Abstract
Type 2 diabetes mellitus remains a critical metabolic disorder requiring novel therapeutic approaches. In this work, a library of 1-deazapurine derivatives was evaluated as α-glucosidase inhibitors through molecular docking with MOE software. The three top-ranked ligands—Methyl 6-(2-hydroxybenzoyl)-3-(2-phenylethyl)imidazo[4,5-b] pyridine-5-carboxylate (–6.1247 kcal/mol), 5-(furan-2-yl)-3-(4-methoxybenzyl)-2-phenyl-7- (trifluoromethyl)imidazo[4,5-b]pyridine (–5.7030 kcal/mol), and 3-[2-phenylethyl]-5-thio phen-2-yl-7-(trifluoromethyl)imidazo[4,5-b]pyridine (–5.5403 kcal/mol)—were further validated by molecular dynamics simulations. ADMET and drug-likeness predictions confirmed favourable pharmacokinetic behaviour, gastrointestinal absorption, and oral bioavailability. These findings highlight 1-deazapurines as promising scaffolds for developing new α-glucosidase inhibitors targeting type 2 diabetes.
1. Introduction
Type 2 diabetes mellitus (T2DM) is the predominant form of diabetes, characterised by chronic hyperglycaemia that predisposes patients to cardiovascular disease, renal dysfunction, neuropathy, and retinopathy. Global projections indicate that more than 700 million people may be affected by 2045 [1]. Vascular complications contribute significantly to mortality, making T2DM a major public health concern [2].
Among therapeutic strategies, alpha-glucosidase inhibitors (AGIs) are widely used to slow carbohydrate hydrolysis in the gastrointestinal tract, thereby moderating glucose absorption and postprandial glycaemia [3]. Compounds such as acarbose and miglitol are effective and generally safe, with additional cardiovascular benefits reported for acarbose [4]. However, their gastrointestinal side effects often limit patient adherence [5]. Other oral antidiabetic agents, although effective, are associated with hepatic toxicity, hypoglycaemia, or gastrointestinal discomfort after prolonged use [6]. These drawbacks emphasise the demand for alternative molecules with improved tolerability and efficacy.
The 1-deazapurine scaffold (imidazo[4,5-b]pyridine) has gained attention as a pharmacophore with diverse biological properties, including anticancer and anti-inflammatory activity [7]. Its structural analogy with purines explains its broad activity spectrum and highlights its potential as a basis for drug development [8].
In this context, fifteen derivatives of 1-deazapurines were evaluated as potential α-glucosidase inhibitors. Computational techniques—molecular docking, molecular dynamics, and ADMET analyses—were employed to identify ligands with strong binding affinity, assess stability within the enzyme active site, and predict pharmacokinetic suitability compared with existing therapeutic options.
2. Materials and Methods
2.1. Ligand Preparation
Fifteen derivatives of 1-deazapurines (L1–L15) were designed and sketched using MOE 2014 software [9]. Their chemical diversity arises from variations such as methyl, tert-butyl, phenyl, methoxybenzyl, hydroxyphenyl, and thiophenyl substituents placed on the imidazo[4,5-b]pyridine core (Figure 1). Energy minimisation of all ligands was performed using MOPAC with the AM1 method.
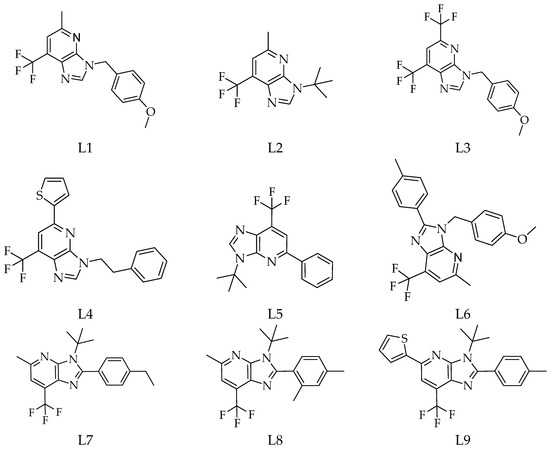
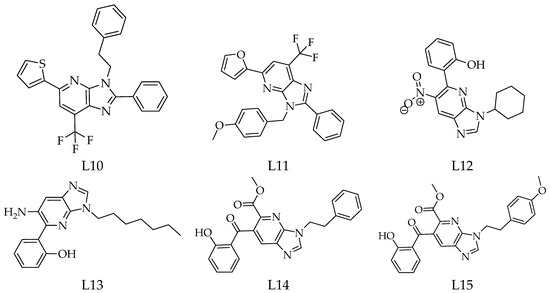
Figure 1.
