Abstract
Information about the food sources for fish is important not only for predation and food competition, but also for the direct impact on organism interactions, relationships, and biodiversity within aquatic ecosystems. We analyzed the food sources of 21 fish species in 9 families of 6 orders based on the literature data from brackish ecosystems in South Korea to improve the understanding of the estuarine ecosystem. The food sources of the 21 fish species contained 173 families, 86 orders, 39 classes, and 22 phyla. The 21 fish species were classified into 4 groups using hierarchical analysis based on their food sources: Group 1 fishes mainly consumed Amphipoda; Group 2 fishes were zooplanktivores; Group 3 were omnivorous; and Group 4 consumed small food sources, such as Nematoda, Corophiidae, and Harpacticidae. The feeding competition index was relatively high within each group, but the competition index for Group 3, the omnivorous fishes, was low. Network analysis and hub scores represented the importance of food sources consumed by each fish species by showing the connections between the fish and their prey. Polychaeta, Actinopterygii, and Decapoda were the most commonly consumed food sources. Food source diversity was high for zooplanktivores (group 2). Additionally, the food contents of each fish species were classified into freshwater and marine sources for identifying the habitats of the food sources. Trophic level measuring values using the data were similar to the reported literature data. This study can be utilized for understanding estuarine ecosystems and identifying food relationships.
Key Contribution:
The 21 brackish fish species inhabiting South Korea are divided into 4 groups according to their food sources and form competitive relationships for food. Fish that inhabit the same location have different primary food sources, minimizing competition for food.
1. Introduction
Producers and consumers are connected through feeding activities, especially in aquatic ecosystems. Fish, in particular, as top predators within the aquatic food web, exhibit various trophic guilds, and understanding their food sources is an important aspect of trophic ecology [1,2,3]. Information about the food sources for fish is important not only for predation and food competition, but also for the direct impact on organism interactions, relationships, and biodiversity within aquatic ecosystems [4,5]. Therefore, information about the food sources for fish, the top predators in the food chain, is essential for understanding trophic interrelations and community structures in aquatic ecosystems.
Although various studies have been conducted using the Index of Relative Importance (IRI) to estimate the relative importance of fish food sources, integrated information on primary food sources and food competition for fish is still lacking [6,7,8]. Even the FishBase platform, which integrates a variety of information on fish worldwide, only reports information on the diet type (carnivorous, herbivorous, and omnivorous) and trophic level for fish, with no information available on their primary food sources [3]. Since most studies on fish feeding habits have been conducted on a species-by-species basis, integrating data on food sources is necessary to comprehensively consider and manage fish resources in ecosystems.
Estuaries are areas where freshwater and seawater mix and are characterized by significant changes in the food source communities of aquatic organisms, such as phytoplankton, zooplankton, and benthic invertebrates [9]. Many studies have reported that epilithic algae were important food sources for estuary fishes to survive and develop. Gal et al. [10] reported that the contribution of epilithic algae, which showed a heavy δ13C value in the estuary of the Tamjin River, increased as a food source. Stable isotope studies in mangrove, marsh, and reed habitats showed that juvenile fishes ate mainly epilithic diatoms and algae rather than macrophytes [11]. While many fish food source studies focus on potential food source estimation due to the simplicity of the stable isotope method, they lack practical ecosystem food web connectivity and analysis [11,12]. Indeed, few studies have attempted to arrange the entire fish-food assemblage into an ecosystem perspective with an ecological network analysis [13,14,15].
However, all fish cannot survive by feeding only on attached algae, and downstream, benthic invertebrates will still be a preferred food source for fish [12]. Fish do not rely on a single food source for growth and development [11]. In particular, benthic invertebrates have been underestimated regarding their role in the food web as food sources for fish. Despite the high preference and competition for benthic invertebrates as food sources, freshwater fish can coexist due to high-density food sources, such as Chironomidae [12,16]. However, in estuaries, aquatic insect tolerance to salt is lower, resulting in a significant decrease in their population density. Macroinvertebrates are still assumed to play an essential role as food sources for fish even though, when compared to freshwater areas, benthic invertebrate diversity decreases and phytoplankton diversity increases in estuarine areas [17,18,19]. Howe et al. [20] showed that macroinvertebrates, such as crustaceans, were highly important food sources, even though they were planktivorous and larvivorous fishes. In a comparative analysis of the gut contents and stable isotope values for Acanthopagrus australis and Sillago ciliata, the feeding rate of fish, decapods, bivalves, and polychaetes was higher than that for algae in both species [21].
The dynamic physicochemical changes inherent in an estuary can affect the potential food sources for fish, such as macroinvertebrates and plankton [22,23]. Therefore, a comprehensive analysis is needed to investigate the feeding ecology of fish in estuarine areas. However, most studies on the food sources for brackish fish have been conducted on individual species or are limited to estimating food sources within a specific range, such as using stable isotope analysis [24,25,26]. Thus, there are limitations to accurately estimating the food sources for brackish fish and understanding their estuarine ecosystems. It is necessary to perform a meta-analysis by compiling the existing literature on the topic to gain a comprehensive understanding of the overall food sources for fish and estuarine ecosystems. The meta-analysis method has been proven to be useful for studying trophic level variation in fish and researching competition among fish in aquatic ecosystems [27,28]. Therefore, this study aims to provide important information on the feeding habits of fish inhabiting the target area by compiling and quantifying the results from a stomach content analysis of fish species in Korea and analyzing their food preferences and competitive relationships regarding their feeding habits.
