Drug Resistance Molecular Markers of Plasmodium falciparum and Severity of Malaria in Febrile Children in the Sentinel Site for Malaria Surveillance of Melen in Gabon: Additional Data from the Plasmodium Diversity Network African Network
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Study Population
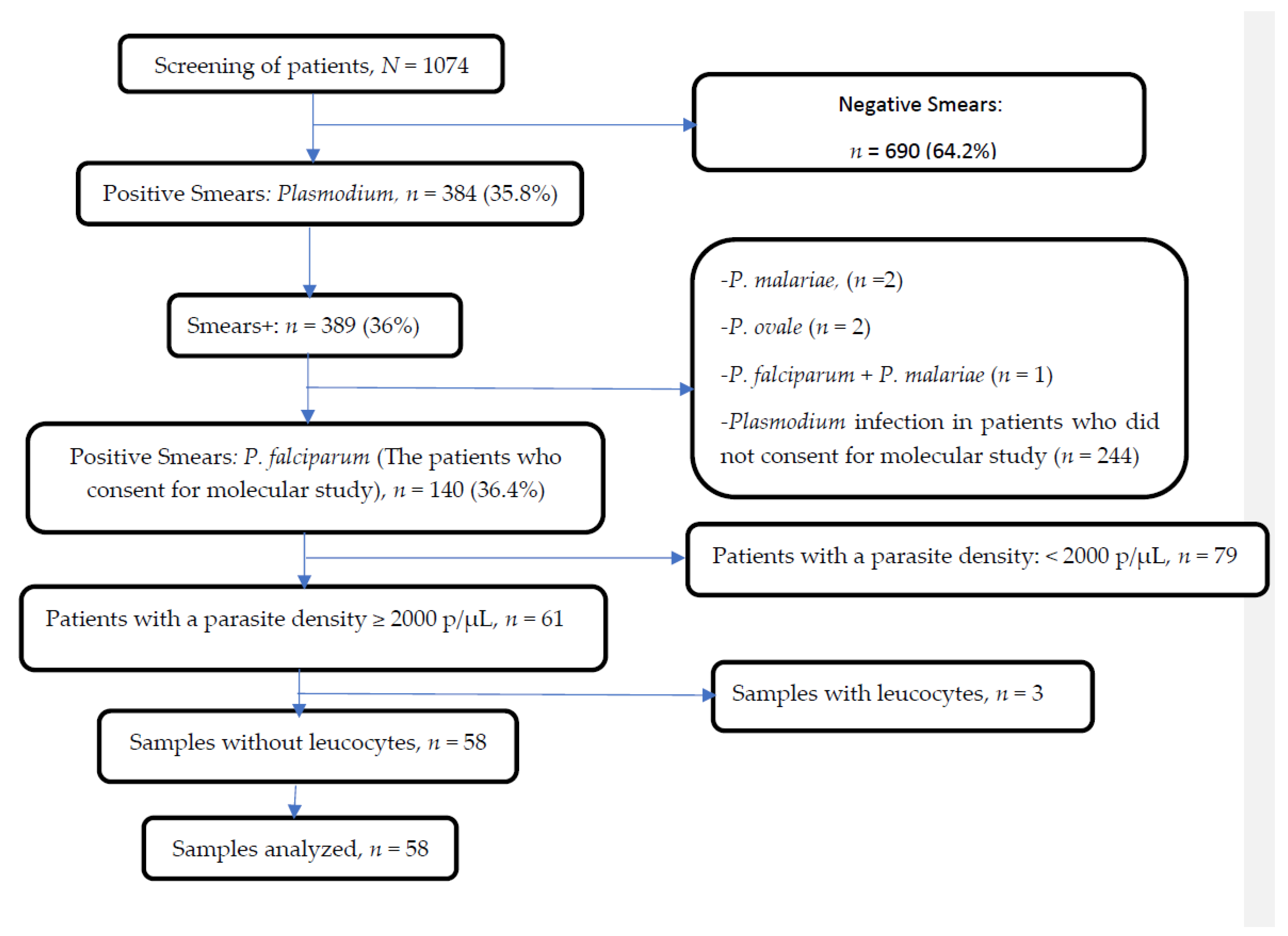
2.3. Malaria Diagnosis
2.4. Leukocyte Depletion
2.5. Molecular Analysis
2.5.1. Genetic Report Card User Guide
2.5.2. Drug Resistance Genes and Haplotypes
2.5.3. Sample Barcodes
2.5.4. Species Co-Infections
2.5.5. Artemisinin Drug Resistance
2.5.6. Chloroquine Drug Resistance
2.5.7. Amodiaquine Drug Resistance
2.5.8. Mefloquine and Lumefantrine Drug Resistance
2.5.9. Piperaquine Drug Resistance
2.6. Ethical Considerations
2.7. Statistical Analysis
3. Results
3.1. Characteristics of the Study Population
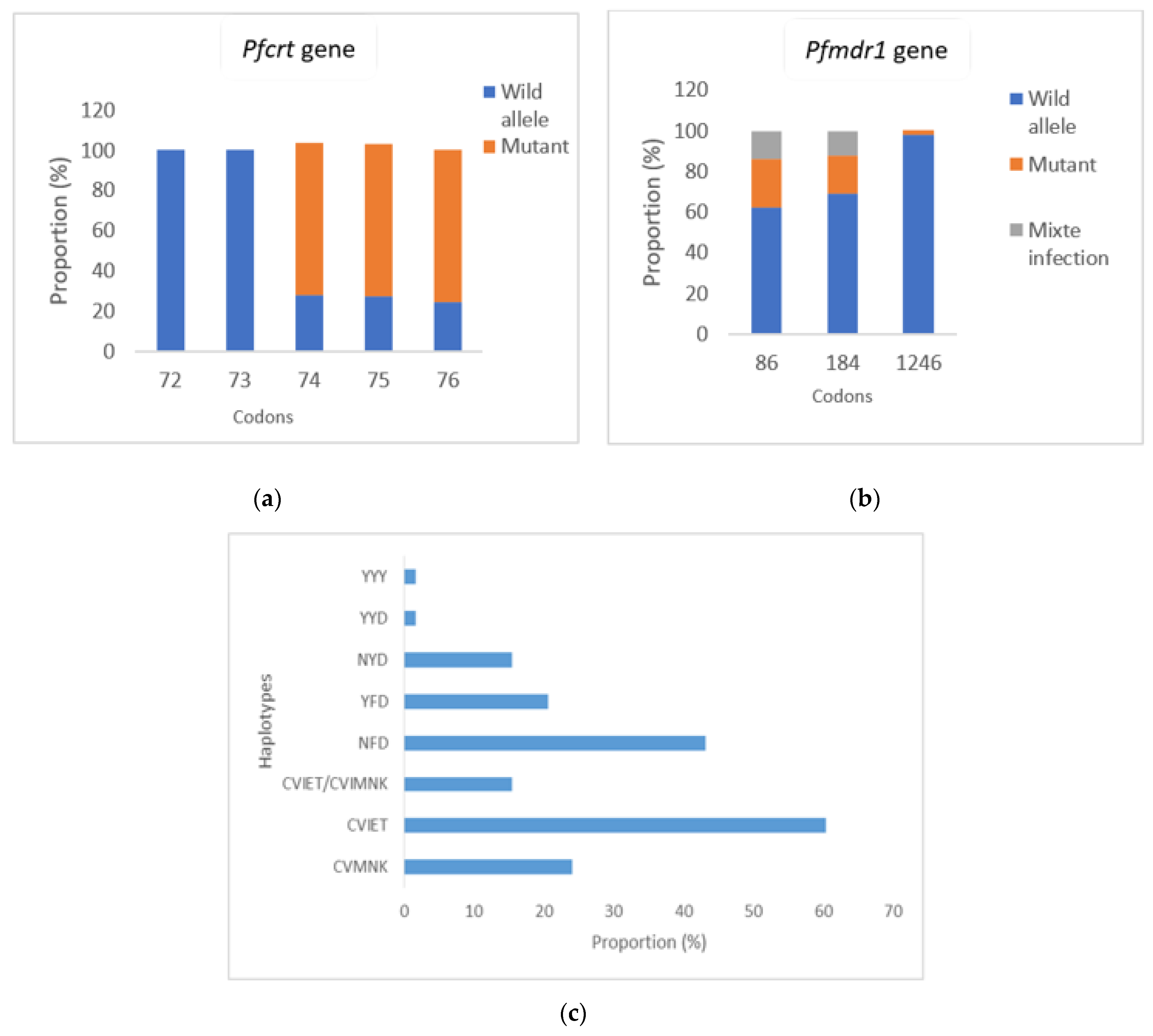
3.2. Artemisinin Resistance Related Molecular Markers Prevalence
3.3. Piperaquine, Amodiaquine and Lumefantrine Resistance Related Molecular Markers Prevalence
3.4. Frequency of Haplotypes According to Malaria Clinical Forms
3.5. Haplotypes and Clinical and Biological Signs of Severe Malaria
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Multiplex | Chromosome | Position | Mutation | Multiplex | Chromosome | Position | Mutation |
|---|---|---|---|---|---|---|---|
| W36960 | 2 | 376222 | K1929E | W36962 | 7 | 704373 | 389E |
| W36960 | 2 | 470013 | G75E | W36962 | 7 | 1066698 | G483S |
| W36960 | 3 | 656861 | 129V | W36962 | 7 | 1213486 | S543N |
| W36960 | 4 | 110442 | G285E | W36962 | 8 | 339406 | 1283C |
| W36960 | 4 | 881571 | 1081R | W36962 | 8 | 701557 | 394G |
| W36960 | 5 | 350933 | 1369N | W36962 | 8 | 1313202 | 799F |
| W36960 | 5 | 369740 | 907L | W36962 | 9 | 452690 | 1018I |
| W36960 | 6 | 900278 | P696S | W36962 | 9 | 599655 | E654D |
| W36960 | 7 | 1044052 | 686K | W36962 | 10 | 1383789 | N114H |
| W36960 | 8 | 413067 | 1044V | W36962 | 10 | 1385894 | 815P |