Chemical structures of the 1-deazapurine derivatives (L1–L15).
2.2. Protein Preparation and Docking Validation
The crystal structure of alpha-glucosidase (PDB ID: 3L4Y; resolution 1.80 Å) was retrieved from the Protein Data Bank. Missing residues were refined with UCSF Modeller [10]. All water molecules and extraneous chains were removed, and the protein was protonated at pH 7.0 before energy minimisation using the MMFF94x force field.
Binding sites were identified with the MOE Site Finder module. Method validation was achieved by re-docking the native ligand (NR4), which reproduced the experimental pose with an RMSD of 1.33 Å, confirming the robustness of the docking protocol [11,12].
2.3. Prediction of Pharmacokinetic Properties
ADMET features were estimated using the pkCSM server [13], covering gastrointestinal absorption, blood–brain barrier passage, cytochrome P450 inhibition, and toxicity endpoints [14]. Drug-likeness was further checked via SwissADME according to Lipinski [15], Veber [16], Ghose [17], Egan [18], and Muegge [19] rules. The BOILED-Egg model predicted both intestinal absorption and brain penetration [20].
2.4. Molecular Dynamics Simulations
The stability of ligand–protein complexes was evaluated through molecular dynamics (MD) simulations. For the best-ranked ligands, trajectories were analysed to assess conformational stability, hydrogen-bond persistence, and dynamic behaviour within the binding pocket [21].
3. Results and Discussion
3.1. Docking Outcomes and Binding Affinities
Molecular docking analyses demonstrated pronounced differences in the binding affinities of the 1-deazapurine derivatives towards α-glucosidase (PDB: 3L4Y). Calculated binding energies ranged from −6.12 to −4.22 kcal/mol, with three compounds—L14, L11, and L4—consistently emerging as the top performers. Among them, L14 (methyl 6-(2-hydroxybenzoyl)-3-(2-phenylethyl)imidazo[4,5-b]pyridine-5-carboxylate) exhibited the most stable complex (−6.12 kcal/mol), followed by L11 (−5.70 kcal/mol) and L4 (−5.54 kcal/mol). These binding energies surpass those of several established α-glucosidase inhibitors reported in the literature [21,22]. Stabilisation of these ligand–enzyme complexes was primarily driven by π–π stacking interactions, extensive hydrogen bonding with key residues, and hydrophobic contacts within the active site.
The validation step, based on re-docking of the native ligand NR4, yielded an RMSD of 1.33 Å, confirming methodological reliability. This agreement ensures that the docking protocol can capture both orientation and binding strength with acceptable accuracy [23].
3.2. Key Interactions at the Binding Pocket
Comprehensive mapping of the binding interactions revealed that the most potent derivatives strategically engaged key residues within the active site. L14, for example, formed robust hydrogen bonds with Met444 and Thr205 (Figure 2), whereas both L11 and L4 established contacts with Asp542; notably, L4 additionally hydrogen-bonded to Gln603, reinforcing its anchoring. Although these contacts lie outside the classical catalytic triad (Asp202, Glu276, Asp340), they contribute substantially to ligand retention and orientation within the pocket and thus to inhibitory potency. By contrast, low-activity compounds such as L2 and L8 were largely confined to superficial, predominantly van der Waals interactions, consistent with their poorer energy scores.
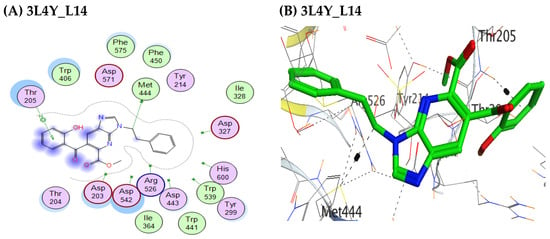
Figure 2.