2. Materials and Methods
In this study, we collected information on the food sources for 21 fish species found in brackish waters in Korea. We obtained data from 19 literature reports, including 12 research papers and 7 dissertations, by searching the Research Information Sharing Service (RISS) and Google Scholar academic databases (Table 1). All studies on fish diets classified the food sources using microscopy. We excluded research that used isotope analysis, which cannot always identify food sources using taxonomic groups (e.g., genus, family, and order).

Table 1.
List of the brackish fish species whose food sources have been reported in the literature. Species are abbreviated by the first letters of the genus and the species epithet.
Recent studies on fish gut contents have used the Index of Relative Importance (IRI) to estimate the contribution of different food sources [48,49,50]. However, the fish gut content analysis differs depending on the researcher or method (e.g., DNA metabarcoding and microscopy), and some previous studies have reported only the number of food sources or the food source ratio (%N). We quantified gut contents using the %N and utilized median values represented by intervals such as phytoplankton to analyze as many fish food sources as possible.
To classify the feeding habits of fish according to their food sources, the Bray–Curtis distance [51] was calculated, and a hierarchical cluster analysis (HCA) was performed using the Ward linkage method [52]. A network analysis was used to investigate the relationships between 21 brackish fish species and their food sources [53]. In the network analysis, fish and food sources were designated as nodes, and the thickness of the connecting lines represented each food source’s feeding rate. All analyses were performed using R studio (version 2021.09.1, Posit PBC, Redmond, WA, USA), with the hierarchical cluster analysis conducted using the vegan package [54] and the network analysis performed using igraph [53]. Data at the family level were used as the input data for the hierarchical cluster analysis and network analysis; for families that were not classified at the family level, data at the order, class, and phylum levels were used. The hub score was calculated based on Fang and Huang [55].
To measure the diversity of prey for brackish fish, we used the dietary breadth index (Bi), which is based on Levin’s standardized niche breadth () [56]. The Bi value ranges from 0 to 1, with higher values indicating greater prey diversity and 0 indicating a single prey type. Equation (1) was used, which utilizes Levin’s measure of niche breadth (). In this equation, pj represents the probability of consuming a specific prey type (j), while n represents the total number of prey types.
To analyze food competition in brackish fish, Horn’s index [57] was used (Equation (2)), where is the ecological overlap index between two species (j and k), and and represent the proportion of the food source i consumed by species j and k, respectively. The higher the calculated value of , the more intense the competition between the two species for the same food source can be interpreted.
The food sources for each fish species were compared by dividing them into freshwater, marine, and universal categories using the World Register of Marine Species (WoRMS; www.marinespecies.org, accessed on 13 March 2023) as a reference. The trophic level was calculated using the formula proposed by Cortes [58], and the resulting trophic levels were compared to those of fish species registered on the FishBase [3] platform.
3. Results
3.1. Compositions of Food Sources for Brackish Fishes
Taxonomic analysis of the food sources for the 21 brackish fish species revealed 176 families, 89 orders, 40 classes, and 21 phyla (Table 2). Arthropoda was the most commonly consumed phylum (by 20 out of the 21 brackish fish species), thus accounting for approximately 72.2%. Annelida and Chordata were also heavily consumed, with 17 and 11 species determined, respectively, but their proportions were low at 4.6% and 7.0%. Only four fish species consumed Bacillariophyta, but the proportion was high at 30%.

Table 2.
Food sources for 21 brackish fish species based on the literature in Table 1. ‘No. of fish’ indicate the number of fish species that consumed the food sources.
At the class level, the food sources that were consumed by more than 10 species of fish were Malacostraca (19 fish species), unidentified Polychaeta (17 fish species), unidentified Actinopterygii (16 fish species), Maxillopoda (13 fish species), and Bivalvia (10 fish species). Species that did not feed on Malacostraca were identified as Kp (Konosirus punctatus) and Bp (Boleophthalmus pectinirostris). Fewer species consumed Maxillopoda compared to Polychaeta and Actinopterygii, but its proportion as a food source was high at 44.1%.
At the order level, the top ten food sources consumed by fish consisted of six macroinvertebrates and two each of fish and zooplankton. The most consumed food sources were Amphipoda and Decapoda, both macroinvertebrates, which were eaten by 19 of the 21 fish species examined. The proportion of fish species that consumed Amphipoda was higher (20.5%) than that of Decapoda (12.4%). Calanoida, a type of zooplankton, was eaten by only nine fish species, yet it represented the highest proportion (25.9%).
At the family level, the food sources that were eaten by 10 or more fish species were divided into 3 groups: unidentified Polychaeta (15 fish species), unidentified Actinopterygii (14 fish species), and unidentified Decapoda (13 fish species). Although Amphipoda had a high feeding ratio at the order level, it was classified as different organisms at the family level, resulting in a lower ratio. On the other hand, the unclassified groups (Polychaeta, Actinopterygii, and Decapoda) still had high ratios at higher classification levels.
The fish that consumed the most prey items was Pleuronichthys cornutus (Pc), which consumed 33 different food sources. Acanthogobius elongatus (Ae) and Platichthys stellatus (Ps) consumed 27 food sources each, while Acanthogobius luridus (Al) consumed 25. On the other hand, Periophthalmus magnuspinnatus (Pm) and Hemibarbus labeo (Hl) had the lowest number of food sources consumed, with only eight food sources each.