| W36960 | 8 | 1314831 | 1342K | W36962 | 11 | 1006911 | D124E |
| W36960 | 9 | 900277 | 1534E | W36962 | 11 | 1295068 | E405K |
| W36960 | 11 | 1018899 | 1199L | W36962 | 11 | 1802201 | 450S |
| W36960 | 11 | 1815412 | E765Q | W36962 | 12 | 858501 | Q469K |
| W36960 | 13 | 1056452 | 1234D | W36962 | 12 | 1667593 | 2381N |
| W36960 | 13 | 1466422 | N66K | W36962 | 12 | 1934745 | 241L |
| W36960 | 14 | 137622 | 1179V | W36962 | 13 | 159086 | 21R |
| W36960 | 14 | 2164225 | 2830S | W36962 | 13 | 388365 | S1236R |
| W36961 | 1 | 145515 | 294I | W36962 | 13 | 1419519 | Q208R |
| W36961 | 3 | 548178 | R2L | W36962 | 13 | 2161975 | D252V |
| W36961 | 4 | 139051 | K438N | W36962 | 13 | 2573828 | I1153M |
| W36961 | 4 | 286542 | H586N | W36962 | 14 | 438592 | N348T |
| W36961 | 4 | 529500 | 1477Y | W36962 | 14 | 2625887 | M238I |
| W36961 | 4 | 1102392 | E808D | W36962 | 14 | 3126219 | S628F |
| W36961 | 5 | 796714 | 396K | W36963 | 1 | 179347 | 311G |
| W36961 | 7 | 461139 | M361I | W36963 | 1 | 180554 | D714N |
| W36961 | 7 | 619957 | 675R | W36963 | 1 | 283144 | H664D |
| W36961 | 7 | 1256331 | L321F | W36963 | 1 | 535211 | 2521F |
| W36961 | 8 | 417335 | R244K | W36963 | 2 | 839620 | 260L |
| W36961 | 8 | 417335 | R244K | W36963 | 4 | 426436 | D560A |
| W36961 | 10 | 317581 | 311I | W36963 | 4 | 531138 | A992E |
| W36961 | 10 | 336274 | I1677V | W36963 | 4 | 891732 | R4468S |
| W36961 | 11 | 477922 | H147Y | W36963 | 5 | 172801 | E218K |
| W36961 | 11 | 1020397 | G700E | W36963 | 6 | 574938 | I2934L |
| W36961 | 11 | 1294107 | 84A | W36963 | 7 | 635985 | T598A |
| W36961 | 11 | 1935227 | R73S | W36963 | 7 | 1308383 | G1945R |
| W36961 | 12 | 1663492 | 1014E | W36963 | 7 | 1358910 | K286E |
| W36961 | 12 | 2171901 | V140D | W36963 | 7 | 1359218 | K388N |
| W36961 | 13 | 1233218 | N277S | W36963 | 8 | 150033 | 1315I |
| W36961 | 13 | 1867630 | M4911I | W36963 | 8 | 399774 | 421K |
