Abstract
Text is the prevalent medium for conveying research findings and developments within and beyond the domain of agriculture. Mining information from text is important for the (research) community to keep track of the most recent developments and identify solutions to major agriculture-related challenges. The task of Named Entity Recognition (NER) can be a first step in such a context. The work presented in this paper relates to a custom NER model for the automated identification and extraction of agricultural terms from text, built on Python’s spaCy library. The model has been trained on a manually annotated text corpus taken from the AGRIS database, and its performance depending on different model configurations is presented. We note that due to the domain ambiguity, inter-annotator agreement and model performance can be improved.
1. Introduction
Named Entity Recognition (NER) or Mention Detection (MD) is a Natural Language Processing (NLP) task that focuses on the identification and extraction of named entities mentioned in text, as well as their classification based on predefined named entity-related categories [1,2]. Depending on the number of the categories required for the classification of the named entity mentions found in text, NER can be considered as a binary or multi-class categorization task. Named entities to be captured may span across one or more word tokens and typically concern person names, names of places and organizations, numerical values indicating order (ordinal values) or quantity, dates, etc. [3]. However, the detection of domain-specific terms is also of importance, and in such cases, NER can be considered a form of the Automatic Term Extraction (ATE) task relating to the use of automated methods for the identification of single-/multi-token domain-specific terms/concepts in text [4,5].
NER can be performed as a standalone task or situated within a broader Information Extraction (IE) context by being the first in a pipeline of NLP tasks for the implementation of semantic text annotation, semantic search, or automatic Knowledge Base (KB) creation and update. The methods for executing NER can be broadly separated into rule-based and statistical-based [1,6]. In the first case, the named entities of interest are detected and classified into the appropriate category based on manually crafted pattern- and/or string-matching rules. In the latter case, named entities are automatically recognized by means of machine trainable language models. The advent of Deep Neural Networks (DNNs) has given rise to the exploitation of statistical NER methods with several pre-trained models being available (e.g., Python’s spaCy, Apache’s OpenNLP, TensorFlow) [3]. Such models have been trained on large, general-purpose text corpora (e.g., news articles), and do not perform well when used to identify domain-specific terms. Customization is needed for the detection of domain-specific terms and this is a custom NER task.
The focus of this paper is on the execution of a custom NER aiming to identify and extract agricultural terms from text. Agriculture is a significant economic sector and it will need to find solutions to the major environmental and societal challenges it faces. Technologies based on NLP and NER can provide helpful, data-driven insights for future research. The identification of agricultural terms in texts is a significant step enabling insights into domain knowledge and contributing to keeping it up to date. This paper focuses on the creation of a custom NER model, based on Python’s spaCy NLP library (https://spacy.io/usage/spacy-101 (accessed on 30 November 2021)), for identifying agricultural terms in texts, and its preliminary evaluation results. Section 2 provides an overview of related work, and Section 3 provides the methods and process involved in the creation of the custom NER model. Section 4 details the results that have been obtained and Section 5 provides some concluding remarks with an emphasis on future research.
2. Related Work
Relatively little work has been done in applying NER to the domain of agriculture. Considerable work on NER has been implemented as part of NLP pipelines in various other disciplines. Beyond the classic work of the 1990s on management succession events, most of the work in this field has been done in the bio-medical domain focusing on the detection of drug and/or disease names, names of active substances used in drugs, disease symptoms, and drug effects by using datasets from various sources. A custom NER tool named Micromed and based on Conditional Random Fields (CRFs) was created in [7]. A set of correctly annotated tweets was the gold standard dataset used. The tool was compared to other existing NER tools (MetaMap and Stanford’s NER tagger) and the F1-scores obtained from the experimentation process ranged from 55% to 66%. A hybrid approach for the identification of bio-medical terms in relevant literature based on a spaCy custom NER model, combined with a dictionary mapping specific entity types to their potential surface forms (i.e., entity type mentions in text), is reported in [8]. The best F1-score-related performance obtained in the experimentation process was 73.79%. A model for custom NER based on transfer learning in combination with a pre-trained language model, trained on a limited amount of texts extracted from electronic health records, is described in [9]. The performance of the proposed model was compared to that of a spaCy-based custom NER model not combined with a language model. Using half of the training data than that required for the spaCy model, the F1-score achieved was 73.4% which was better than the F1-score of the competing model (70.4%). In study [10], a custom NER model based on CRFs has been built to identify agricultural terms in text and classify them in one out of 19 empirically defined entity types. The performance results reported are 84.25% (precision), 79.62% (recall), and 82.23% (F1-score).