Arthropoda was the most commonly consumed phylum-level food source by brackish fish, with 20 out of the 21 fish species feeding on them, thus accounting for approximately 72.2%. Annelida and Chordata were also heavily consumed, with 17 and 11 species determined as predators, respectively, but their proportions were low at 4.6% and 7.0%. Only four species of fish consumed Bacillariophyta, but the proportion was high at 30%.
3.2. Classification of Feeding Groups and Important Food Sources for Brackish Fishes
The 21 brackish fish were divided into four groups according to their food sources using hierarchical clustering analysis (Figure 1). The horizontal and vertical axes of the hierarchical clustering represented the food sources and brackish fish groups, respectively. Group 1, which included Opsariichthys uncirostris amurensis (Ou), Hl, Pm, Syngnathus schlegeli (Sc), and Favonigobius gymnauchen (Fg), was correlated with unidentified Amphipoda (Amph). Group 2, which included Al, Ae, and Nuchequula nuchalis (Nn), fed on Decapoda (Dec), Calanidae (Cal), and Corycaeidae (Cory) and formed clusters, all of which were zooplankton. Group 3, which contained the largest number of fish species, consisted of omnivorous fish that consume various food sources. Group 4 consisted of Lateolabrax japonicus (Lj), Tridentiger bifasciatus (Tb), Periophthalmus modestus (Po), and Takifugu niphobles (Tn) and were categorized by their consumption of Harpacticidae (Har), Corophiidae (Cor), and Nematoda (Nem). Although the fish in Group 4 had different higher-level food source classifications (e.g., Malacostraca, zooplankton, and benthos) that they mainly consumed, these food sources were small in size, and thus, Group 4 fish were characterized as preferring small food sources.
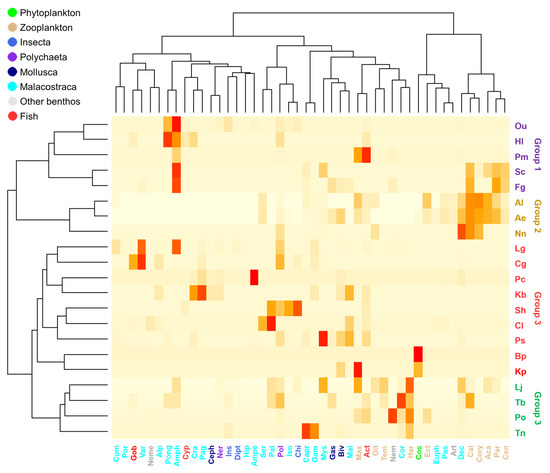
Figure 1.
Hierarchical cluster analysis of brackish fishes according to their food sources. The abbreviations for food sources are as follows: Aca, Acartiidae; Ani, Anisogammaridae; Ant, Anthozoa; Aph, Aphroditidae; Ath, Anthuridae; Aty, Atyidae; Bae, Bacillariophyceae; Bao, Bacillariaceae; Biv, Bivalvia; Bra, Branchiopoda; Ceph, Cephalopoda; Cod, Codonellidae; Col, Coleoptera; Cory, Corycaeidae; Cyp, Cyprinidae; Dex, Dexaminidae; Eugl, Euglyphidae; Eup, Euglenophyceae; Euph, Euphausiacea; Harp, Harpacticoida; Ins, Insecta; Lil, Liliaceae; Lot, Lottiidae; Mac, Macrophthalmidae; Mag, Magelonidae; Max, Maxillopoda; Mys, Mysidae; Neme, Nemertea; Ophu, Ophiuridae; Ost, Ostracoda; Pas, Pasiphaeidae; Pha, Phacaceae; Pho, Pholidae; Rot, Rotifera; Sabe, Sabellidae; Sem, Semelidae; Sph, Sphaeromatidae; Sto, Stomatopoda; Ter, Terebellidae; Tha, Thalassiosiraceae; Uri, Uristidae; Var, Varunidae. The abbreviation for brackish fishes can be found in Table 1.
In the network analysis between brackish fishes and food sources, the food sources at the family level were represented by the top nine classification groups (Figure 2a). Bp and Kp, which are planktivorous fish, were located on the right side of the network, with Bp feeding only on phytoplankton, while Kp was found to consume phytoplankton and zooplankton. Pc, Kareius bicoloratus (Kb), and Chaenogobius gulosus (Cg) were located at the top of the network and showed a strong connection to Polychaeta and Mollusca. Malacostraca and animal plankton were the food sources consumed by most of the fish species.
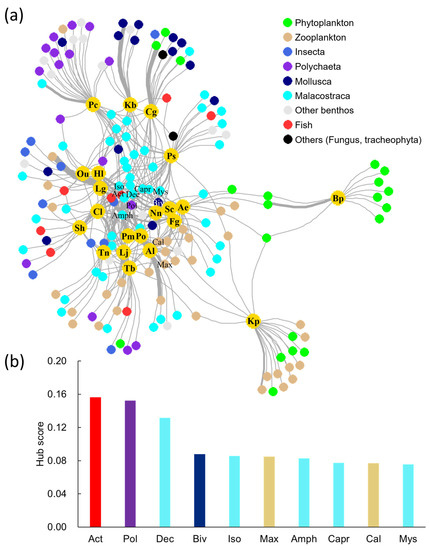
Figure 2.