| W36961 | 13 | 2377887 | 2002S | W36963 | 8 | 1056829 | L474I |
| W36961 | 14 | 2355751 | H1589Q | W36963 | 9 | 1379145 | R398Q |
| W36961 | 14 | 3046108 | 417V | W36963 | 10 | 1386850 | 927K |
| W36962 | 2 | 529709 | F487L | W36963 | 11 | 408668 | 1058I |
| W36962 | 2 | 714480 | D258G | W36963 | 11 | 828596 | K240E |
| W36962 | 3 | 155697 | 150P | W36963 | 11 | 1935031 | I139L |
| W36962 | 4 | 648101 | 51V | W36963 | 12 | 857245 | E50G |
| W36962 | 4 | 1037656 | 2776I | W36963 | 14 | 107014 | 215K |
| W36962 | 5 | 1204155 | 1338I | W36963 | 14 | 1757603 | D1365G |
| W36962 | 6 | 1282691 | 803K | W36963 | 14 | 2733656 | 557C |
| W36962 | 6 | 1289212 | 125T |
| ID | Species | CRT | MDR1 | PGB | EXO |
|---|---|---|---|---|---|
| QW0009-CW | Pf | CVMNK | NYD | VDNTI | E |
| QW0010-CW | Pf | CVIET | YYY | VDNTT | E |
| QW0010-CxW | Pf | CVIET | YFD | VDNTT | E |
| QW0014-CW | Pf | CVMNK | NYD | VDNTI | E |
| QW0016-CW | Pf | CVMNK | NYD | VDNTI | E |
| QW0018-CW | Pf | CVIET,CVMNK | **D | VDNT* | E |
| QW0019-CW | Pf | CVIET,CVMNK | NYD | VDNT* | E |
| QW0020-CW | Pf | CVMNK | NFD | VDNTI | E |
| QW0021-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0022-CW | Pf | CVIET | NYD | VDNTT | E |
| QW0023-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0025-CW | Pf | CVIET | YFD | VDNTT | E |
| QW0027-CW | Pf | CVIET | YYD | VDNTT | E |
| QW0031-CW | Pf | CVIET,CVMNK | YFD | VDNT* | E |
| QW0033-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0035-CW | Pf | CVIET | **D | VDNTT | E |
| QW0038-CW | Pf | CVMNK | YFD | VDNTI | E |
| QW0041-CW | Pf | CVIET | **D | VDNTT | E |
| QW0043-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0046-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0049-CW | Pf | CVIET | YFD | VDNTT | E |
| QW0050-CW | Pf | CVIET | YFD | VDNTT | E |
| QW0057-CW | Pf | CVMNK | NYD | VDNTI | E |
| QW0058-CW | Pf | CVIET | NFD | VDNTI | E |
| QW0064-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0067-CW | Pf | CVMNK | NFD | VDNTI | E |