Representative binding conformation of L14 within the catalytic pocket, showing hydrogen bonds with Met 444 and Thr 205 and hydrophobic contacts with aromatic residues.
Examination of substituent effects clarified structure–activity relationships: phenethyl and furyl substituents (L14, L11) enhance polar contact networks and hydrogen-bonding capacity, while thiophene and phenethyl motifs in L4 promote pronounced π–π stacking with aromatic residues, augmenting hydrophobic stabilisation. These observations demonstrate that subtle electronic and steric modifications to the imidazo[4,5-b]pyridine core can finely tune both enthalpic and entropic components of binding, offering clear leads for rational optimisation of potency and pharmacokinetic profile [24].
3.3. Pharmacokinetic Predictions and ADMET Profile
The pkCSM and SwissADME servers provided valuable insights into the pharmacological behaviour of the designed molecules. Most ligands demonstrated high predicted gastrointestinal absorption and satisfactory permeability across intestinal barriers. Notably, L14 and L11 adhered to both Lipinski and Veber’s rules, suggesting their suitability for oral bioavailability. The BOILED-Egg model positioned L14 and L4 in the “white region”, reflecting good human intestinal absorption and a limited risk of blood–brain barrier penetration—an advantageous property for antidiabetic agents, as central side effects are undesirable [25].
Metabolic stability predictions revealed that several ligands are unlikely to inhibit major cytochrome P450 isoforms (CYP3A4, CYP2D6), thereby reducing the risk of drug–drug interactions. Toxicity analysis further indicated low hepatotoxicity and mutagenic risks for L14 and L11, in contrast to some derivatives bearing bulky aromatic substituents [26]. Overall, the ADMET data reinforce the view that a subset of compounds, notably L14 and L11, combine high enzyme affinity with favourable pharmacokinetic behaviour.
3.4. Molecular Dynamics Simulations
To complement static docking, molecular dynamics (MD) simulations were conducted on the three top-ranked ligands over a 100 ns timescale. Root mean square deviation (RMSD) trajectories revealed that initial deviations in all complexes were followed by rapid stabilisation. The L11 complex remained particularly stable throughout the simulation, and the L14 complex also displayed a stable trajectory, in contrast to L4, which showed more significant fluctuations.
Hydrogen-bond analysis provided further evidence, with L14 maintaining a consistent number of bonds (0–1), while L11 exhibited a slightly higher range (0–2). The radius of gyration (Rg) and solvent-accessible surface area (SASA) values indicated that the ligands did not induce significant structural changes in the protein. Notably, the L11 complex demonstrated the lowest SASA value, suggesting a more compact and less solvent-exposed structure compared to L14 and L4.
Collectively, the MD data validate the docking predictions and reinforce the position of both L14 and L11 as the most promising lead candidates [27].
3.5. Comparative Interpretation with Reference Drug
For benchmarking, the interaction patterns of acarbose, a clinically used α-glucosidase inhibitor, were compared with the best-performing derivatives. While acarbose achieved comparable hydrogen bonding, its bulky saccharide structure rendered its oral absorption prediction unfavourable in pkCSM. In contrast, L14 and L11 combined compact heteroaromatic scaffolds with acceptable ADMET characteristics, making them promising small-molecule alternatives. Furthermore, the binding free energies of L14 (−6.12 kcal/mol) and L11 (−5.70 kcal/mol) were close to or better than reported values for acarbose analogues [28,29]. This highlights the therapeutic potential of imidazo[4,5-b]pyridine scaffolds to serve as next-generation α-glucosidase inhibitors.
3.6. Structure–Activity Relationship Insights
Analysis across the series revealed important structure–activity relationship (SAR) trends. Substitution at the C-6 position with hydroxybenzoyl moieties (as in L14 and L15) enhanced hydrogen bonding and polar interactions, translating into higher affinity. Incorporation of thiophene rings (L4, L9) contributed to π–π stacking but sometimes compromised solubility. Bulky tert-butyl substituents (L2, L5, L7, L8 and L9) tended to reduce activity, possibly due to steric hindrance. The overall pattern confirms that balanced polarity and aromaticity are critical determinants of activity within this scaffold family [30].