The network analysis between brackish fishes and food sources. (a) Colored nodes with abbreviations in the network represent fish species, and links appear in the association between fishes and food sources. The link width is proportionate to the feeding frequency. (b) Hub score of networks for the top 10 food sources. The abbreviation can be found in Table 1.
The hub scores were calculated for the top 10 food sources based on the strength of the connection between brackish fish species and food sources, as determined by the network analysis (Figure 2b). Malacostraca had the highest frequency among the top ten food sources with five classification groups, followed by two zooplankton groups and one each of fish, Polychaeta, and Mollusca. The highest hub score was observed for Actinopterygii (Act) at 0.156, followed by Polychaeta (Pol) at 0.152, Dec at 0.132, Bivalvia (Biv) at 0.088, and Isopoda (Iso) at 0.086. Insecta and phytoplankton, which were among the top nine classification groups in the network analysis, did not have high hub scores.
3.3. Diversity and Competition for Food Sources in Brackish Fish Species
Among the 21 brackish fish species studied, Ae exhibited the highest niche breadth with a Bi value of 0.254 (Figure 3). Bi values for Hl (0.226), Nn (0.219), and Al (0.214) were relatively high with values above 0.2, indicating high dietary diversity. On the other hand, nine species, including Ps (0.100), Tn (0.085), Cl (0.068), Ou (0.063), Po (0.051), Kb (0.041), Pc (0.041), Cg (0.033), and Lg (0.001), showed a low niche breadth with Bi values below 0.1.
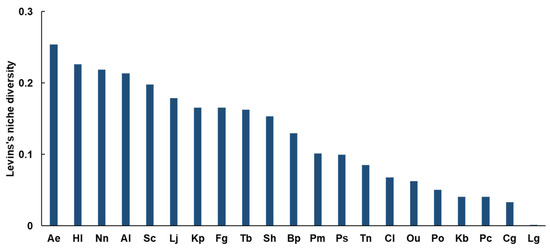
Figure 3.
Levin’s niche diversity index for brackish fishes. The abbreviation for brackish fishes can be found in Table 1.
In the Horn’s index analysis, the species with a possibility for food competition greater than 0.5 were Hl-Ou (0.68) and Fg-Sc (0.76) in Group 1 and Al-Ae (0.82), Al-Nn (0.55), and Ae-Nn (0.52) in Group 2 (Figure 4). Pontogeneiidae (Pong) and Amph were the common food sources for Hl-Ou, while Amphi and zooplankton were the common food sources for Fg-Sc. The food competition between Ae and Al, which had the highest niche overlap index, was mostly for zooplankton, while Nn competed for Dec, Cal, and Cory with Al and Ae. Groups 3 and 4 had relatively low niche overlap indexes of 0.5 or less. In addition, the within-group niche overlap index for Groups 1, 2, and 4, except for Group 3, showed significant results with values greater than 0.2.
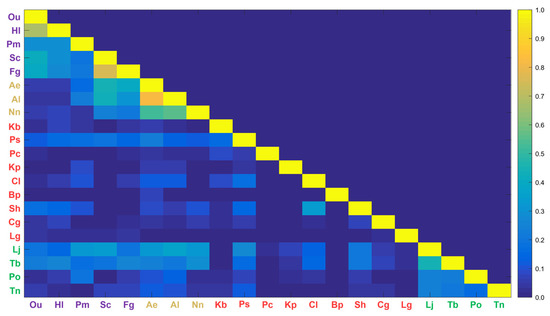
Figure 4.
Horn’s niche overlap index for the 21 brackish fish species. The abbreviations for brackish fishes can be found in Table 1.
3.4. Origin of Food Sources and Trophic Level of Brackish Fishes
The food sources for fish were categorized at the family level as freshwater, marine, and universal according to their habitat (Figure 5). Although all fish species consumed marine food sources, the proportion varied by species. Pc had the highest proportion of marine food sources at 99.2%, and more than 80% of the food sources for Kb, Po, and Cl were marine. On the other hand, a total of 11 species consumed freshwater food sources, with Sh consuming the most at 43.9%. Among the 11 species that consumed freshwater food sources, Sc, Fg, Al, Tn, and Lg obtained less than 1% of their diet from freshwater, with most of their food sources being marine or universal.
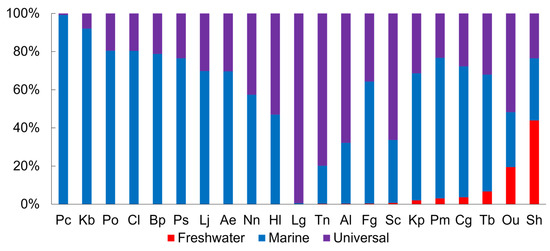
Figure 5.
Proportions of freshwater and marine habitats of food sources for brackish fishes. The abbreviation for brackish fishes can be found in Table 1.
The trophic level range for the 21 fish species studied in this research was between 2.0 and 3.7, which appeared to be similar to the trophic level range reported in FishBase (2.0–3.9) (Figure 6). However, there were differences in the trophic levels between the species. The species with the highest trophic level in this study was Pm, with a calculated trophic level of 3.7, which was reported as 3.2 in FishBase. The species with the lowest trophic level was Bp, which was 2.0 in both this study and FishBase. The species that showed the greatest difference in trophic levels between this study and FishBase was Ou, with a trophic level of 2.9 in this study and 3.9 in FishBase.