| QW0071-CW | Pf | CVIET,CVMNK | *FD | VDNT* | E |
| QW0072-CW | Pf | CVIET | N*D | VDNTI | E |
| QW0074-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0076-CxW | Pf | CVIET | NFD | VDNTT | E |
| QW0077-CW | - | - | --- | ----- | - |
| QW0077-CxW | Pf | CVIET | NFD | VDNTT | E |
| QW0081-CW | Pf | CVIET | N*D | VDNTT | E |
| QW0083-CxW | Pf | CVIET | NFD | VDNTT | E |
| QW0084-CxW | Pf | CVIET | YFD | VDNTT | E |
| QW0087-CW | Pf | CVIET | **D | VDNTT | E |
| QW0088-CW | Pf | CVMNK | YFD | VDNTI | E |
| QW0089-CxW | Pf | CVIET | NFD | VDNTT | E |
| QW0090-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0090-CxW | Pf | CVIET | NFD | VDNTT | E |
| QW0091-CxW | Pf | CVIET,CVMNK | **D | VDNTT | E |
| QW0092-CxW | Pf | CVIET | NFD | VDNTT | E |
| QW0096-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0100-CW | Pf | CVMNK | NFD | VDNTI | E |
| QW0101-CW | Pf | CVIET | YFD | VDNTT | E |
| QW0107-CW | Pf | CVMNK | NYD | VDNTI | E |
| QW0109-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0115-CW | Pf | CVMNK | NFD | VDNTI | E |
| QW0118-CW | Pf | CVMNK | YFD | VDNTI | E |
| QW0119-CxW | Pf | CVIET | NYD | VDNTT | E |
| QW0120-CxW | Pf | CVMNK,CVIET | NYD | VDNT* | E |
| QW0125-CxW | Pf | CVIET | YFD | VDNTT | E |
| QW0127-CW | Pf | CVIET,CVMNK | *FD | VDNTT | E |
| QW0127-CxW | Pf | CVIET,CVMNK | YFD | VDNTT | E |
| QW0129-CW | Pf | CVIET | NFD | VDNTT | E |
| QW0139-CxW | Pf | CVIET | *FD | VDNTT | E |
| QW0144-CxW | Pf | CVMNK | NFD | VDNTI | E |
| QW0146-CW | Pf | CVMNK | NFD | VDNTI | E |
| QW0148-CW | Pf | CVMNK,CVIET | NFD | VDNT* | E |
References
- WHO. World Malaria Report 2022; WHO: Geneva, Switzerland, 2022; ISBN 9789240064898.
- Lalloo, D.G.; Shingadia, D.; Bell, D.J.; Beeching, N.J.; Whitty, C.J.M.; Chiodini, P.L. UK Malaria Treatment Guidelines 2016. J. Infect. 2016, 72, 635–649. [Google Scholar] [CrossRef] [PubMed]
- WHO. Word Malaria Report 2021; WHO: Geneva, Switzerland, 2021; ISBN 9789240040496.
