Abstract
High temperatures are important environmental stressors affecting the metabolism, growth, immunity, and mortality of Chinese mitten crabs (Eriocheir sinensis). In this study, Chinese mitten crabs were divided into two groups and exposed to temperatures of 35 °C (thermal stress group) or 25 °C (control group) for 24 h, and the transcriptome of the heart was analyzed. There were 4007 differentially expressed genes (DEGs) between the thermal stress and the control groups, including 2660 upregulated and 1347 downregulated genes. Heat shock proteins (HSPs) and transcription factors (TFs) were temperature-sensitive DEGs in Chinese mitten crabs. DEGs mainly focused on protein processing in the endoplasmic reticulum, ribosome biogenesis, glycine, serine, and threonine metabolism, protein export, and insect hormone biosynthesis pathways. A total of 28,916 SSRs and 59 TF families, including 851 TFs, were detected among all unigenes of E. sinensis transcripts. The qRT-PCR results for the HSPs and apoptotic DEGs from the heart exhibited the same trends as those in the E. sinensis transcriptome data. Results of light microscopy analyzing histological sections of the heart indicated that most myocardial fibers were lysed, and the number of nuclei and the connective tissue contents between the myocardial layers were both reduced following 35 °C exposure for 24 h.
Key Contribution:
Four thousand and seven DEGs and protein processing in the endoplasmic reticulum pathway are modulated at high temperatures in Chinese mitten crabs, thus indicating their importance in crab adaptation to high-temperature stress.
1. Introduction
Temperature is an important environmental factor that affects the physiological processes of aquatic organisms [1]. With global warming, extremely high temperatures occur in China and other regions during the summer [2]. Aquatic organisms are ectothermic animals whose temperatures fluctuate depending on the water temperature [3]. Heat stress commonly occurs in aquaculture, ultimately resulting in high mortality rates. However, the mechanisms underlying thermal stress in aquatic organisms are not fully understood. When water temperature fluctuates, the synthesis and release of stress hormone-related genes in European sea bass (Dicentrarchus labrax) can be affected [4]. Furthermore, acute or chronic thermal stress can alter the stress axis functions and other stress responses [5]. Once the water temperature exceeds the normal range for crustaceans and fish, their growth, survival, and immunity are negatively affected [6,7,8,9].
The Chinese mitten crab Eriocheir sinensis is one of the most popular crustaceans in China due to its economic and nutritional properties and its pleasant aroma [10]. The production of E. sinensis reached 815,318 tons in 2022 [11]. With the Earth getting warmer in recent years, water temperatures have also increased and this may significantly affect the immunity, metabolism, and survival of E. sinensis. In China, most E. sinensis are cultured in earthen ponds filled with submerged macrophytes (Hydrilla verticillata) and Nuttall’s waterweed (Elodea nuttallii), and the depth of the water is approximately 1.5 m. Chinese mitten crabs rest at the bottom of the pond and are surrounded by aquatic weeds. Although the water temperature on the surface of the pond can reach 37 °C or even higher in the summer, the water temperature at the bottom of the pond surrounding aquatic weeds can be 35 °C or even lower. A previous study demonstrated that E. sinensis could molt normally at 35 °C, and the changes in diel water temperature from 28 °C to 35 °C did not affect molting [12]. Thus, 35 °C can be used as the thermal stress temperature. According to Li et al. [13], immune-related enzymes peaked at 12 h or 24 h and then decreased following thermal stress. When the temperature was elevated from 18 °C to 30 °C, E. sinensis grew faster, the molting period shortened, and the survival rate decreased from 100% to 97.2% [14]. Recent research revealed that all E. sinensis died after 10 min in 40 °C water, while in response to 35 °C water temperature stress, they began to die at 3 d [15]. Thus, water temperature should be controlled in E. sinensis aquaculture during the high-temperature season.
The heart is an important organ in fish and crustaceans and pumps blood or hemolymph containing cytokines, nutrition, and oxygen to the entire body [16]. To adapt to heat or cold stress, heart morphology may change to maintain its function. When male rainbow trout (Oncorhynchus mykiss) were cultured in warm water (20 °C) for eight weeks, the thickness of the compact myocardium increased, and connective tissue contents and spongy myocardium number decreased [17]. Temperature can also alter the heart rate and stroke volume. As the temperature increases, the heartbeat strength decreases and the frequency increases [18,19]. A previous study demonstrated that the stroke volume of Dungeness crabs (Cancer magister) decreased as the temperature changed from 4 °C to 12 °C but remained stable between 12 °C and 20 °C [20].
Transcriptome analysis is widely used to analyze differentially expressed genes (DEGs) in various biological processes [21,22]. A previous study revealed that EsTreh transcript levels are inhibited under hot or cold stress in Chinese mitten crabs [23]. To date, transcriptome analysis in response to thermal stress remains scarce, and DEGs responding to heat stress in the heart require further study. Therefore, it is essential to conduct transcriptome analyses to identify DEGs in response to heat stress.
In this study, the effects of heat stress on the transcriptome of the Chinese mitten crab heart were investigated. Eight DEGs were used to validate the transcriptome results by qRT-PCR, and histological sections of the heart were analyzed.
2. Materials and Methods
2.1. Crabs
The experiment was conducted at the Fisheries Institute, Anhui Academy of Agricultural Sciences, in June 2022. Healthy male E. sinensis individuals (60.1 ± 2.6 g) were obtained by cage traps from our experiment station ponds and acclimated in 20,000 L plastic tanks containing recycling aerated tap water. The temperature of the water was 24 °C, and one-quarter of the water volume was changed daily. The crabs were fed with sinking pellets twice daily. This study was conducted according to the Experimental Animal Welfare and Ethical Review Board of Anhui Academy of Agricultural Sciences guidelines of animals for research (AAAS2022-20).