3. Methods
3.1. Text Corpus Construction
The process of collecting and annotating text to use it for the training, validation, and testing of our model is shown in Figure 1 below.
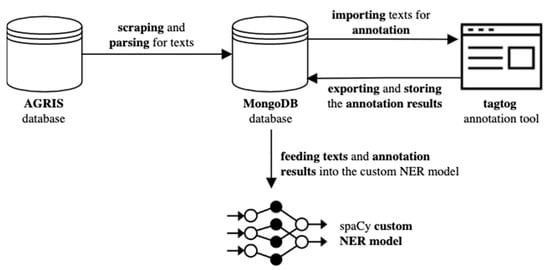
Figure 1.
The process of collecting and annotating text to use it as input for the training, validation, and testing of our model.
For assembling our corpus of agricultural texts, the AGRIS database was scraped and the retrieved HTML parsed to extract content from it. A Python script was used to submit a search query to the AGRIS database by specifying query parameters like the subject and language of the search results, as well as the range of text publication dates. To build our query, “agriculture” was specified as our texts’ subject and “English” as their language. The range of text publication years was set from 2000 to 2021. The webpages were parsed to extract the text and some metadata. The texts were pre-processed to remove any URLs and HTML tags still remaining in them, and stored into a MongoDB (https://www.mongodb.com/ (accessed on 2 December 2021)) database. Each text was then imported into tagtog (https://tagtog.net/ (accessed on 4 December 2021)), the tool used for the annotation of the texts by a team of human annotators. The annotation results (available in JSON and TSV formats) were also stored into the MongoDB database by updating the respective records. The texts and their annotations were used to feed into our custom NER model.
3.2. Dataset
The dataset used for the purpose of training, validating, and testing our custom NER model consisted of 966 agricultural texts. The texts selected for our dataset were abstracts of publications available from the United Nations Food and Agriculture Organization’s (FAO) AGRIS database (https://agris.fao.org/agris-search/index.do (accessed on 9 December 2021)). The total number of words in our texts was 340,379 (our calculations have been based on considering an average length of 5 words per English word). Some quantitative, dataset-related characteristics are shown in Table 1.

Table 1.
Quantitative characteristics of our dataset.
3.3. Model Setup
Python’s spaCy is a popular library for NLP tasks. It allows for building NLP pipelines of pre-trained components (e.g., word and sentence tokenizer, POS tagger, dependency parser, NER model), or training NLP components from scratch. The creation of our custom NER model for identifying agricultural terms in text was based on spaCy’s Tok2Vec (https://spacy.io/api/tok2vec (accessed on 10 December 2021)) (for word token vectorization) and NER (https://spacy.io/api/entityrecognizer (accessed on 10 December 2021)) components. The NER component was trained from scratch. The model has a configuration file specifying the training, validation, and test dataset directories, the Tok2Vec and NER components (Tok2Vec architectures: https://spacy.io/api/architectures (accessed on 10 December 2021); NER architecture: https://spacy.io/api/architectures#parser (accessed on 10 December 2021)), as well as all the training hyperparameters. The custom NER model developed in our work has been based on spaCy’s default Tok2Vec and NER architectures (spacy.Tok2Vec.v2 and spacy.TransitionBasedParser.v2 respectively).