- Dondorp, A.M.; Nosten, F.; Yi, P.; Das, D.; Phyo, A.P.; Tarning, J.; Lwin, K.M.; Ariey, F.; Hanpithakpong, W.; Lee, S.J.; et al. Artemisinin Resistance in Plasmodium Falciparum Malaria. N. Engl. J. Med. 2009, 361, 455–467. [Google Scholar] [CrossRef]
- Fairhurst, R.M.; Dondorp, A.M.; Medicine, M.T.; Kingdom, U. Artemisinin-Resistant Plasmodium falciparum Malaria. Microbiol. Spectr. 2016, 4, 1–25. [Google Scholar] [CrossRef]
- Duru, V.; Khim, N.; Leang, R.; Kim, S.; Domergue, A.; Kloeung, N.; Ke, S.; Chy, S.; Eam, R.; Khean, C.; et al. Plasmodium Falciparum Dihydroartemisinin-Piperaquine Failures in Cambodia Are Associated with Mutant K13 Parasites Presenting High Survival Rates in Novel Piperaquine in Vitro Assays: Retrospective and Prospective Investigations. BMC Med. 2015, 13, 305. [Google Scholar] [CrossRef]
- Plasmodium, M. Genomic Epidemiology of Artemisinin Resistant Malaria. Epidemiol. Glob. Health Microbiol. Infect. Dis. 2016. [CrossRef]
- Straimer, J.; Gnädig, N.F.; Witkowski, B.; Amaratunga, C.; Duru, V.; Ramadani, A.P.; Dacheux, M.; Khim, N.; Zhang, L.; Lam, S.; et al. K13-Propeller Mutations Confer Artemisinin Resistance in Plasmodium Falciparum Clinical Isolates. Science 2015, 347, 428–431. [Google Scholar] [CrossRef]
- Ariey, F.; Witkowski, B.; Amaratunga, C.; Beghain, J.; Ma, L.; Lim, P.; Leang, R.; Duong, S.; Sreng, S. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2016, 505, 50–55. [Google Scholar] [CrossRef]
- Miotto, O.; Amato, R.; Ashley, E.A.; Macinnis, B.; Dhorda, M.; Imwong, M.; Woodrow, C.; Manske, M.; Stalker, J. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015, 47, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Nyunt, M.H.; Wang, B.; Aye, K.M.; Aye, K.H.; Han, J.H.; Lee, S.K.; Han, K.T.; Htut, Y.; Han, E.T. Molecular Surveillance of Artemisinin Resistance Falciparum Malaria among Migrant Goldmine Workers in Myanmar. Malar. J. 2017, 16, 97. [Google Scholar] [CrossRef]
- Nyunt, M.H.; Soe, M.T.; Myint, H.W.; Oo, H.W.; Aye, M.M.; Han, S.S.; Zaw, N.N.; Cho, C.; Aung, P.Z.; Kyaw, K.T.; et al. Clinical and Molecular Surveillance of Artemisinin Resistant Falciparum Malaria in Myanmar (2009–2013). Malar. J. 2017, 16, 333. [Google Scholar] [CrossRef]
- Otienoburu, S.D.; Suay, I.; Garcia, S.; Thomas, N.V.; Srisutham, S.; Björkman, A.; Humphreys, G.S. An Online Mapping Database of Molecular Markers of Drug Resistance in Plasmodium Falciparum: The ACT Partner Drug Molecular Surveyor. Malar. J. 2019, 18, 12. [Google Scholar] [CrossRef] [PubMed]
- Wootton, J.C.; Feng, X.; Ferdig, M.T.; Cooper, R.A.; Mu, J.; Baruch, D.I.; Magill, A.J.; Su, X.Z. Genetic Diversity and Chloroquine Selective Sweeps in Plasmodium Falciparum. Nature 2002, 418, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.P.; Stein, W.D.; Lanzer, M. Is PfCRT a Channel or a Carrier? Two Competing Models Explaining Chloroquine Resistance in Plasmodium Falciparum. Trends Parasitol. 2007, 23, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Amato, R.; Lim, P.; Miotto, O.; Neal, A.T.; Sreng, S.; Suon, S.; Drury, E. Europe PMC Funders Group Genetic Markers Associated with Dihydroartemisinin—Piperaquine Failure in Plasmodium Falciparum Malaria in Cambodia: A Genotype-Phenotype Association Study. Lancet Infect. Dis. 2017, 17, 164–173. [Google Scholar] [CrossRef]
- Otienoburu, S.D.; Ascofaré, O.M.; Schramm, B.; Jullien, V.; Jones, J.J.; Zolia, Y.M.; Houzé, P.; Ashley, E.A.; Kiechel, J.R.; Guérin, P.J. Selection of Plasmodium Falciparum Pfcrt and Pfmdr1 Polymorphisms after Treatment with Artesunate—Amodiaquine Fixed Dose Combination or Artemether—Lumefantrine in Liberia. Malar. J. 2016, 15, 452. [Google Scholar] [CrossRef]