2.2. Experimental Design and Sampling
After acclimation for seven days, the crabs were randomly divided into two groups, including the thermal stress group (TSG) and the control group (CG), each with three replicate tanks (185 L) with 20 individuals each. The water temperatures in the thermal stress group and the control group were maintained at 35 °C and 25 °C, respectively, using aquarium heaters and air conditioners. Water temperature was increased at a rate of 1 °C/24 h. The temperature of the water in the control group was increased from 24 °C to 25 °C and maintained at 25 °C for 24 h. In the thermal stress group, water temperature was increased from 24 °C to 35 °C and maintained at each integer temperature for 24 h. One-quarter of the water in the six experimental tanks was replaced with same-temperature water each day and aerated continuously. Food was not provided during the study period. The complete hearts of 5 individuals from each tank were collected from the control group and the thermal stress group 24 h after the water temperature reached 25 °C and 35 °C, respectively. Five hearts from each group were separately placed in liquid nitrogen and then stored at −80 °C for transcriptome analysis. The other hearts were placed in 4% paraformaldehyde for histology.
2.3. Total RNA Extraction and cDNA Library Construction
Total RNA was extracted from the hearts of E. sinensis using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and the quantity was assessed using a NanoPhotometer spectrophotometer and agarose gel electrophoresis. RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Oligo(dT) beads were used to extract total RNA. Total mRNA was fragmented using ultrasound, and the fragmented mRNA was reverse-transcribed into first-strand cDNA using M-MuLV reverse transcriptase (Promega, Madison, WI, USA) and random primers (Promega, Madison, WI, USA). Total RNA was then degraded using RNaseH. Second-strand cDNA was synthesized using DNA polymerase I (Promega, Madison, WI, USA), and the double-stranded cDNAs were purified. After adding poly(A), the cDNAs were connected to sequencing adapters and approximately 200 bp ligations were acquired and amplified.
2.4. RNA Sequencing and Transcriptome De Novo Assembly
The amplified cDNA was sequenced using an Illumina sequencing platform at GeneDenovo Biotechnology Co., Ltd. (Guangzhou, China). Fastp (version 0.18.0) [24] was used to filter the original data of adapters, unknown nucleotides exceeding 10%, or low-quality bases (q value ≤ 20) exceeding 50% before assembly and analyses. After filtering, high-quality reads were prepared to assemble a de novotranscriptome using Trinity (version v2.8.4) [25]. BUSCO (version 3) [26] was used to evaluate the completeness of the E. sinensis de novo transcriptome.
2.5. Transcription Factor (TF) and Structure Analysis
Protein-coding sequences in unigenes were aligned to the Animal TFdb (http://www.bioguo.org/AnimalTFDB/, accessed on 2 March 2023) to predict TF families using BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome, accessed on 2 March 2023). Simple sequence repeats (SSR) in the transcriptome were predicted using the MIcroSAtellite (v2.1) (http://pgrc.ipk-gatersleben.de/misa/, accessed on 2 March 2023).
2.6. Correlation and PCA Analysis
Pearson’s correlation coefficient and principal component analysis (PCA) for sample replicates and sample stability were performed using the cor() function [27] in the R environment.
2.7. DEGs and Their Functional Analysis
Unigenes were identified using BLASTx (http://www.ncbi.nlm.nih.gov/BLAST/, accessed on 3 March 2023) with an e-value < 0.00001, and this aligned sequence was queried against databases of nonredundant proteins (Nr) in NCBI, Kyoto Encyclopedia of Genes and Genomes (KEGG), COG/KOG, and Swiss-Prot protein. Protein functional annotation was then conducted according to the best alignment results. Unigenes and their expression levels were calculated and normalized to RPKM (reads per kilobase per million reads) [28]. Differential RNA expression between the thermal stress and the control groups was analyzed using DESeq [29]. The genes with a parameter of false discovery rate (FDR) below 0.05 and absolute fold-change (FC) ≥ 2 were considered to be differentially expressed genes (DEGs). To further analyze the biological functions of the DEGs, all DEGs were blasted to the Gene Ontology (GO) terms in the GO database (http://www.geneontology.org/, accessed on 3 March 2023). Significantly enriched GO terms for the DEGs were analyzed using the Omicsshare platform (https://www.omicshare.com/, accessed on 3 March 2023).
2.8. Quantitative Real-Time PCR (qRT-PCR)
Eight DEGs associated with immunity or apoptosis were selected to verify the RNA-seq results using qRT-PCR. RNA used for heart qRT-PCR was the same as that used for transcriptome analysis. qRT-PCR was performed using a SYBR Premix Ex Taq kit (Invitrogen) according to the manufacturer’s instructions. All reactions were performed in triplicate. β-actin was selected as a housekeeping gene. The relative expression of each DEG was calculated using the 2−ΔΔCt method [30]. The primer sequences for the eight DEGs and β-actin are listed in Table 1.

Table 1.
Primer sequences used for qRT-PCR.