3.4. Model Training, Validation, and Testing
To use our dataset for training, validating and testing our spaCy custom NER model, and evaluating its performance in the automatic identification of agricultural terms in text, a manual annotation task was undertaken. The annotation task was done by a team of five human annotators (all graduates of the Agricultural University of Athens) using the web-based version of the tagtog annotation tool. Each team member was assigned nearly 200 texts and was instructed to identify text spans relating to mentions of agricultural terms, and label them using the “agriculture” entity type defined with the help of the annotation tool. The annotation results include the annotation offsets (i.e., the labeled text span, the starting character of each agricultural term mention, and the entity type) in a JSON format, as well as a mapping of the annotated text spans to their entity type in a TSV format. The dataset was split into training and test texts by using the 80/20 ratio (i.e., 80% of the texts were randomly assigned to the training set and the remaining 20% to the test set). The training set was further divided into training and validation texts using again the 80/20 splitting ratio. From the 966 texts in our dataset, 617 texts were used for training, 155 texts for validation, and 194 texts for testing the model. A computer system having a 6-Core CPU synchronizing at 3 GHz and 16 GBs of DDR4 RAM memory synchronizing at 2667 MHz was used in our work.
4. Results
4.1. Insights into the Manual Annotation of the Dataset
A sample of 29 texts were randomly selected from the entire set of agricultural texts to measure the consensus of the annotation team members regarding their manual annotation of texts, based on the annotation tool’s affordances for calculating Inter Annotator Agreement (IAA). The IAA was 40.75% on average and illustrates the challenges inherent to manual text annotation for identifying agriculture-related terms. As made evident from the obtained results, classifying a text string as an agricultural term depends on annotator decisions about the degree of the text string’s relatedness to the agricultural domain. Thus, there is room for different interpretations (e.g., “climate change” may not be classified as an agricultural term). The detailed IAA results are shown in Table 2 below.

Table 2.
Annotation team member agreement measured on a sample of our dataset’s texts.
4.2. Model Performance
Our experimentation settings have been based on different model configurations related to: (i) the spaCy language model used (namely, “en_core_web_sm” or “en_core_web _lg”) (https://spacy.io/models/en (accessed on 2 December 2021)); (ii) the batch size (64 or 128), and (iii) the learning rate (0.0001, 0.001, or 0.01). For each combination, at least two experiments were conducted based on different training, validation, and test datasets sampled from our set of agricultural texts (created using the 80/20 splitting ratio). Table 3 below shows the min, max, and average precision, recall, and F1-score overall achieved during the model’s testing, as well as their standard deviations.

Table 3.
Precision, recall, and F1-score metric-related values achieved during testing.
The min, max, and average precision, recall, and F1-score achieved within the context of the experimental settings using the “en_core_web_sm” language model, as well as their standard deviations, are reported in Table 4.

Table 4.
Precision, recall and F1-score linked to the en_core_web_sm language model-based settings.
Table 5 shows the results of the experiments based on the use of “en_core_web_lg”.

Table 5.
Precision, recall and F1-score linked to the en_core_web_lg language model-based settings.
Finally, Table 6 below summarizes the best precision, recall, and F1-score values that have been achieved, and the model configurations giving those results.

Table 6.
Best precision, recall, and F1-score values and the associated model configurations.
5. Discussion and Conclusions
The best of the preliminary results achieved as part of our work are close to the range of results reported in [7], yet lower than those in [8,9] and what is the state-of-the-art. In regards to the difference with the results provided in [10], this can be attributed to the different texts used and the fact that in [10], custom NER is implemented as a multi-class categorization task. The results obtained are indicative of the complexities inherent to the development of our model approaching the identification of agricultural terms in text as a binary classification problem. This is also evident from the IAA score achieved. Specifically, the manual classification of a text string as an agricultural term has a great degree of vagueness, and consequently subjectivity, leaving room for different interpretations by human annotators.