3.7. Broader Pharmacological Implications
Beyond their antidiabetic potential, deazapurine derivatives are known to exhibit diverse pharmacological activities. The present results extend their relevance to carbohydrate metabolism, reinforcing the versatility of this scaffold in drug discovery. The relatively simple synthetic accessibility of these compounds, coupled with their favourable pharmacokinetic predictions, provides a strong rationale for further preclinical exploration [15]. Moreover, their compact aromatic frameworks may facilitate derivatisation into multifunctional agents capable of addressing not only glycaemic control but also oxidative stress, which often accompanies diabetes progression [31].
3.8. Limitations and Perspectives
While computational approaches provide valuable insights, certain limitations must be acknowledged. The sample size of 15 derivatives, though adequate for preliminary screening, does not cover the full structural diversity of deazapurines. Moreover, in silico predictions cannot fully substitute for experimental pharmacokinetic and toxicity assessments. Sensitivity of docking scores to algorithmic parameters is another source of uncertainty, although validation with the co-crystallised ligand mitigated this concern. Future work should therefore involve in vitro enzymatic assays to confirm inhibitory activity, followed by in vivo models to assess pharmacodynamics and safety. Extending the structural library with tailored modifications—such as halogen substitutions or fused heterocycles—may further improve potency and selectivity [32].
4. Conclusions
The computational evaluation of 1-deazapurine derivatives against α-glucosidase revealed that a subset of compounds, particularly L14 and L11, exhibit strong binding affinities, stable ligand–enzyme interactions, and favourable pharmacokinetic predictions. Molecular dynamics confirmed the persistence of key hydrogen bonds, while ADMET analyses highlighted their suitability for oral administration with limited toxicity risks. These outcomes suggest that selected deazapurines represent promising scaffolds for the design of novel α-glucosidase inhibitors with potential applications in type 2 diabetes management. Nonetheless, experimental validation remains essential to confirm efficacy and safety, and future optimisation may enhance potency and broaden therapeutic relevance.
Author Contributions
Conceptualization, H.A.; methodology, H.A.; software, F.B.-H., W.S. and S.A.C.; validation, F.B.-H., W.S., S.G. and S.A.C.; formal analysis, F.B.-H.; investigation, F.B.-H., W.S. and S.A.C.; resources, H.A.; data curation, F.B.-H.; writing—original draft preparation, H.A.; writing—review and editing, H.A.; visualization, H.A. and F.B.-H.; supervision, H.A. and F.B.-H.; project administration, H.A.; funding acquisition, H.A. All authors have read and agreed to the published version of the manuscript.
Funding
Supported by the PRFU project B00L01UN130120220004 (Ministry of Higher Education and Scientific Research, Algeria; Abou BekrBelkaïd University, Tlemcen). No other funding was received.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All relevant data are contained within the article.
Acknowledgments
Institutional support from the Ministry of Higher Education and Scientific Research and Abou BekrBelkaïd University, Tlemcen, is gratefully acknowledged.
Conflicts of Interest
No potential conflicts of interest are associated with this publication.