2.9. Analyses of Histological Sections of the Heart
Chinese mitten crab hearts that were used for histological sections were fixed in 4% paraformaldehyde for 12 h, embedded in paraffin wax, and serially sectioned using a microtome (Leica Microsystems, Ankara, Turkey). Cut sections (5 μm thick) were immersed in sequence in Environmentally Friendly Dewaxing Transparent Liquid I (Servicebio, Wuhan, China) for 20 min, Environmentally Friendly Dewaxing Transparent Liquid II (Servicebio, Wuhan, China) for 20 min, Anhydrous ethanol I for 5 min, anhydrous ethanol II for 5 min, 75% ethyl alcohol for 5 min, and finally rinsed with tap water. The sections were then placed in a hematoxylin solution for 3–5 min and then rinsed with tap water. We then placed the sections, in sequence, in 85% ethanol for 5 min, 95% ethanol for 5 min, and eosin dye for 5 min. Sections were then placed in absolute ethanol I for 5 min, absolute ethanol II for 5 min, absolute ethanol III for 5 min, xylene I for 5 min, and xylene II for 5 min. All sections were mounted on glass slides with neutral gum. An Olympus light microscope (BX51) was used to microscopically examine all stained sections, and digital images were photographed using a cellSens Entry (Olympus Corporation, Tokyo, Japan).
3. Results and Discussion
3.1. Transcriptome and Unigene Annotation
When low-quality sequences were removed from the thermal stress group and the control group libraries, the raw reads number ranged from 37.73 M to 55.73 M, the clean reads numbers ranged from37.33 M to 55.48 M, and clean bases ranged from 5.57 G to 8.28 G. Values for Q20 and Q30 of the sequenced libraries ranged from 96.82% to 98.14% and from 91.67% to 94.31% individually, and the GC content ranged from 38.54% to 43.95% (Table S1), thus indicating that the sequence data were high quality and reliable for further study.
High-quality clean sequences were first assembled into contigs, and the assembled contigs were further assembled into unigenes. In this study, 43,606 unigenes were identified. The maximum, minimum, and average unigene lengths were 36,785 bp, 201 bp, and 1190 bp, respectively. The number and length of the N50 for unigenes were 6491 and 2190 bp, respectively. Of all unigenes, 14,800 (33.93%) surpassed 1 kb in length and 7353 (16.86%) surpassed 2 kb in length (Figure S1). In the E. sinensis transcriptomic assembly, BUSCO (Benchmarking Universal Single-Copy Orthologs) analysis revealed 904 (92.4%) complete, 34 (3.5%) fragmented, and 40 (4.1%) missing genes (Table 2).

Table 2.
BUSCO analysis of the Eriocheir sinensis transcriptome.
The sequences of 43,606 unigenes were compared to those of genes in four protein databases: Swiss-Prot, KEGG, KOG, and Nr. There were 11,687 (26.80%), 17,141 (39.31%), 10,395 (23.84%), and 17,048 (39.10%) unigenes annotated using the Swiss-Prot, KEGG, KOG, and Nr databases, respectively (Figure 1). Raw reads from E. sinensis have been deposited in the NCBI database (accession number: PRJNA1064616).

Figure 1.
Distribution of 43,606 E. sinensis unigenes in four protein databases: Swiss-Prot, KEGG, KOG, and Nr.
3.2. SSR
SSR markers are codominant, multi-allelic, and useful tools for genetic, evolutionary, and polymorphism studies [31]. In this study, 28,916 SSRs were detected within E. sinensis unigenes. The most abundant repeat motifs were dinucleotides (13,356, 46.16%), followed by trinucleotides (11,925, 41.08%), tetranucleotides (2834, 9.8%), and hexanucleotides (136, 0.66%). Among the detected SSRs, the AC/GT motif was the most abundant (31.36%), followed by AG/CT (13.34%), AGG/CCT (11.37%), and ACC/GGT (7.32%) motifs (Figure 2).
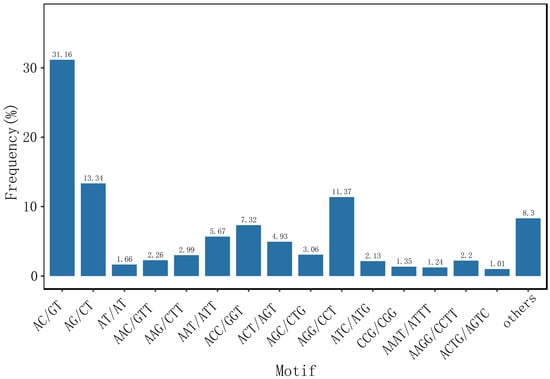
Figure 2.
Frequenciesof detected SSRs in Eriocheir sinensis transcripts.
3.3. TFs
In this study, 59 TF families, including 851 TFs, were detected among all unigenes of E. sinensis transcripts. The most abundant TF family was zf-C2H2 (473), followed by bHLH (35), HMG (30), ZBTB (29), MYB (28), TF_bZIP (24), THAP (23), homeobox (19), GCNF-like (18), and ETS proteins (12).
