.; Garza-González, E.; Waksman de Torres, N.; Arrasate, S.; Sotomayor, N.; Lete, E.; González-Díaz, H. Modeling Antibacterial Activity with Machine Learning and Fusion of Chemical Structure Information with Microorganism Metabolic Networks. J. Chem. Inf. Model. 2019, 59, 1109–1120. [Google Scholar] [CrossRef]
- Kanehisa-Laboratories. KEGG Kyoto Encyclopedia of Genes and Genomes. Available online: https://www.genome.jp/kegg/ (accessed on 1 July 2025).
- Bento, A.P.; Gaulton, A.; Hersey, A.; Bellis, L.J.; Chambers, J.; Davies, M.; Krüger, F.A.; Light, Y.; Mak, L.; McGlinchey, S.; et al. The ChEMBL Bioactivity Database: An Update. Nucleic Acids Res. 2014, 42, D1083–D1090. [Google Scholar] [CrossRef]
- Papadatos, G.; Overington, J.P. The ChEMBL Database: A Taster for Medicinal Chemists. Future Med. Chem. 2014, 6, 361–364. [Google Scholar] [CrossRef]
- Martí-Carreras, J.; Gener, A.; Miller, S.; Brito, A.; Camacho, C.; Connor, R.; Deboutte, W.; Glickman, C.; Kristensen, D.; Meyer, W.; et al. NCBI’s Virus Discovery Codeathon: Building “FIVE”—The Federated Index of Viral Experiments API Index. Viruses 2020, 12, 1424. [Google Scholar] [CrossRef]
- Morales, J.; Pujar, S.; Loveland, J.E.; Astashyn, A.; Bennett, R.; Berry, A.; Cox, E.; Davidson, C.; Ermolaeva, O.; Farrell, C.M.; et al. A Joint NCBI and EMBL-EBI Transcript Set for Clinical Genomics and Research. Nature 2022, 604, 310–315. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Kim, T.; Park, H. Computational Prediction of Octanol–Water Partition Coefficient Based on the Extended Solvent-Contact Model. J. Mol. Graph. Model. 2015, 60, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Danishuddin; Khan, A.U. Descriptors and Their Selection Methods in QSAR Analysis: Paradigm for Drug Design. Drug Discov. Today 2016, 21, 1291–1302. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.-L. Statistical Analysis: With Business and Economic Applications; Holt, Rinehart and Winston: Fort Worth, TX, USA, 1975. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Bediaga, H.; Arrasate, S.; González-Díaz, H. PTML Combinatorial Model of ChEMBL Compounds Assays for Multiple Types of Cancer. ACS Comb. Sci. J. 2018, 20, 621–632. [Google Scholar] [CrossRef]


| Model | Best Hyperparameters (Randomized/Grid Search) | Train Accuracy | Test Accuracy | Precision | Recall | F1-Score | ROC AUC |
|---|---|---|---|---|---|---|---|
| Random Forest | n_estimators = 610; max_depth = 26; criterion = entropy; max_features = sqrt; min_samples_split = 7; min_samples_leaf = 2 | 0.947 | 0.877 | 0.872 | 0.890 | 0.881 | 0.947 |
| SVM (RBF) | C = 10; gamma = scale; kernel = rbf; degree = 3; probability = True | 0.827 | 0.788 | 0.751 | 0.875 | 0.808 | 0.864 |
| Decision Tree | criterion = entropy; max_depth = 32; min_samples_split = 7; min_samples_leaf = 5; max_features = sqrt | 0.930 | 0.837 | 0.844 | 0.836 | 0.840 | 0.891 |
| KNN | n_neighbors = 15; weights = distance; p = 2 (Euclidean) | 0.996 | 0.833 | 0.827 | 0.851 | 0.839 | 0.912 |
| Gradient Boosting | n_estimators = 310; learning_rate = 0.14; max_depth = 5; subsample = 0.75; max_features = sqrt | 0.995 | 0.895 | 0.892 | 0.903 | 0.898 | 0.958 |
| XGBoost | n_estimators = 160; learning_rate = 0.27; max_depth = 12; subsample = 1.0; colsample_bytree = 0.55; gamma = 1.7; reg_alpha = 0.3; reg_lambda = 1.5 | 0.973 | 0.889 | 0.891 | 0.893 | 0.892 | 0.959 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baltasar-Marchueta, M.; López, N.; Arrasate, S.; Montemore, M.M.; González-Díaz, H. LIFE.PTML Model Development Targeting Calmodulin Pathway Proteins. Chem. Proc. 2025, 18, 38. https://doi.org/10.3390/ecsoc-29-26890
Baltasar-Marchueta M, López N, Arrasate S, Montemore MM, González-Díaz H. LIFE.PTML Model Development Targeting Calmodulin Pathway Proteins. Chemistry Proceedings. 2025; 18(1):38. https://doi.org/10.3390/ecsoc-29-26890
Chicago/Turabian StyleBaltasar-Marchueta, Maider, Naia López, Sonia Arrasate, Matthew M. Montemore, and Humberto González-Díaz. 2025. "LIFE.PTML Model Development Targeting Calmodulin Pathway Proteins" Chemistry Proceedings 18, no. 1: 38. https://doi.org/10.3390/ecsoc-29-26890
APA StyleBaltasar-Marchueta, M., López, N., Arrasate, S., Montemore, M. M., & González-Díaz, H. (2025). LIFE.PTML Model Development Targeting Calmodulin Pathway Proteins. Chemistry Proceedings, 18(1), 38. https://doi.org/10.3390/ecsoc-29-26890