References
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef] [PubMed]
- Pallot, G. Inflammation et maladies métaboliques: Analyse du métabolisme des lipopolysaccharides d’origine intestinale ou péritonéale et de leur prise en charge par les lipoprotéines. Ph.D. Thesis, Université de Bourgogne, Dijon, France, 2021. [Google Scholar]
- Maurya, A.K.; Mulpuru, V.; Mishra, N. Discovery of Novel Coumarin Analogs against the α-Glucosidase Protein Target of Diabetes Mellitus: Pharmacophore-Based QSAR, Docking, and Molecular Dynamics Simulation Studies. ACS Omega 2020, 5, 16700–16716. [Google Scholar] [CrossRef] [PubMed]
- Hanefeld, M.; Cagatay, M.; Petrowitsch, T.; Neuser, D.; Petzinna, D.; Rupp, M. Acarbose reduces the risk for myocardial infarction in type 2 diabetic patients: Meta-analysis of seven long-term studies. Eur. Heart J. 2004, 25, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Khwaja, N.U.D.; Arunagirinathan, G. Efficacy and cardiovascular safety of alpha-glucosidase inhibitors. Curr. Drug Saf. 2021, 16, 122–128. [Google Scholar] [CrossRef]
- Undis, R.; Loizzo, M.R.; Menichini, F. Natural products as α-amylase and α-glucosidase inhibitors and their hypoglycaemic potential in the treatment of diabetes: An update. Mini-Rev. Med. Chem. 2010, 10, 315–331. [Google Scholar] [CrossRef]
- Bavetsias, V.; Sun-Bouloc, C.; Reynisson, N.J.; Workman, P.; Linardopoulos, S.; McDonald, E. Novel purine derivatives as CDK inhibitors. Bioorg. Med. Chem. Lett. 2007, 17, 6567. [Google Scholar]
- Iaroshenko, V.O.; Ali, I.; Mkrtchyan, S.; Semeniuchenko, V.; Ostrovskyi, D.; Langera, P. Transition-metal-catalyzed arylation of 1-deazapurines via C–H bond activation. Synlett 2012, 23, 2603–2608. [Google Scholar] [CrossRef]
- Molecular Operating Environment (MOE); Chemical Computing Group Inc.: Montreal, QC, Canada, 2014.
- Šali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Hevener, K.E.; Zhao, W.; Ball, D.M.; Babaoglu, K.; Qi, J.; White, S.W.; Lee, R.E. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 2009, 49, 444–460. [Google Scholar] [CrossRef]
- Bajda, M.; Więckowska, A.; Hebda, M.; Guzior, N.; Sotriffer, C.A.; Malawska, B. Structure-based search for new inhibitors of cholinesterases. Int. J. Mol. Sci. 2013, 14, 5608–5632. [Google Scholar] [CrossRef]
- Lin, J.; Sahakian, D.C.; de Morais, S.M.; Xu, J.J.; Polzer, R.J.; Winter, S.M. The role of absorption, distribution, metabolism, excretion and toxicity in drug discovery. Curr. Top. Med. Chem. 2003, 3, 1125–1154. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1, A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Kukol, A. Lipid Membranes for Membrane Proteins; Humana Press: New York, NY, USA, 2014; pp. 73–90. [Google Scholar]
- Meng, X.Y.; Zhang, H.X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Kitchen, D.B.; Decornez, H.; Furr, J.R. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.G.; dos Santos, R.N.; Oliva, G. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Ogboye, R.M.; Patil, R.B.; Famuyiwa, S.O.; Faloye, K.O. Novel α-amylase and α-glucosidase inhibitors from selected Nigerian antidiabetic plants: An in silico approach. J. Biomol. Struct. Dyn. 2021, 40, 6340–6349. [Google Scholar] [CrossRef]
- Janiak, C. A critical account on π–π stacking in metal complexes with aromatic nitrogen-containing ligands. J. Chem. Soc. Dalton Trans. 2000, 21, 3885–3896. [Google Scholar] [CrossRef]
- Burley, S.K.; Petsko, G.A. Aromatic–aromatic interaction: A mechanism of protein structure stabilization. Science 1985, 229, 23–28. [Google Scholar] [CrossRef]
- Piovesan, D.; Minervini, G.; Tosatto, S.C.E. The RING 2.0 web server for high-quality residue interaction networks. Nucleic Acids Res. 2016, 44, W367–W374. [Google Scholar] [CrossRef]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Gore, M.; Jagtap, U.B. (Eds.) Computational Drug Discovery and Design; Springer: New York, NY, USA, 2018. [Google Scholar] [CrossRef]
- Kola, I.; Landis, J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004, 3, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Schuster, D.; Laggner, C.; Langer, T. Why drugs fail—A study on side effects in new chemical entities. Curr. Pharm. Des. 2005, 11, 3545–3559. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).