3.4. DEG Analysis
Genes that were differentially expressed between the thermal stress group (35 °C) and the control group (25 °C) were selected according to p-value < 0.05 and |log2 foldchange| ≥ 1. A total of 4007 DEGs (Table S2) were identified between the two groups, and these included 2660 upregulated and 1347 downregulated genes. When the temperature was elevated to 35 °C for 24 h, DEGs of heat shock proteins and transcription factors were shown to be sensitive to temperature. DEGS mainly focused on protein processing in the endoplasmic reticulum, ribosome biogenesis in eukaryotes, glycine, serine, and threonine metabolism, protein export, and the insect hormone biosynthesis pathway. Molting and metamorphosis are essential processes in the growth and development of Chinese mitten crabs. In insects, molting and metamorphosis are primarily controlled by two hormones: ecdysone and juvenile hormones. Ecdysone and juvenile hormones are conserved between crustaceans and insects [32,33]. In this study, ecdysone and juvenile hormones in the insect hormone biosynthesis pathway were downregulated under thermal stress, possibly due to the observation that more energy was used for survival and not growth. When grass carp (Ctenopharyngodon idellus) were cultured at 34 °C for 48 h and infected with Aeromonas hydrophila, 3355 DEGs in the spleen were identified, including heat shock proteins and immune-related genes [34]. Transcriptome analysis demonstrated that when turbot (Scophthalmus maximus) was maintained at normal temperature (14 °C) or three different high temperatures (20 °C, 25 °C, 28 °C) for 24 h, DEGs were enriched in seven different pathways, and the numbers of DEGs increased with the increase in temperature [35]. After E. sinensis was exposed to hot (32 °C) or cold water (4 °C) for 0, 2, 6, 12, or 24 h, EsTreh expression was gradually downregulated [23].
Heat shock proteins (HSPs) are conserved proteins that are important for protein folding, refolding, translocation, and degradation. HSP expression is regulated by thermal stress [36], pH, ammonia–N stress [37], salinity, and bacteria [38]. In the present study, the DEGs associated with HSPs included HSP60A, HSC70-3, HSP90AA1, HSP22, HSP110, AHSA1, HSPA8, HSPe1, HSPbp1, andenpl-1. HSP60A, HSP90AA1, HSP110, HSP22, Hsc70-3, AHSA1, HSPA8, HSPe1, HSPbp1, and enpl-1 (Table S2) were significantly upregulated (p <0.05) in the thermal stress group, and the log2fold-changes were 4.94, 4.76, 7.61, 4.95, 3.27, 5.32, 7.62, 4.33, 3.65, and 3.14, respectively. According to Oksala et al. [39], HSPs (HSP60, HSP70, HSP90, and HSC70) from doctor fish (Garra rufa) muscle were more highly elevated in high-temperature water (34.4 °C) than in normal-temperature water (25.4 °C). When pool barb (Puntius sophore) were fed curcumin and thermal-treated (36 °C) for 6 h, the expressions of HSP60, HSP70, HSP90, and HSP110 were elevated in the gills, while the expressions of HSP70 and HSP 110 were elevated in the liver [40]. When rainbow trout (Oncorhynchus mykiss) were stressed by high temperature (24 °C), the expressions of HSPa4L, HSPa8a, HSPa5, and HSP70a in the liver and HSPa4, HSPa4L, HSPa5, HSPa8b, HSPa8a, HSPa9 and HSP70a in the head kidney were upregulated compared with levels in the normal temperature group (18 °C) [41]. HSP22 is a small heat shock protein that protects organisms against high temperature [42] and apoptosis [43]. AHSA1 is a cofactor of the HSP90 that can be upregulated by heat stress (24 °C) in rainbow trout [44]. HSPe1 is a member of the HSP10 family and ismore highly expressed in the liver, brain, and head kidney than in the gills, heart, and spleen under heat shock stress [45]. HSPbp1 is an HSP70-binding protein that affects the expression of chaperones by inhibiting HSP70 proteasome degradation and ubiquitylation [46]. HSPbp1 can be significantly upregulated in zebrafish (Danio rerio) during gonadal differentiation when they are exposed to high temperatures (35 °C) [47].
A previous study demonstrated that high temperature upregulates the expression of apoptotic genes [48]. In this study, Cyt-c-p, CYC, and eIF5 (Table S2) were detected as apoptotic DEGs and were significantly upregulated in the thermalstress group, with log2fold-changes of 3.07, 2.09, and 1.62, respectively. Cyt-c-p belongs to the cyt c family of proteins. Deficiency of cyt-c-p in Drosophila can result in embryonic death due to its respiratory function in the mitochondria [49]. Cytc is released from mitochondria and binds to the apoptotic protease activating factor in the cytoplasm to activate the apoptosome [50]. Cytochrome c (CYC) is part of the respiratory chain and can be up-regulated in heart tissue. For example, this can be observed in rainbow trout maintained at 4 °C for four weeks compared with levels in the control group (18 °C) [51]. Eukaryotic translation initiation factor 5 (eIF5) is a GTPase responsible for the initiation of protein translation, and reduced eIF5B expression may disrupt proteostasis and trigger cellular processes associated with stress responses. eIF5 was upregulated when eIF5B was knocked down in 293 T and HepG2 cells using the CRISPR/cas9 system [52].
3.5. PCA
After quality control, Pearson’s correlation analysis was conducted on the samples to compare the thermal stress and control groups, and the results revealed that the samples were reliable (Table S3). PCA demonstrated that five samples in the same group clustered together and that the two groups were clearly separated (Figure 3).

Figure 3.
Principal component analysis of Eriocheir sinensis in the thermal stress group (35 °C) and the control group (25 °C). TSG and CG indicate the thermal stress group and the control group, respectively.
3.6. Functional Annotation
DEGs in the hearts of samples in the thermal stress group were significantly enriched (p < 0.05) for 1377 GO terms. The three most-enriched biological process (BP) items were “cellular process”, “metabolic process”, and “biological regulation”. The three most-enriched cellular component (CC) items were “cellular anatomical entity”, “protein-containing complex”, and “virion component”. A comprehensive sequence and structure analysis of major virion proteins indicate that they evolved on about 20 independent occasions, and in some of these cases, likely ancestors are identifiable among the proteins of cellular organisms. Although the replication modules of at least some classes of viruses might descend from primordial selfish genetic elements, bona fide viruses evolved on multiple independent occasions throughout the course of evolution by the recruitment of diverse host proteins that became major virion components [53]. The three most-enriched molecular function (MF) items were “binding”, “catalytic activity”, and “transporter activity” (Figure 4).