Specific steps will be taken as a follow-up to our work for improving both IAA and our model’s performance. A broader dataset, not necessarily limited to AGRIS abstracts, will be used. This way, different contexts of use of agricultural terms will be considered. Another approach will be to use more explicit, granular categories, and thus hope to raise the inter-annotator agreement. This task can be further enhanced by an automated text pre-annotation process based on the use of agriculture-specific, broadly-known controlled vocabularies (e.g., AGROVOC (https://www.fao.org/agrovoc/) (accessed on 2 January 2022)) and ontologies (e.g., FoodOn (https://foodon.org/) (accessed on 2 January 2022)). In addition, we intend to leverage the power of state-of-the-art transformer-based architectures (also supported in spaCy) for creating a model version capable of making predictions with less training data.
Author Contributions
Conceptualization, H.P. and C.B.; methodology, H.P.; formal analysis, B.E.-G.; investigation, H.P.; resources, H.P.; data curation, H.P.; writing—original draft preparation, H.P.; writing—review and editing, C.B.; visualization, H.P.; supervision, C.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially supported by the H2020 EUREKA project agreement no. 862790.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The code developed as part of the work reported in this paper is available on GitHub (https://github.com/herculespan/customNERforAgriEntities (accessed on 12 January 2022)). The text corpus used for the training, validation, and testing of the custom NER model and the manual annotation results are stored in a MongoDB database. Access can be granted upon request to the paper’s lead author.
Acknowledgments
The authoring team would like to deeply thank the members of the annotation team who contributed to the task of manual text annotation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wu, G.; He, Y.; Hu, X. Entity linking: An issue to extract corresponding entity with knowledge base. IEEE Access 2018, 6, 6220–6231. [Google Scholar] [CrossRef]
- Kolitsas, N.; Ganea, O.E.; Hofmann, T. End-to-end neural entity linking. In Proceedings of the 22nd Conference on Computational Natural Language Learning (CoNLL 2018), Brussels, Belgium, 31 October–1 November 2018. [Google Scholar]
- Shelar, H.; Kaur, G.; Heda, N.; Agrawal, P. Named entity recognition approaches and their comparison for custom ner model. Sci. Technol. Libr. 2020, 39, 324–337. [Google Scholar] [CrossRef]
- Zhang, Z.; Iria, J.; Brewster, C.; Ciravegna, F. A Comparative Evaluation of Term Recognition Algorithms. In Proceedings of the Sixth International Conference on Language Resources and Evaluation (LREC’08), Marrakech, Morocco, 26 May–1 June 2008. [Google Scholar]
- Zhang, Z.; Petrak, J.; Maynard, D. Adapted TextRank for Term Extraction: A Generic Method of Improving Automatic Term Extraction Algorithms. Procedia Comput. Sci. 2018, 137, 102–108. [Google Scholar] [CrossRef]
- Popovski, G.; Seljak, B.K.; Eftimov, T. A survey of named-entity recognition methods for food information extraction. IEEE Access 2020, 8, 31586–31594. [Google Scholar] [CrossRef]
- Jimeno-Yepes, A.; MacKinlay, A.; Han, B.; Chen, Q. Identifying Diseases, Drugs, and Symptoms in Twitter. Stud. Health Technol. Inf. 2015, 216, 643–647. [Google Scholar]
- Ramachandran, R.; Arutchelvan, K. Named entity recognition on bio-medical literature documents using hybrid based approach. J. Ambient Intell. Humaniz. Comput. 2021, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tarcar, A.K.; Tiwari, A.; Dhaimodker, V.N.; Rebelo, P.; Desai, R.; Rao, D. Healthcare NER Models Using Language Model Pretraining. In Proceedings of the 13th ACM International WSDM Conference (WSDM 2020), Houston, TX, USA, 3–7 February 2020. [Google Scholar]
- Malarkodi, C.S.; Lex, E.; Devi, S.L. Named Entity Recognition for the Agricultural Domain. Res. Comput. Sci. 2016, 117, 121–132. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).