- Frank, M.; Lehners, N.; Mayengue, P.I.; Gabor, J.; Dal-bianco, M.; Kombila, D.U.; Ngoma, G.M.; Supan, C.; Lell, B.; Ntoumi, F.; et al. A Thirteen-Year Analysis of Plasmodium Falciparum Populations Reveals High Conservation of the Mutant Pfcrt Haplotype despite the Withdrawal of Chloroquine from National Treatment Guidelines in Gabon. Malar. J. 2011, 10, 304. [Google Scholar] [CrossRef]
- Nzondo, S.M.; Kouna, L.C.; Mourembou, G.; Boundenga, L.; Karl, R.; Limoukou, I.; Matsiegui, P.B.; Zoleko, R.M.; Mbatchi, B.; Raoult, D.; et al. Malaria in Urban, Semi-Urban and Rural Areas of Southern of Gabon: Comparison of the Pfmdr 1 and Pfcrt Genotypes from Symptomatic Children. Malar. J. 2016, 15, 420. [Google Scholar] [CrossRef]
- Mawili-mboumba, D.P.; Akotet, M.K.B.; Kendjo, E.; Nzamba, J.; Medang, M.O. Increase in Malaria Prevalence and Age of at Risk Population in Different Areas of Gabon. Malar. J. 2013, 12, 3. [Google Scholar] [CrossRef]
- Ndong Ngomo, M.J.; Abang Ekouaghe, P.L.; M’boundoukowe, N.P.; Koumba Lengongo, J.V.; Mawili-Mboumba, D.P.; Bouyou-Akotet, M.K. Profil Clinico-Biologique Du Paludisme Grave Dans Un Contexte de Transition Épidémiologique, Congress of the Ivorian Society of Parasitology and Mycology, 2nd ed.; Université Félix Houphouet-Boigny: Abidjan, Ivory Coast, 2016; p. 39. [Google Scholar]
- Mourou, J.R.; Coffinet, T.; Jarjaval, F.; Pradines, B.; Amalvict, R.; Rogier, C.; Kombila, M.; Pagès, F. Malaria Transmission and Insecticide Resistance of Anopheles Gambiae in Libreville and Port-Gentil, Gabon. Malar. J. 2010, 9, 321. [Google Scholar] [CrossRef]
- WHO World Malaria Report 2005 (WHO/HTM/MAL/2005.1102). 2005. Available online: https://www.rbm.who.int/wmr2005/html/1----2.htm (accessed on 16 January 2023).
- Planche, T.; Krishna, S.; Kombila, M.; Engel, K.; Faucher, J.F.; Kremsner, P.G.; Kingdom, U.; Hospital, A.S.; Humanparasitologie, S. Comparison of Methods for the Rapid Laboratory Assessment of Children with Malaria. Am. J. Trop. Med. Hyg. 2001, 65, 599–602. [Google Scholar] [CrossRef]
- Chenet, S.M.; Okoth, S.A.; Kelley, J.; Lucchi, N.; Huber, C.S.; Vreden, S.; Macedo De Oliveira, A.; Barnwell, J.W.; Udhayakumar, V.; Adhin, M.R. Molecular Profile of Malaria Drug Resistance Markers of Plasmodium Falciparum in Suriname. Antimicrob. Agents Chemother. 2017, 61, 1–8. [Google Scholar] [CrossRef]
- Price, R.N.; Cassar, C.; Brockman, A.; Duraisingh, M.; White, N.J.; Nosten, F.; Krishna, S.; Al, P.E.T.; Hemother, A.N.A.G.C. The Pfmdr1 Gene Is Associated with a Multidrug-Resistant Phenotype in Plasmodium Falciparum from the Western Border of Thailand. Antimicrob. Agents Chemother. 1999, 43, 2943–2949. [Google Scholar] [CrossRef] [PubMed]
- Veiga, M.I.; Dhingra, S.K.; Henrich, P.P.; Straimer, J.; Gna, N.; Uhlemann, A.; Martin, R.E.; Lehane, A.M.; Fidock, D.A. Globally Prevalent PfMDR1 Mutations Modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat. Commun. 2016, 7, 11553. [Google Scholar] [CrossRef] [PubMed]
- Fidock, D.A.; Nomura, T.; Talley, A.K.; Cooper, R.A.; Dzekunov, S.M.; Ferdig, M.T.; Ursos, L.M.B.; Sidhu, S.; Deitsch, K.W.; Su, X.; et al. Mutations in the P. falciparum Digestive Vacuole Transmembrane Protein PfCRT and Evidence for Their Role in Chloroquine Resistance. Mol. Cell 2000, 6, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Kamau, E.; Campino, S.; Amenga-etego, L.; Drury, E.; Ishengoma, D.; Johnson, K.; Mumba, D.; Kekre, M.; Yavo, W.; Mead, D.; et al. K13-Propeller Polymorphisms in Plasmodium Falciparum Parasites from Sub-Saharan Africa. J. Infect. Dis. 2015, 211, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Wellems, T.E.; Plowe, C. V Chloroquine-Resistant Malaria; Nature Publishing Group: Berlin, Germany, 1990; pp. 770–776. [Google Scholar]