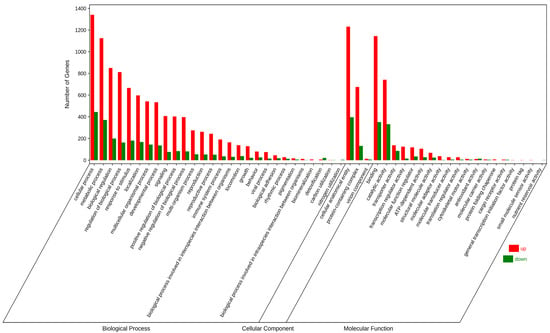
Figure 4.
Gene ontology (GO) terms for DEGs between the thermal stress group (35 °C) and the control group (25 °C).
The top 20 KEGG pathways included protein processing in the endoplasmic reticulum, ribosome biogenesis in eukaryotes, peroxisome, glycine serine and threonine metabolism, aminoacyl-tRNA biosynthesis, arginine and proline metabolism, pyruvate metabolism, N-glycan biosynthesis, and tryptophan metabolism (Figure 5).

Figure 5.
Top 20 KEGG pathways ofDEGs. The x and y axes indicate the percentage of transcripts and pathway names, respectively.
3.7. qRT-PCR
Eight DEGs (Cyt-c-p, eIF5, api5-b, CYC, Hsp60A, Hsc70-5, Lamp1, and Tspan11) involving apoptosis or heat stress proteins were selected for qRT-PCR analysis. The qRT-PCR results revealed that the expression levels of the eight DEGs in the heart (Figure 6B) were in accordance with the results of RNA-Seq analysis (Figure 6A). These results indicated that the E. sinensis transcriptome data were reliable.
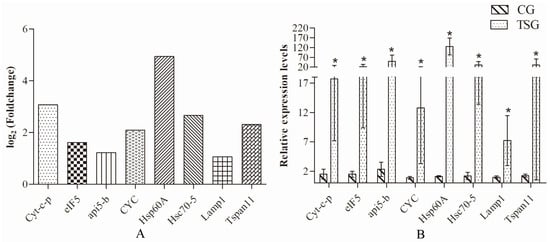
Figure 6.
RNA-seq and qRT-PCR analyses of eight differentially expressed genes (DEGs). (A) fold-change of eight apoptosis or immune-related DEGs between thethermal stress group (35 °C) and the control group (25 °C)based onRNA-Seq analysis. (B) qRT-PCR expression of the Eriocheir sinensis DEGs in the heart; * indicates p < 0.05; TSG and CG represent the thermal stress group (35 °C) and the control group (25 °C), respectively.
3.8. Correlation between RN-seq and qRT-PCR Analyses
The correlation between the RNA-seq and qRT-PCR results for the eight DEGs in E. Sinensis heart tissue was analyzed using the cor() function in R. The R2 values were 0.50, 0.63, 0.47, 0.66, 0.44, 0.63, 0.70, and 0.63 for Cyt-c-p, eIF5, api5-b, CYC, Hsp60A, Hsc70-5, Lamp1, and Tspan11, respectively. All eight DEGs in the E. sinensis heart demonstrated a linear correlation between qRT-PCR and RNA-seq (Figure 7).
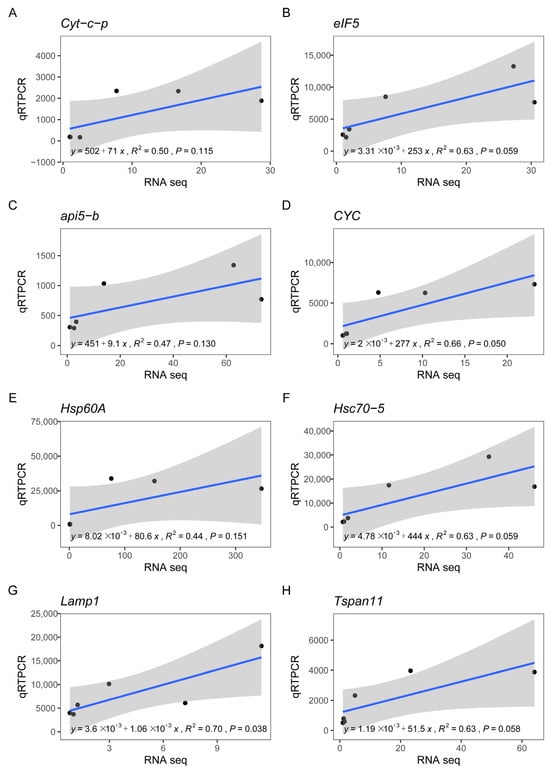
Figure 7.
Correlation between qRT-PCR and RNA-seqresults for the eight DEGs in the Eriocheir sinensis heart. The x axis and y axis indicate RNA-seq and qRT-PCR results, respectively. (A), Cyt-c-p. (B), eIF5. (C), api5-b. (D), CYC. (E), Hsp60A. (F), Hsc70-5. (G), Lamp1. (H), Tspan11.
3.9. Heart Tissue Histology
For male E. sinensis cultured at 25 °C for 24 h, horizontal strips of myocardial fibers and nuclei were clear in histological sections of heart tissue (Figure 8A). Most myocardial fibers were lysed, and the number of nuclei and the connective tissue contents between the myocardial layers reduced after 24 h at 35 °C (Figure 8B). This result indicates that high temperatures damaged myocardial fibers and cells. When rainbow trout were acclimated in warm water (17 °C) for eight weeks, cardiac muscle cross-sectional areas decreased 0.8-fold and the thickness of the myocardium increased [17].
