Abstract
Odorant receptors (ORs) in Drosophila melanogaster represent important proteins of the insect’s olfactory system, enabling the detection of environmental cues such as food sources, host plants, and mating signals. Their modulation by natural ligands offers a sustainable strategy for pest management, particularly through the use of bioactive compounds obtained from agricultural crop and food production residues (ACFPR). In this study, as a model we employed the AlphaFold-predicted structure of the odorant receptor Q9W1P8 for structure-based virtual screening. Molecular docking was carried out using GNINA, a deep learning-enhanced docking tool. Screening of 164 ACFPR-derived compounds from different sources revealed several strong binders, including α-tomatine, peonidin 3-rutinoside, and cinnamtannin B1. Predicted binding modes support the role of plant-derived molecules as candidate modulators of insect olfactory receptors. These findings highlight the utility of integrating AlphaFold models with advanced docking platforms to support the development of sustainable pest management strategies.
1. Introduction
The agricultural sector worldwide is experiencing increasing challenges associated with the growing impact of agricultural pests. Reliance on chemical pesticides as the main control strategy has raised major concerns related to environmental pollution, food chain contamination, and health risks for farmers and consumers alike [1,2]. These limitations emphasize the need to develop more sustainable solutions that can reduce negative impacts while ensuring food security and supporting agricultural growth. The fruit fly, Drosophila melanogaster, is often attracted to overripe or spoiled fruits, a behaviour driven by its complex olfactory system that enables the recognition and discrimination of a broad spectrum of odorants [3,4]. The ability to detect chemical cues from the environment is essential for survival, directing behaviours including foraging, mate selection, and habitat choice [5,6]. Odorant receptors (ORs) are encoded by large, diverse gene families in Drosophila and they are present in the olfactory sensory neurons (OSNs), located primarily on the antennae and maxillary palps [7,8]. Drosophila melanogaster features a comparatively simple olfactory system, containing only 60 odorant receptor (Or) genes, in contrast to the ~1000 identified in mice [9]. D. melanogaster serves as a model organism for olfactory research due to its genetic accessibility and fully sequenced genome [3]. Studies of its odorant receptors provide insights that may support the development of eco-friendly strategies against related pest species such as D. suzukii, which causes severe damage to soft fruit crops [4]. Modulation of the olfactory system of insect pests offers a sustainable approach to disrupt behaviours such as host seeking and mating [7,10]. Essential oils and plant-derived extracts, particularly from agricultural and food production residues, represent promising sources of biodegradable, low-toxicity compounds with insecticidal activity [11,12,13]. These bioactive molecules may also act synergistically across multiple pathways, enabling eco-friendly alternatives that support crop protection and promote a circular economy in agriculture. Herein, our research explores in silico screening of natural compounds from agricultural residues to identify potential modulators of Drosophila melanogaster odorant receptors, as environmentally friendly alternatives to chemical pesticides.
2. Materials and Methods
2.1. Database Search of Odorant Receptor and Structure Retrieval
The UniProt database was queried to identify relevant odorant receptor in D. melanogaster (https://www.uniprot.org/) [14]. UniProt is a freely accessible database that provides a centralized resource of protein sequences and functional information, curating data from many sources. Protein selection was based on the availability of an AlphaFold-predicted model for D. melanogaster already reported in the literature (odorant receptor Or59b, Uniprot ID: Q9W1P8) [15]. The 3D structure of this odour receptor was retrieved as models predicted by AlphaFold [16] and stored in AlphaFold Protein Structure Database [16].
2.2. Molecular Docking Studies
2.2.1. Library Preparation
A small molecule library composed of components from agricultural crop and food production residues (ACFPR) was previously prepared [17]. The SMILES strings of these compounds were retrieved from PubChem database [18], and used to generate the three-dimensional (3D) conformations using Open Babel [19]. Open Babel is a chemical toolbox designed to interconvert between various chemical file formats and to perform tasks like generating 3D structures from 2D representations (SMILES strings). These models included hydrogen atoms to reflect the ionizable groups’ charge state pH 7.4 and were minimized using molecular mechanics and MMFF94 force field implemented in OpenBabel. The final 3D conformations of both sets were saved as mol2 file.
2.2.2. Virtual Screening and Analyses
The blind molecular docking was performed using DockM8 platform [20], employing docking engine GNINA [21], to explore potential binding sites across the entire receptor surface. AutoDock Vina (version 1.1.2) is a widely used open-source programme for molecular docking, known for its speed and accuracy. Specifically, GNINA is a deep learning-enhanced version of Vina. The docking protocol utilized a comprehensive approach to explore protein–ligand interactions, with engine applied to ensure robust and diverse binding pose predictions with high computational exhaustiveness (set to 32) across 28 CPU cores. Protein preparation was conducted using Protoss approach [22], while ligand conformer and protonation state generation was achieved using GypsumDL [23]. GypsumDL is a tool that prepares ligands for docking by generating various 3D conformers and considering different protonation states relevant to the physiological pH, as a ligand’s shape and charge can influence its binding. Binding site identification and validation were carried out using DoGSiteScorer and JAMDA tools implemented in ProteinPlus platform [24].