- Venkatesan, M.; Gadalla, N.B.; Stepniewska, K.; Dahal, P.; Nsanzabana, C.; Moriera, C.; Price, R.N.; Rosenthal, P.J.; Dorsey, G.; Sutherland, C.J.; et al. Polymorphisms in Plasmodium Falciparum Chloroquine Resistance Transporter and Multidrug Resistance 1 Genes: Parasite Risk Factors That Affect Treatment Outcomes for P. falciparum Malaria after Artemether-Lumefantrine and Artesunate-Amodiaquine. Am. J. Trop. Med. Hyg. 2014, 91, 833–843. [Google Scholar] [CrossRef]
- Malmberg, M.; Ferreira, P.E.; Tarning, J.; Ursing, J.; Ngasala, B.; Björkman, A.; Mårtensson, A.; Gil, J.P. Plasmodium Falciparum Drug Resistance Phenotype as Assessed by Patient Antimalarial Drug Levels and Its Association with Pfmdr1 Polymorphisms. J. Infect. Dis. 2013, 207, 842–847. [Google Scholar] [CrossRef]
- Leroy, D.; Macintyre, F.; Adoke, Y.; Ouoba, S.; Barry, A.; Mombo-Ngoma, G.; Ndong Ngomo, J.M.; Varo, R.; Dossou, Y.; Tshefu, A.K.; et al. African Isolates Show a High Proportion of Multiple Copies of the Plasmodium Falciparum Plasmepsin-2 Gene, a Piperaquine Resistance Marker. Malar. J. 2019, 18, 1–11. [Google Scholar] [CrossRef]
- Mawili-Mboumba, D.P.; Ngomo, J.M.N.; Maboko, F.; Guiyedi, V.; Mbina, J.R.M.; Kombila, M.; Akotet, M.K.B. Pfcrt 76T and Pfmdr1 86Y Allele Frequency in Plasmodium Falciparum Isolates and Use of Self-Medication in a Rural Area of Gabon. Trans. R. Soc. Trop. Med. Hyg. 2014, 108, 729–734. [Google Scholar] [CrossRef]
- Zhou, R.; Zhang, H.; Yang, C.; Liu, Y.; Zhao, Y.; Li, S.; Qian, D.; Xu, B. Molecular Mutation Profile of Pfcrt in Plasmodium Falciparum Isolates Imported from Africa in Henan Province. Malar. J. 2016, 15, 265. [Google Scholar] [CrossRef]
- Ishengoma, D.S.; Mandara, C.I.; Francis, F.; Talundzic, E.; Lucchi, N.W.; Ngasala, B.; Kabanywanyi, A.M.; Mahende, M.K.; Kamugisha, E.; Kavishe, R.A.; et al. Efficacy and Safety of Artemether-Lumefantrine for the Treatment of Uncomplicated Malaria and Prevalence of Pfk13 and Pfmdr1 Polymorphisms after a Decade of Using Artemisinin—Based Combination Therapy in Mainland Tanzania. Malar. J. 2019, 18, 88. [Google Scholar] [CrossRef] [PubMed]
- Voumbo-Matoumona, D.F.; Kouna, L.C.; Madamet, M.; Maghendji-Nzondo, S.; Pradines, B.; Lekana-Douki, J.B. Prevalence of Plasmodium Falciparum Antimalarial Drug Resistance Genes in Southeastern Gabon from 2011 to 2014. Infect. Drug Resist. 2018, 11, 1329–1338. [Google Scholar] [CrossRef]
- Koussounda, F.K.; Jeyaraj, S.; Nguetse, C.N.; Nkonganyi, C.N.; Kokou, K.C.; Beka, M.K.E.; Ntoumi, F. Molecular Surveillance of Plasmodium Falciparum Drug Resistance in the Republic of Congo: Four and Nine Years after the Introduction of Artemisinin - Based Combination Therapy. Malar. J. 2017, 16, 155. [Google Scholar] [CrossRef] [PubMed]
- Mbaye, A.; Dieye, B.; Ndiaye, Y.D.; Bei, A.K.; Muna, A.; Deme, A.B.; Yade, M.S.; Diongue, K.; Gaye, A.; Ndiaye, I.M.; et al. Selection of N86F184D1246 Haplotype of Pfmrd1 Gene by Artemether—Lumefantrine Drug Pressure on Plasmodium Falciparum Populations in Senegal. Malar. J. 2016, 15, 433. [Google Scholar] [CrossRef]
- Nguetse, C.N.; Adegnika, A.A.; Agbenyega, T.; Ogutu, B.R.; Krishna, S.; Kremsner, P.G.; Velavan, T.P. Molecular Markers of Anti-Malarial Drug Resistance in Central, West and East African Children with Severe Malaria. Malar. J. 2017, 16, 217. [Google Scholar] [CrossRef] [PubMed]