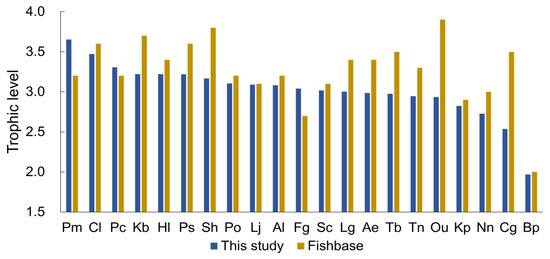
Figure 6.
Comparison of trophic levels between this study and FishBase. The abbreviation for brackish fishes can be found in Table 1.
4. Discussion
Research was conducted on the food sources for 21 species of brackish fish in Korea, including 11 species of Gobiidae, 3 species of Pleuronectidae, 2 species of Cyprinidae, and 1 species each of Moronidae, Sciaenidae, Leiognathidae, Clupeidae, Syngnathidae, and Tetraodontidae. Although the Gobiidae fish has the most species and is a representative species of brackish fish that inhabits estuaries, other fish also inhabit estuaries for various reasons, such as spawning, food, and growth. For example, juvenile Pleuronectidae fish live near shallow estuaries and move to deeper waters as they mature, while Lj migrates up to river estuaries in the spring and down to the coast in the fall. In this way, they inhabit estuaries for various reasons and compete for food resources, thus forming an ecosystem.
Gobiidae fish are widely distributed in freshwater, estuaries, and seawater, and have diverse feeding and habitat preferences [59]. Furthermore, since the presence of multiple species within the same ecosystem has been reported, it is necessary to analyze niche relationships among similar species. The species investigated in this study, Pm, Po, and Bp, were collected in the same period from Beolgyo, South Korea [35]. In the network analysis, Bp, which mainly feeds on phytoplankton, showed differences from other species, while Pm and Po were located close together and appeared to share the same food source. However, the feeding competition index between the two species was low at 0.2, indicating that their competitive relationship was not significant. In two-way clustering, Pm mainly consumed Act, while Po mainly consumed Nem; thus, the two species were clustered into different groups. Even though they inhabit the same ecosystem, differences in the primary food source between species can create different niches, allowing them to coexist without competition [60,61].
Out of 45 species in a study analyzing the food sources for freshwater fish, 39 were found to consume Diptera (e.g., Chironomidae), which was a significant food source with a %N of 82.6% [16]. However, due to the low salinity tolerance of Diptera, it was assumed to be replaced with other food sources in estuaries. Indeed, only 7 out of 21 species consumed Diptera, with a %N of only 1.69%, resulting in a low contribution of Diptera as a food source in estuaries. Instead, among the 21 brackish fish species, Pol, Act, and Dec were the most important food sources, with 15, 14, and 12 species feeding on each, respectively. These prey types have become more important as food sources compared to those in freshwater. These results suggest that aquatic insects, the primary food source for freshwater fish, have been replaced by fish and macroinvertebrates in brackish water.
The niche overlap index shows the competition between species regarding diet but does not indicate which food resources are shared. However, in the two-way clustering analysis, fish were clustered based on shared food resources, so it is possible to infer which resources are being shared and competed for. Therefore, by conducting both analyses, we can determine which common food sources the fish consume and their food competition index values. The two analyses found that Ou and Hl mainly feed on Amph and Pong, with a food competition index of 0.68, while Sc and Fg mainly feed on Amph and zooplankton, respectively, showing a food competition index of 0.76. Furthermore, both analyses showed high intragroup competition among fish in Group 2 (Al, Ae, and Nn) with a competition index of 0.5 or higher, indicating that they are zooplanktivorous fish. In particular, Ae and Al, which belong to the same genus Acanthogobius, showed a high overlap in zooplankton food resources despite being collected at different times and places and having the highest interspecies competition index of 0.82. Copepods (Cal, Cory, Aca) are highly preferred as food sources for fish and not only dominate Korean estuaries but also exhibit high species diversity. Zooplanktivorous fish species (Ae, Al, Nn) categorized as such consume a variety of animal plankton, resulting in relatively high values for Levin’s niche index (Figure 3). However, despite consuming multiple species, Cal and Cory showed a higher feeding preference, leading to higher food competition indices compared to other groups. These results indicate that fish species with a high preference for specific food resources, such as planktivores, piscivores, and herbivores, have higher competition indices in the ecosystem [62]. However, Group 3, classified as omnivorous fish, had an inter-species feeding competition index of less than 0.2. This result suggests that omnivorous fish, with their low selectivity in food sources, can avoid food competition by consuming alternative food sources [63]. Due to the large physical and chemical variations in estuaries, omnivorous fish with low niche overlap can contribute to the stability of the ecosystem by increasing the efficiency of resource utilization and coexisting without food competition [64,65].