3. Results and Discussion
3.1. Molecular Docking
3.1.1. GNINA Binding Affinity
The virtual screening was utilized for evaluating the interactions between Alphafold model of odour receptor available in UniProt database (Q9W1P8) and the components of ACFPRs. AlphaFold version 3 is an artificial intelligence system developed by DeepMind that predicts the 3D structure of proteins from their amino acid sequence, significantly advancing our ability to understand protein function. The blind docking results, represented as GNINA binding affinities, showed that the top ten ACFPR molecules exhibited values ranging from −11.5 to −8.5 kcal/mol, highlighting the possibility of favourable ligand–receptor interactions (Table 1).

Table 1.
The top ten compounds from ACFPR sources with top GNINA affinity score against odour receptor target Q9W1P8 from D. Melanogaster obtained using AlphaFold model.
3.1.2. Interactions of ACFPRs with Model Odour Receptor Q9W1P8
Additionally, Figure 1 illustrates the workflow applied to explore ligand–receptor interactions. In part (a), the DoGSiteScorer web server (http://dogsite.zbh.uni-hamburg.de, accessed on 29 July 2025) [25] was used to predict the binding pocket within the odorant receptor Q9W1P8 (yellow surface representation). Into this pocket, the known repellent DEET was docked using AutoDock Vina ver. 1.1.2 software, as shown in part (b). Finally, part (c) demonstrates how a compound derived from agricultural crop and food production residues (peonidin 3-rutinosid) occupied a very similar region to DEET [15], suggesting that both ligands may share comparable binding modes within the receptor.
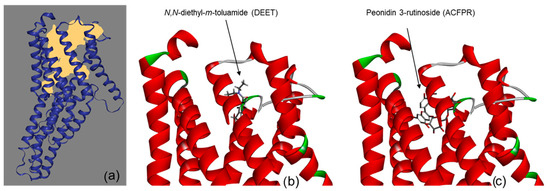
Figure 1.
The predicted binding pocket of odorant receptor Q9W1P8 by DoGSiteScorer tool within ProteinPlus framework (a), docking poses of known repellent N,N-diethyl-m-toluamide (DEET) (b) and ACFPR compound peonidin 3-rutinoside (c).
The molecular docking analysis of the odorant receptor from Drosophila melanogaster with the well-known repellent DEET revealed hydrophobic interactions, primarily involving Trp168 (π–alkyl/alkyl interactions), as well as Val87 and Tyr160 (Figure 2). In contrast, docking with peonidin-3-rutinoside, one of top ten ACFPR compounds according to GNINA affinity, demonstrated a hydrogen bond interaction with Trp168. This suggests a different binding mode, where the polar hydroxyl groups of the glycosylated anthocyanin establish stronger polar contacts with the receptor compared to the predominantly hydrophobic contacts observed for DEET. The involvement of the same key residue (Trp168) in both cases highlights the potential importance of this interaction in ligand recognition by odorant receptor Q9W1P8.
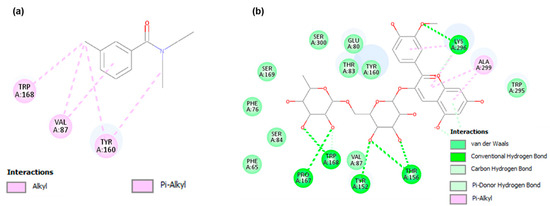
Figure 2.
Two-dimensional ligand-odorant receptor Q9W1P8 interaction diagrams: (a) interaction with the reference repellent DEET; (b) interaction with peonidin-3-rutinoside (ACFPR molecule).
In addition to the presented two-dimensional interaction diagrams, the binding sites of all evaluated ACFPR molecules were analyzed. Figure 3a provides the enlarged view of the binding pocket that had been previously predicted as the preferred site for approximately fifty compounds derived from agricultural crop and food production residues. Within this pocket, peonidin-3-rutinoside is shown in a space-filling CPK (Corey–Pauling–Koltun) representation, highlighting its binding mode relative to the helical structure of the receptor.