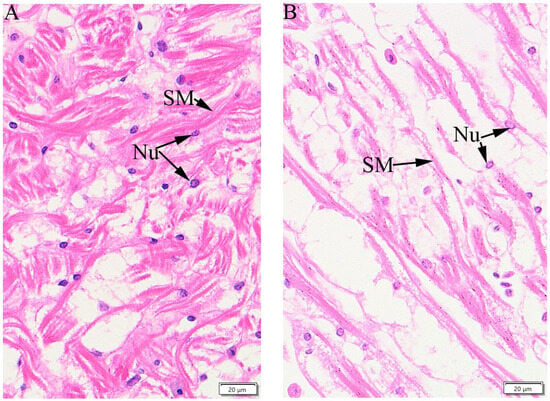
Figure 8.
Light microscopic image of a stained Eriocheir sinensis histological cardiac section (HE-staining). (A), crab myocardium at 25 °C, horizontal strips of myocardial fiber and nuclei are clear; (B), crab myocardium at 35 °C, some myocardial fibers appear degraded, and horizontal strips are not clear. Bar markers = 20 μm in (A,B). SM: striated muscle; Nu: nuclei.
4. Conclusions and Prospects
In conclusion, transcriptome data from the hearts of male E. sinensis were analyzed at 35 °C and 25 °C. There were 4007 DEGs between the thermal stress group and the control group, and these included 2660 upregulated and 1347 downregulated genes. Heart DEGs in the thermal stress group were significantly enriched for 1377 GO terms, and the top 20 KEGG pathways included protein processing in the endoplasmic reticulum. Transcriptome quality, SSR, and TFs were also analyzed. RNA-seq and qRT-PCR revealed a significant (p < 0.05) correlation for the eight DEGs. This study demonstrated that DEGs of heat shock proteins, transcription factors, and pathways of protein processing in the endoplasmic reticulum were sensitive to high temperatures in male E. sinensis. These results will contribute to our understanding of the adaptation of Chinese mitten crabs to high temperatures.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes9030092/s1, Table S1: Quality of heart transcriptome in thermal stress group (35 °C) (TSG) and the control group (25 °C) (CG).; Table S2: DEGs identified between thermal stress group (35 °C) and the control group (25; Table S3: Sample correlation between the thermal stress group (35 °C) and the control group (25 °C). Figure S1: Length distribution of the assembled unigenes.
Author Contributions
T.P.: methodology, data curation, and writing—original draft preparation. T.L.: methodology and writing—review and editing. M.Y. and H.J.: writing—review and editing. J.L.: project administration, supervision, writing—review and editing. Q.G.: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Ministry of Agriculture and Rural Affairs of the People’s Republic of China earmarked fund (grant number: CARS-48) and Anhui Province innovation platform of major science and technology projects (grant number: S202305a12020001).
Institutional Review Board Statement
All crab experiments were conducted under the national regulations on laboratory animals of China and approved by the Experimental Animal Welfare and Ethical Review Board of Anhui Academy of Agricultural Sciences guidelines of use of animals for research (Approval Code: AAAS2022-20).
Data Availability Statement
All the data generated or used during the study appear in the submitted article.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of the work described in this manuscript.
References
- Bowden, T. Modulation of the immune system of fish by their environment. Fish Shellfish. Immunol. 2008, 254, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zeng, G.; Yang, X.; Jiang, Z. Future changes in extreme high temperature over china at 1.5 C–5 C global warming based on cmip6 simulations. Adv. Atmos. Sci. 2021, 38, 253–267. [Google Scholar] [CrossRef]
- Gotthard, K. Growth strategies of ectothermic animals in temperate environments. Environ. Anim. Dev. 2001, 287–304. Available online: https://xueshu.baidu.com/usercenter/paper/show?paperid=5ca32de23cc3ed5113925ea0fe9c0aa7&site=xueshu_se&hitarticle=1 (accessed on 8 January 2024).
- Goikoetxea, A.; Sadoul, B.; Blondeau-Bidet, E.; Aerts, J.; Blanc, M.-O.; Parrinello, H.; Barrachina, C.; Pratlong, M.; Geffroy, B. Genetic pathways underpinning hormonal stress responses in fish exposed to short-and long-term warm ocean temperatures. Ecol. Indic. 2021, 120, 106937. [Google Scholar] [CrossRef]
- Alfonso, S.; Gesto, M.; Sadoul, B. Temperature increase and its effects on fish stress physiology in the context of global warming. J. Fish Biol. 2021, 98, 1496–1508. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.-b.; Su, Y.q.; Mao, Y.; You, X.-x.; Ding, S.; Wang, J. Dietary supplementation with bacillus can improve the growth and survival of the kuruma shrimp Marsupenaeus japonicus in high-temperature environments. Aquac. Int. 2013, 22, 607–617. [Google Scholar] [CrossRef]
- Sanda, T.; Shimizu, T.; Iwasaki, T.; Dan, S.; Hamasaki, K. Effect of temperature on survival, intermoltperiod, and growth of juveniles of two mud crab species, Scylla paramamosain and Scylla serrata (Decapoda: Brachyura: Portunidae), under laboratory conditions. Nauplius 2022, 30, e2022012. [Google Scholar] [CrossRef]
- Yu, K.; Shi, C.; Ye, Y.; Li, R.; Mu, C.; Ren, Z.; Wang, C. The effects of overwintering temperature on the survival of female adult mud crab, Scylla paramamosain, under recirculating aquaculture systems as examined by histological analysis of the hepatopancreas and expression of apoptosis-related genes. Aquaculture 2022, 565, 739080. [Google Scholar] [CrossRef]
- Zarco-Perello, S.; Pratchett, M.; Liao, V. Temperature-growth performance curves for a coral reef fish, Acanthochromis polyacanthus. Galaxea 2012, 14, 97–103. [Google Scholar] [CrossRef][Green Version]
- Chen, D.-W.; Zhang, M.; Shrestha, S. Compositional characteristics and nutritional quality of Chinese mitten crab (Eriocheir sinensis). Food Chem. 2007, 103, 1343–1349. [Google Scholar] [CrossRef]
- China Fishery Bureau. China Fisheries Yearbook; Chinese Agriculture Express: Beijing, China, 2023; p. 24. Available online: https://www.doc88.com/p-99029850248323.html (accessed on 8 January 2024).