The differentiation of fish food sources into freshwater, brackish, and marine using microscopy can provide insights into the habitat and surrounding environment of the fish. For example, Sh, which had the highest proportion of freshwater food sources, is a predator that inhabits a wide range of habitats (from freshwater to seawater) and consumes various organisms, such as crabs, small fish, and shrimp [66]. In addition, it migrates to estuaries for spawning during periods of rising water temperature and moves upstream when the water temperature drops [67]. In Paik’s [38] study, the proportion of freshwater food sources for Sh was 69.9%, the highest among 21 brackish fish species. Moreover, the area where Sh was collected was located near a dam and a sluice gate, which artificially blocked the influence of seawater. Since the dam and sluice gate were opened only twice a month and the water flow was almost stagnant, the high proportion of freshwater food sources compared to marine food sources is likely due to this.
In this study, we compared the trophic level reported in FishBase with the trophic level derived from brackish fish. The top three species that showed the greatest difference in trophic level between this study and FishBase were Ou, Cg, and Sh, with all three having a difference of 0.6 or more. The trophic level can generally vary depending on the habitat or analysis method. Particularly for omnivorous fish, such as those that eat a variety of small invertebrates, fish, and crustaceans, the trophic level can vary depending on the prey consumed, even for the same species [68]. For example, Ou is a top predator that mainly feeds on fish in freshwater [69]. Piscivores fish typically belong to the upper trophic level (with a value of 3.5 to 4.0) within an ecosystem [70]. However, in this study, the trophic level of Ou was calculated to be 2.9, which is relatively low. The reason Ou was collected in the estuary is thought to be due to heavy rainfall, which lowered the estuary’s salinity, and Ou temporarily moved to the estuary during low tide when the freshwater expanded [71]. In addition, Ou had a standard length of 2.6–9.1 (5.2 ± 1.2 cm) and mainly fed on Amphipoda (49.6%) instead of fish, which is considered to have resulted in a lower trophic level. Methodological differences can also result in varying trophic levels. In studies [72,73] that calculated the trophic level of Sh using stable isotope analysis, the range was 2.79–2.95 and 2.61–2.65, respectively, which was lower than the trophic level calculated in FishBase (3.8) and this study (3.2). When observing feeding habits by dissecting the gut contents, the trophic level is calculated based on the food sources utilized within a few hours of feeding. However, stable isotope analysis to determine the trophic level reflects the food sources consumed over a more extended period, which can result in differences in trophic levels [74].
Research on fish food webs plays an important role in understanding ecological characteristics and analyzing competition, cooperation, ecosystem stability, and biodiversity maintenance within ecosystems [75,76,77]. In addition, conducting meta-analyses on species-specific food webs allows for the analysis of energy flow in aquatic food webs. Some publications have presented information on food sources as the %N, but it is essential to have information on individual numbers and biomass for a more advanced understanding [12]. Studying food sources by analyzing gut contents can be challenging in terms of identifying digested prey, and there may be differences in the sub-classification of taxonomic groups at the genus and species levels depending on the researcher. Therefore, if new analysis methods, such as DNA metabarcoding, are introduced for difficult-to-classify (e.g., plant and animal fragments) or digested food sources, an accurate analysis of food sources is expected to be possible, and these results are expected to be helpful in the future management of estuarine resources as well as the analysis and understanding of food webs in aquatic ecosystems.
5. Conclusions
The feeding sources for 21 brackish fish species in Korea were analyzed using a meta-analysis. The fish species were divided into four groups based on their feeding sources using two-way clustering analysis. Group 1 primarily consumed amphipods, Group 2 relied on zooplankton, Group 3 exhibited generalist feeding behavior, and Group 4 consumed small prey items. The generalist feeding group (Group 3) showed lower competition indices than the other groups, indicating that they had different primary food sources and experienced less intense food competition. The network analysis and hub scores demonstrated the importance of polychaetes and Actinopterygii as key prey organisms. Furthermore, even when occupying the same habitat, the fish species could coexist using different primary food sources, avoiding intense competition. The information on food sources provides insights into the food web, species interactions, and habitat characteristics within the ecosystem. This study analyzed the feeding relationships between fish species inhabiting estuaries using the results of their feeding sources, which can be valuable for future understanding of estuarine ecosystems.
Author Contributions
Conceptualization, C.W.J. and I.-S.K.; methodology, T.-S.Y., C.W.J. and I.-S.K.; formal analysis, T.-S.Y. and C.W.J.; writing–original draft preparation, T.-S.Y.; writing–review and editing, all authors; visualization, T.-S.Y. and C.W.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research Foundation of Korea (grant number NRF-2018R1A6A1A03024314).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Brose, U.; Archambault, P.; Barnes, A.D.; Bersier, L.F.; Boy, T.; Canning-Clode, J.; Conti, E.; Dias, M.; Digel, C.; Dissanayake, A.; et al. Predator traits determine food-web architecture across ecosystems. Nat. Ecol. Evol. 2019, 3, 919–927. [Google Scholar] [CrossRef]
- Estes, J.A.; Terborgh, J.; Brashares, J.S.; Power, M.E.; Berger, J.; Bond, W.J.; Carpenter, S.R.; Essington, T.E.; Holt, R.D.; Jackson, J.B.C.; et al. Trophic downgrading of planet earth. Science 2011, 333, 301–306. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. FishBase. Available online: https://fishbase.se/ (accessed on 5 March 2023).