| Protein | Antimalarial | Gene | Amino-Acid Position | Wild | Mutant | References |
|---|---|---|---|---|---|---|
| PGB Artemisinin genetic background | ART AS AL DHA | Pfarps10 Ferredoxine Pfcrt Pfmdr2 | 127 193 326 356 484 | V D N I T | M Y S T I | [11] [26] [12] |
| EXO | PQ | Exonuclease | 415 | E | [17] | |
| MDR1 | CQ | Pfmdr1 | 86 | N | Y | [27] [28] |
| AQ | 184 | F | Y | |||
| LUM | 1246 | D | Y | |||
| MQ | ||||||
| CRT | CQ AQ | Pfcrt | 72 | C | S | [29] |
| 73 | V | M | ||||
| 74 | M | I | ||||
| 75 | N | E | ||||
| 76 | K | T |
| Characteristics | All Participants N = 58 | Uncomplicated Malaria N = 41 | Complicated Malaria N = 17 | p-Value |
|---|---|---|---|---|
| Median age, months | 69 (66–72) | 48 (40–60) | 60 (36–66) | <0.01 |
| Gender (Male/Female) ratio | 1.23 | 0.48 | 2.6 | <0.01 |
| Median parasite density (µ/L) | 32,200 (25,900–39,200) | 21,000 (19,600–25,200) | 105,700 (87,500–156,100) | <0.01 |
| Mean T °C (±SD) | 37.5 ± 0.77 | 37.8 ± 0.76 | 39.5 ± 0.85 | 0.02 |
| PGB (Aminos Acids) | Pfcrt (Aminos Acids) | Total N = 58 n(%) | Median PD (25–75%) | |
|---|---|---|---|---|
| Haplotype 0 (H0) | VDNTI | CVMNK | 14 (24.2) | 9100 (7700–10,500) |
| Haplotype 1 (H1) | VDNTI | CVIET | 2 (3.4) | 87,675 (4900–170,450) |
| Haplotype 2 (H2) | VDNTT | CVIET | 34 (58.6) | 39,900 (39,200–40,600) |
| Haplotype 3 (H3) | VDNTT/I * | CVIET, CVMNK | 5 (8.6) | 67,200 (42,000–133,000) |
| Haplotype 4 (H4) | VDNTT | CVIET, CVMNK | 3 (5.2) | 49,000 (9100–101,500) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ndong Ngomo, J.M.; Mawili-Mboumba, D.P.; M’Bondoukwé, N.P.; Ditombi, B.M.; Koumba Lengongo, J.V.; Batchy Ognagosso, F.B.; Bouyou-Akotet, M.K. Drug Resistance Molecular Markers of Plasmodium falciparum and Severity of Malaria in Febrile Children in the Sentinel Site for Malaria Surveillance of Melen in Gabon: Additional Data from the Plasmodium Diversity Network African Network. Trop. Med. Infect. Dis. 2023, 8, 184. https://doi.org/10.3390/tropicalmed8040184
Ndong Ngomo JM, Mawili-Mboumba DP, M’Bondoukwé NP, Ditombi BM, Koumba Lengongo JV, Batchy Ognagosso FB, Bouyou-Akotet MK. Drug Resistance Molecular Markers of Plasmodium falciparum and Severity of Malaria in Febrile Children in the Sentinel Site for Malaria Surveillance of Melen in Gabon: Additional Data from the Plasmodium Diversity Network African Network. Tropical Medicine and Infectious Disease. 2023; 8(4):184. https://doi.org/10.3390/tropicalmed8040184
Chicago/Turabian StyleNdong Ngomo, Jacques Mari, Denise Patricia Mawili-Mboumba, Noé Patrick M’Bondoukwé, Bridy Moutombi Ditombi, Jeanne Vanessa Koumba Lengongo, Fanny Bertrande Batchy Ognagosso, and Marielle Karine Bouyou-Akotet. 2023. "Drug Resistance Molecular Markers of Plasmodium falciparum and Severity of Malaria in Febrile Children in the Sentinel Site for Malaria Surveillance of Melen in Gabon: Additional Data from the Plasmodium Diversity Network African Network" Tropical Medicine and Infectious Disease 8, no. 4: 184. https://doi.org/10.3390/tropicalmed8040184
APA StyleNdong Ngomo, J. M., Mawili-Mboumba, D. P., M’Bondoukwé, N. P., Ditombi, B. M., Koumba Lengongo, J. V., Batchy Ognagosso, F. B., & Bouyou-Akotet, M. K. (2023). Drug Resistance Molecular Markers of Plasmodium falciparum and Severity of Malaria in Febrile Children in the Sentinel Site for Malaria Surveillance of Melen in Gabon: Additional Data from the Plasmodium Diversity Network African Network. Tropical Medicine and Infectious Disease, 8(4), 184. https://doi.org/10.3390/tropicalmed8040184