- Zhang, T.; Li, T. Ecological observations on molting of juveniles of the Chinese mitten crab, Eriocheir sinensis. J. Lake Sci. 1999, 11, 333–337. [Google Scholar]
- Li, Z.; Zhao, Z.; Luo, L.; Wang, S.; Zhang, R.; Guo, K.; Yang, Y. Immune and intestinal microbiota responses to heat stress in Chinese mitten crab (Eriocheir sinensis). Aquaculture 2023, 563, 738965. [Google Scholar] [CrossRef]
- Yuan, Q.; Wang, Q.; Zhang, T.; Li, Z.; Liu, J. Effects of water temperature on growth, feeding and molting of juvenile Chinese mitten crab Eriocheir sinensis. Aquaculture 2017, 468, 169–174. [Google Scholar] [CrossRef]
- Peng, J.; Zhao, Y.; Xu, Z.; Liu, B.; Duan, C.; Tang, Y. Effect of temperature stress on the survival of juvenile Chinese mitten crab (Eriocheir sinensis). Iran. J. Fish. Sci. 2019, 18, 763–774. [Google Scholar]
- McMahon, B.R. Control of cardiovascular function and its evolution in Crustacea. J. Exp. Biol. 2001, 204, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Keen, A.N.; Klaiman, J.M.; Shiels, H.A.; Gillis, T.E. Temperature-induced cardiac remodeling in fish. J. Exp. Biol. 2017, 220, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Camacho, J.; Qadri, S.A.; Wang, H.; Worden, M.K. Temperature acclimation alters cardiac performance in the lobster Homarus americanus. J. Comp. Physiol. A 2006, 192, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Kushinsky, D.; Morozova, E.O.; Marder, E. In vivo effects of temperature on the heart and pyloric rhythms in the crab Cancer borealis. J. Exp. Biol. 2019, 222, jeb199190. [Google Scholar] [CrossRef] [PubMed]
- DeWachter, B.; Wilkens, J.L. Comparison of temperature effects on heart performance of the Dungeness crab, Cancer magister, in vitro and in vivo. Biol. Bull. 1996, 190, 385–395. [Google Scholar] [CrossRef]
- Duan, P.; Tian, Y.; Li, Z.; Chen, S.; Li, L.; Wang, X.; Wang, L.; Liu, Y.; Zhai, J.; Li, W. Comparative transcriptome analysis of hybrid jinhu grouper (Epinephelus fuscoguttatus♀ × Epinephelus tukula♂) and Epinephelus fuscoguttatus under temperature stress. Aquaculture 2024, 578, 740037. [Google Scholar] [CrossRef]
- Pan, T.; Yang, M.; Jiang, H.; Li, T.; Duan, G.; Ling, J.; Gao, Q. Effect of Astragalus membranaceus on transcriptome and survival of hybrid yellow catfish (Pseudobagrus vachellii♂ × Tachysurus fulvidraco♀) inresponse to Aeromonas hydrophila challenge. Fishes 2023, 8, 454. [Google Scholar] [CrossRef]
- Bao, J.; Wang, X.; Feng, C.; Li, X.; Jiang, H. Trehalose metabolism in the Chinese mitten crab Eriocheir sinensis: Molecular cloning of trehalase and its expression during temperature stress. Aquac. Rep. 2021, 20, 100770. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.; Haas, B.; Yassour, M.; Levin, J.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from rna-seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. Busco update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Becker, R.A.; Chambers, J.M.; Wilks, A.R. The New S Language; Wadsworth&Brooks/Cole: London, UK, 1988. [Google Scholar]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by rna-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression of rna-seq data at the gene level the deseq package. Eur. Mol. Biol. Lab. (EMBL) 2012, 10, f1000research. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Guo, X.-F.; Liu, M.; Zhou, Y.-L.; Wei, W.-Y.; Li, Z.; Zhou, L.; Wang, Z.-W.; Gui, J.-F. Genetic diversity evaluation and population structure analysis of red swamp crayfish (Procambarus clarkii) from lakes and rice fields by ssr markers. Fishes 2022, 7, 142. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, M.; Deng, Y.; Yang, Y.; Li, X.; Lu, Q.; Ge, J.; Pan, J.; Xu, Z. Molecular cloning, characterization and expression analysis of two juvenile hormone esterase-like carboxylesterase cDNAs in Chinese mitten crab, Eriocheir sinensis. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2017, 205, 46–53. [Google Scholar] [CrossRef]
- Shen, H.; Ma, Y.; Hu, Y.; Zhou, X. Cloning of the ecdysone receptor gene from the Chinese mitten crab, Eriocheir sinensis, and sexually dimorphic expression of two splice variants. J. World Aquac. Soc. 2015, 46, 421–433. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, H.; Li, H.; Wang, A.; Yu, H.-y. Effect of high temperature on immune response of grass carp (Ctenopharyngodon idellus) by transcriptome analysis. Fish Shellfish. Immunol. 2016, 58, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Ma, A.; Yang, S.; Huang, Z. Integrated metabolome and transcriptome analyses revealing the effects of thermal stress on lipid metabolism in juvenile turbot Scophthalmus maximus. J. Therm. Biol. 2021, 99, 102937. [Google Scholar] [CrossRef] [PubMed]