- Kristensen, E.; Delefosse, M.; Quintana, C.O.; Flindt, M.R.; Valdemarsen, T. Influence of benthic macrofauna community shifts on ecosystem functioning in shallow estuaries. Front. Mar. Sci. 2014, 1, 41. [Google Scholar] [CrossRef]
- Ohore, O.E.; Wei, Y.J.; Wang, J.H.; Wang, Y.W.; Ifon, B.E.; Liu, W.H.; Wang, Z. Vertical characterisation of phylogenetic divergence of microbial community structures, interaction, and sustainability in estuary and marine ecosystems. Sci. Total Environ. 2022, 851, 158369. [Google Scholar] [CrossRef]
- El-Naggar, H.A.; Allah, H.M.M.K.; Masood, M.F.; Shaban, W.M.; Bashar, M.A.E. Food and feeding habits of some Nile River fish and their relationship to the availability of natural food resources. Egypt. J. Aquatic Res. 2019, 45, 273–280. [Google Scholar] [CrossRef]
- Kara, C.; Alp, A. Feeding habits and diet composition of brown trout (Salmo trutta) in the upper streams of River Ceyhan and River Euphrates in Turkey. Turk. J. Vet. Anim. Sci. 2005, 29, 417–428. [Google Scholar]
- Sandhya, K.M.; Chakraborty, S.K.; Jaiswar, A.K.; Shah, T.H. Food and feeding habits of Otolithes cuvieri (Trewavas, 1974) from Ratnagiri, Maharashtra. Indian J. Fish. 2014, 61, 99–102. [Google Scholar]
- Cloern, J.E.; Foster, S.Q.; Kleckner, A.E. Phytoplankton primary production in the world’s estuarine-coastal ecosystems. Biogeosciences 2014, 11, 2477–2501. [Google Scholar] [CrossRef]
- Gal, J.-K.; Kim, M.-S.; Lee, Y.-J.; Seo, J.-W.; Shin, K.-H. Foodweb of aquatic ecosystem within the Tamjin River through the determination of carbon and nitrogen stable isotope ratios. KJEE 2012, 45, 242–251. [Google Scholar]
- Whitfield, A.K. The role of seagrass meadows, mangrove forests, salt marshes and reed beds as nursery areas and food sources for fishes in estuaries. Rev. Fish Biol. Fisher. 2017, 27, 75–110. [Google Scholar] [CrossRef]
- Ji, C.W.; Lee, D.S.; Lee, D.Y.; Park, Y.S.; Kwak, I.S. Analysis of food preference and competition based on stomach contents of fish species inhabiting fresh and brackish waters in South Korea. Ecol. Freshw. Fish. 2023, 32, 64–79. [Google Scholar] [CrossRef]
- Baird, D.; Ulanowicz, R. Comparative study on the trophic structure, cycling and ecosystem properties of four tidal estuaries. Mar. Ecol. Prog. Ser. 1993, 99, 221–237. [Google Scholar] [CrossRef]
- de Jonge, V.N.; Schückel, U.; Baird, D. Subsets of food webs cannot be used as a substitute to assess the functioning of entire ecosystems. Mar. Ecol. Prog. Ser. 2019, 613, 49–66. [Google Scholar] [CrossRef]
- Scharler, U.M.; Baird, D. A comparison of selected ecosystem attributes of three South African estuaries with different freshwater inflow regimes, using network analysis. J. Mar. Syst. 2005, 56, 283–308. [Google Scholar] [CrossRef]
- Ji, C.W.; Lee, D.-S.; Lee, D.-Y.; Kwak, I.-S.; Park, Y.-S. Analysis of food resources of 45 fish species in freshwater ecosystems of South Korea (Based on literature data analysis). KJEE 2020, 53, 311–323. [Google Scholar] [CrossRef]
- Chen, W.L.; Guo, F.; Huang, W.J.; Wang, J.G.; Zhang, M.; Wu, Q. Advances in phytoplankton population ecology in the Pearl river estuary. Front. Environ. Sci. 2023, 11, 68. [Google Scholar] [CrossRef]
- Pickwell, A.; Constable, D.; Chadd, R.; Extence, C.; Little, S. The development of a novel macroinvertebrate indexing tool for the determination of salinity effects in freshwater habitats. River Res. Appl. 2022, 38, 522–538. [Google Scholar] [CrossRef]
- Starr, S.M.; Wallace, J.R. Ecology and biology of aquatic insects. Insects 2021, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.R.; Simenstad, C.A.; Toft, J.D.; Cordell, J.R.; Bollens, S.M. Macroinvertebrate prey availability and fish diet selectivity in relation to environmental variables in natural and restoring north San Francisco bay tidal marsh channels. San Franc. Estuary Watershed Sci. 2014, 12, 1–46. [Google Scholar] [CrossRef]
- Hadwen, W.L.; Russell, G.L.; Arthington, A.H. Gut content- and stable isotope-derived diets of four commercially and recreationally important fish species in two intermittently open estuaries. Mar. Freshw. Res. 2007, 58, 363–375. [Google Scholar] [CrossRef]
- Muylaert, K.; Sabbe, K.; Vyverman, W. Spatial and temporal dynamics of phytoplankton communities in a freshwater tidal estuary (Schelde, Belgium). Estuar. Coast. Shelf Sci. 2000, 50, 673–687. [Google Scholar] [CrossRef]