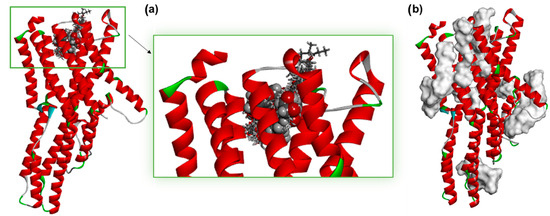
Figure 3.
(a) Previously predicted binding pocket of receptor Q9W1P8 for ~50 ACFPR-derived compounds, showing peonidin-3-rutinoside in CPK representation (enlarged binding pocket). (b) The overlay of top docked poses of all components of ACFPR (grey surface representation) set onto the predicted structure of the odour receptor (ribbon presentation coloured according to secondary structure).
Figure 3b presents the overall binding distribution of all 164 ACFPR compounds, which were found to interact not only within the predicted binding site but also at distinct regions of the receptor. These findings suggest that, in addition to the predicted binding site, the receptor may accommodate ligands at alternative locations, raising the possibility of allosteric modulation.
4. Conclusions
Our results demonstrate that both the reference repellent DEET and the natural compound peonidin-3-rutinoside interact with the odorant receptor Q9W1P8, engaging the same residue (Trp168), but through different interaction types in the binding site. Furthermore, the analysis of all ACFPR compounds revealed binding to alternative receptor surfaces, leading potentially to modulation of OR through allosteric interactions, which should be the subject of further experimental and computational studies.
Author Contributions
Conceptualization, M.Z.; methodology, M.Z. and M.I. (Milena Ivkovic); software, M.Z. and M.I. (Milena Ivkovic); validation, M.Z. and M.I. (Milena Ivkovic); formal analysis, M.I. (Milena Ivkovic), J.N., J.K., M.A., M.I. (Milan Ilic), N.J.L. and M.Z.; investigation, M.I. (Milena Ivkovic), J.N., J.K., M.A., M.I. (Milan Ilic), N.J.L. and M.Z.; resources, M.Z., M.I. (Milan Ilic), and N.J.L.; data curation, M.I. (Milena Ivkovic), J.N., J.K., M.A., M.I. (Milan Ilic), N.J.L. and M.Z.; writing, M.Z. and M.I. (Milena Ivkovic); writing—review and editing, M.I. (Milena Ivkovic), J.N., J.K., M.A., M.I. (Milan Ilic), N.J.L. and M.Z.; visualization, M.Z. and M.I. (Milena Ivkovic); supervision, M.Z.; project administration, M.I. (Milan Ilic); funding acquisition, M.Z., M.I. (Milan Ilic), and N.J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Provincial Secretariat for Higher Education and Scientific Research, Autonomous Province of Vojvodina, Republic of Serbia: 142-451-3008/2023-1.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Deguine, J.-P.; Aubertot, J.-N.; Flor, R.J.; Lescourret, F.; Wyckhuys, K.A.G.; Ratnadass, A. Integrated Pest Management: Good Intentions, Hard Realities. A Review. Agron. Sustain. Dev. 2021, 41, 38. [Google Scholar] [CrossRef]
- Khan, B.A.; Nadeem, M.A.; Nawaz, H.; Amin, M.M.; Abbasi, G.H.; Nadeem, M.; Ali, M.; Ameen, M.; Javaid, M.M.; Maqbool, R.; et al. Pesticides: Impacts on Agriculture Productivity, Environment, and Management Strategies. In Emerging Contaminants and Plants: Interactions, Adaptations and Remediation Technologies; Aftab, T., Ed.; Springer International Publishing: Cham, Switzerland, 2023; pp. 109–134. ISBN 978-3-031-22269-6. [Google Scholar]
- Vosshall, L.B. Olfaction in Drosophila. Curr. Opin. Neurobiol. 2000, 10, 498–503. [Google Scholar] [CrossRef]
- Asplen, M.K.; Anfora, G.; Biondi, A.; Choi, D.-S.; Chu, D.; Daane, K.M.; Gibert, P.; Gutierrez, A.P.; Hoelmer, K.A.; Hutchison, W.D. Invasion Biology of Spotted Wing Drosophila (Drosophila Suzukii): A Global Perspective and Future Priorities. J. Pest Sci. 2015, 88, 469–494. [Google Scholar] [CrossRef]
- Hallem, E.A.; Ho, M.G.; Carlson, J.R. The Molecular Basis of Odor Coding in the Drosophila Antenna. Cell 2004, 117, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Syed, Z.; Pelletier, J.; Flounders, E.; Chitolina, R.F.; Leal, W.S. Generic Insect Repellent Detector from the Fruit Fly Drosophila Melanogaster. PLoS ONE 2011, 6, e17705. [Google Scholar] [CrossRef]