- Colson-Proch, C.; Morales, A.; Hervant, F.; Konecny, L.; Moulin, C.; Douady, C.J. First cellular approach of the effects of global warming on ground water organisms: A study of the hsp70 gene expression. Cell Stress Chaperones 2010, 15, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, J.; Chen, P.; Chang, Z.; He, Y.; Liu, P.; Wang, Q.; Li, J. Cloning of a heat shock protein 90 (hsp90) gene and expression analysis in the ridgetail white prawn Exopalaemon carinicauda. Fish Shellfish. Immunol. 2012, 32, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ye, H.; Huang, H.; Li, S.; Liu, X.; Zeng, X.; Gong, J. Expression of hsp70 in the mud crab, Scylla paramamosain in response to bacterial, osmotic, and thermal stress. Cell Stress Chaperones 2013, 18, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Oksala, N.K.; Ekmekçi, F.G.; Özsoy, E.; Kirankaya, Ş.; Kokkola, T.; Emecen, G.; Lappalainen, J.; Kaarniranta, K.; Atalay, M. Natural thermal adaptation increases heat shock protein levels and decreases oxidative stress. Redox Biol. 2014, 3, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Mahanty, A.; Mohanty, S.; Mohanty, B.P. Dietary supplementation of curcumin augments heat stress tolerance through upregulation of nrf-2-mediated antioxidative enzymes and hsps in Puntius sophore. Fish Physiol. Biochem. 2017, 43, 1131–1141. [Google Scholar] [CrossRef]
- Ma, F.; Luo, L. Genome-wide identification of hsp70/110 genes in rainbow trout and their regulated expression in response to heat stress. PeerJ 2020, 8, e10022. [Google Scholar] [CrossRef]
- Chowdary, T.K.; Raman, B.; Ramakrishna, T.; Rao, C.M. Mammalian hsp22 is a heat-inducible small heat-shock protein with chaperone-like activity. Biochem. J. 2004, 381, 379–387. [Google Scholar] [CrossRef]
- Gober, M.D.; Smith, C.C.; Ueda, K.; Toretsky, J.A.; Aurelian, L. Forced expression of the h11 heat shock protein can be regulated by DNA methylation and trigger apoptosis in human cells. J. Biol. Chem. 2003, 278, 37600–37609. [Google Scholar] [CrossRef]
- Huang, J.; Li, Y.; Liu, Z.; Kang, Y.; Wang, J. Transcriptomic responses to heat stress in rainbow trout Oncorhynchus mykiss head kidney. Fish Shellfish. Immunol. 2018, 82, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Luo, L.T.; Wang, Q. Hsp60/10 and shsp families of heat shock protein genes in rainbow trout (Oncorhynchus mykiss) and their expression under heat stress. Aquac. Int. 2022, 30, 1–18. [Google Scholar] [CrossRef]
- Rogon, C.; Ulbricht, A.; Hesse, M.; Alberti, S.; Vijayaraj, P.; Best, D.; Adams, I.R.; Magin, T.M.; Fleischmann, B.K.; Höhfeld, J. Hsp70-binding protein hspbp1 regulates chaperone expression at a posttranslational level and is essential for spermatogenesis. Mol. Biol. Cell 2014, 25, 2260–2271. [Google Scholar] [CrossRef]
- Wang, C.; Chen, X.; Dai, Y.; Zhang, Y.; Sun, Y.; Cui, X. Comparative transcriptome analysis of heat-induced domesticated zebrafish during gonadal differentiation. BMC Genom. Data 2022, 23, 39. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-H.; Yang, F.-F.; Liao, S.-A.; Miao, Y.-T.; Ye, C.-X.; Wang, A.-L.; Tan, J.-W.; Chen, X.-Y. High temperature induces apoptosis and oxidative stress in pufferfish (Takifugu obscurus) blood cells. J. Therm. Biol. 2015, 53, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Arama, E.; Bader, M.; Srivastava, M.; Bergmann, A.; Steller, H. The two Drosophila cytochrome c proteins can function in both respiration and caspase activation. EMBO J. 2006, 25, 232–243. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, X. Cytochrome c-mediated apoptosis. Annu. Rev. Biochem. 2004, 73, 87–106. [Google Scholar] [CrossRef]
- Vornanen, M.; Hassinen, M.; Koskinen, H.; Krasnov, A. Steady-state effects of temperature acclimation on the transcriptome of the rainbow trout heart. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2005, 289, R1177–R1184. [Google Scholar] [CrossRef][Green Version]
- Jiang, X.; Jiang, X.; Feng, Y.; Xu, R.; Wang, Q.; Deng, H. Proteomic analysis of eif5b silencing-modulated proteostasis. PLoS ONE 2016, 11, e0168387. [Google Scholar] [CrossRef]
- Krupovic, M.; Koonin, E.V. Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. USA 2017, 114, E2401–E2410. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).