- Venthur, H.; Zhou, J.-J. Odorant Receptors and Odorant-Binding Proteins as Insect Pest Control Targets: A Comparative Analysis. Front. Physiol. 2018, 9, 1163. [Google Scholar] [CrossRef] [PubMed]
- Larter, N.K.; Sun, J.S.; Carlson, J.R. Organization and Function of Drosophila Odorant Binding Proteins. elife 2016, 5, e20242. [Google Scholar] [CrossRef]
- Rollmann, S.M.; Wang, P.; Date, P.; West, S.A.; Mackay, T.F.; Anholt, R.R. Odorant Receptor Polymorphisms and Natural Variation in Olfactory Behavior in Drosophila Melanogaster. Genetics 2010, 186, 687–697. [Google Scholar] [CrossRef]
- Anderson, A.R.; Newcomb, R.D. 20—Olfactory Genomics and Biotechnology in Insect Control. In Insect Pheromone Biochemistry and Molecular Biology, 2nd ed.; Blomquist, G.J., Vogt, R.G., Eds.; Academic Press: London, UK, 2021; pp. 645–674. ISBN 978-0-12-819628-1. [Google Scholar]
- Benbrahim, C.; Barka, M.S.; Basile, A.; Maresca, V.; Flamini, G.; Sorbo, S.; Carraturo, F.; Notariale, R.; Piscopo, M.; Khadir, A.; et al. Chemical Composition and Biological Activities of Oregano and Lavender Essential Oils. Appl. Sci. 2021, 11, 5688. [Google Scholar] [CrossRef]
- Chaudhari, A.K.; Singh, V.K.; Kedia, A.; Das, S.; Dubey, N.K. Essential Oils and Their Bioactive Compounds as Eco-Friendly Novel Green Pesticides for Management of Storage Insect Pests: Prospects and Retrospects. Environ. Sci. Pollut. Res. 2021, 28, 18918–18940. [Google Scholar] [CrossRef] [PubMed]
- Pereira, V.; Figueira, O.; Castilho, P.C. Flavonoids as Insecticides in Crop Protection—A Review of Current Research and Future Prospects. Plants 2024, 13, 776. [Google Scholar] [CrossRef]
- The UniProt Consortium UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [CrossRef]
- Renthal, R. Arthropod Repellent Interactions with Olfactory Receptors and Ionotropic Receptors Analyzed by Molecular Modeling. Curr. Res. Insect Sci. 2024, 5, 100082. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Vidović, S.; Ilić, M.; Nakomčić, J.; Nastić, N.; Kvrgić, J.; Song, X.; Jakimov, D.; Galović, A.J.; Jovanović, N.L.; Zloh, M. Mining Bioactive Components in Agricultural Crop and Food Production Residue for Sustainable Solutions: In Silico Screening for Skin Anti-ageing Properties. Int. J. Cosmet. Sci. 2025, 47, 793–806. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2025 Update. Nucleic Acids Res. 2025, 53, D1516–D1525. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Lacour, A.; Ibrahim, H.; Volkamer, A.; Hirsch, A.K. DockM8: An All-in-One Open-Source Platform for Consensus Virtual Screening in Drug Design. ChemRxiv 2024. [Google Scholar] [CrossRef]
- McNutt, A.T.; Francoeur, P.; Aggarwal, R.; Masuda, T.; Meli, R.; Ragoza, M.; Sunseri, J.; Koes, D.R. GNINA 1.0: Molecular Docking with Deep Learning. J. Cheminform. 2021, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Bietz, S.; Urbaczek, S.; Schulz, B.; Rarey, M. Protoss: A Holistic Approach to Predict Tautomers and Protonation States in Protein-Ligand Complexes. J. Cheminform. 2014, 6, 12. [Google Scholar] [CrossRef]
- Ropp, P.J.; Spiegel, J.O.; Walker, J.L.; Green, H.; Morales, G.A.; Milliken, K.A.; Ringe, J.J.; Durrant, J.D. Gypsum-DL: An Open-Source Program for Preparing Small-Molecule Libraries for Structure-Based Virtual Screening. J. Cheminform. 2019, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Schöning-Stierand, K.; Diedrich, K.; Ehrt, C.; Flachsenberg, F.; Graef, J.; Sieg, J.; Penner, P.; Poppinga, M.; Ungethüm, A.; Rarey, M. Proteins Plus: A Comprehensive Collection of Web-Based Molecular Modeling Tools. Nucleic Acids Res. 2022, 50, W611–W615. [Google Scholar] [CrossRef] [PubMed]
- Volkamer, A.; Kuhn, D.; Grombacher, T.; Rippmann, F.; Rarey, M. Combining Global and Local Measures for Structure-Based Druggability Predictions. J. Chem. Inf. Model. 2012, 52, 360–372. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).