- Shervette, V.R.; Gelwick, F. Seasonal and spatial variations in fish and macroinvertebrate communities of oyster and adjacent habitats in a Mississippi estuary. Estuaries Coast. 2008, 31, 584–596. [Google Scholar] [CrossRef]
- Chaudhuri, A.; Mukherjee, S.; Homechaudhuri, S. Diet composition and digestive enzymes activity in carnivorous fishes inhabiting mudflats of Indian Sundarban Estuaries. Turkish J. Fish. Aquat. Sci. 2012, 12, 265–275. [Google Scholar] [CrossRef]
- Herzka, S.Z. Assessing connectivity of estuarine fishes based on stable isotope ratio analysis. Estuar. Coast. Shelf Sci. 2005, 64, 58–69. [Google Scholar] [CrossRef]
- Tran, C.C.; Nguyen, T.H.D.; Nguyen, H.T.T.; Vo, L.T.T.; Dinh, Q.M. Diet composition and feeding habit of Glossogobius sparsipapillus caught from estuarine regions in the Mekong Delta. Egypt. J. Aquat. Res. 2021, 47, 313–319. [Google Scholar] [CrossRef]
- Laub, B.G.; Budy, P. Assessing the likely effectiveness of multispecies management for imperiled desert fishes with niche overlap analysis. Conserv. Biol. 2015, 29, 1153–1163. [Google Scholar] [CrossRef]
- Zengeya, T.A.; Booth, A.J.; Chimimba, C.T. Broad niche overlap between invasive Nile Tilapia Oreochromis niloticus and indigenous congenerics in Southern Africa: Should we be concerned? Entropy 2015, 17, 4959–4973. [Google Scholar] [CrossRef]
- Kang, J.M. Feeding habits of starry flounder (Platichthys stellatus) in middle East sea of Korea. Master’s Thesis, Pukyong National University, Busan, Republic of Korea, 2013. [Google Scholar]
- Nam, K.M. The ecological study of Pleuronectes yokohamae and Pleuronicthys cornutus in the coastal waters off Tongyeong, Korea. Ph.D. Thesis, Pukyong National University, Busan, Republic of Korea, 2013. [Google Scholar]
- Heo, Y.S. Feeding habits of the stone flounder, Kareius bicoloratus in the coastal waters off Tongyeong, Korea. Master’s Thesis, Pukyong National University, Busan, Republic of Korea, 2013. [Google Scholar]
- Yeom, S.D. Diet composition of juvenile sea perch, Lateolabrax japonicus in tidal creek at Sangnae-ri Suncheon, Korea. Master’s Thesis, Gyeongsang National University, Jinju, Republic of Korea, 2015. [Google Scholar]
- Huh, S.-H.; Kwak, S.N. Feeding habits of Favonigobius gymnauchen in the eelgrass (Zostera marina) bed in Kwangyang Bay. Korean J. Fish. Aquat. Sci. 1998, 31, 372–379. [Google Scholar]
- Ye, S.J. Species composition of fishes and feeding habits of dominant species Tridentiger bifasciatus in the tidal creek of Sangnae-ri Suncheon, Korea. Master’s Thesis, Gyeongsang National University, Jinju, Republic of Korea, 2014. [Google Scholar]
- Choi, D.U. Feeding habits of three species of Gobiid Fishes (Gobiidae) from the Beolgyo intertidal zone, Korea. Ph.D. Thesis, Kunsan National University, Kunsan, Republic of Korea, 2017. [Google Scholar]
- Kim, J.-Y.; Noh, Y.-T. Feeding habits of Acanthogobius elongatus from the Kunsan coast intertidal zone, Neacho-do in the West Coast of Korea. Korean J. Fish. Aquat. Sci. 1997, 30, 413–422. [Google Scholar]
- Kim, J. Feeding habits of Acanthogobius luridus inhabiting the intertidal zone of the western coast of Korea. Korean J. Fish. Aquat. Sci. 2000, 13, 309–316. [Google Scholar]
- Paik, E.-I. A study on the food of the goby, Synechogobius hasta. Korean J. Fish. Aquat. Sci. 1969, 2, 47–62. [Google Scholar]
- Baeck, G.-W.; Park, C.-I.; Jeong, J.-M.; Kim, M.-C.; Huh, S.-H.; Park, J.-M. Feeding habits of Chaenogobius gulosus in the coastal waters of Tongyeong, Korea. Korean J. Ichthyol. 2010, 22, 41–48. [Google Scholar]
- Kim, B.G.; Kim, J.H.; Chung, S.W.; Han, K.N. Feeding ecology of Luciogobius guttatus (Pisces; Gobiidae) in the Youngjong tide pool, Incheon, Korea. Korean J. Ichthyol. 2014, 26, 202–211. [Google Scholar]
- Chung, S.-W.; Kim, B.-G.; Kim, J.-H.; Kim, M.-G.; Han, K.-N. Feeding ecology of Collichthys lucidus in the Han River estuary, Korea. Korean J. Ichthyol. 2014, 26, 303–309. [Google Scholar]
- Huh, S.-H.; Kwak, S.-N. Feeding habits of Leiognathus nuchalis in eelgrass (Zostera marina) bed in Kwangyang Bay. Korean J. Ichthyol. 1997, 9, 221–227. [Google Scholar]
- Choi, H.C.; Park, J.M.; Baeck, G.W.; Huh, S.H. The summer diet of a juvenile barbell steed, Hemibarbus labeo, in the surf zone of the Nakdong River estuary, Korea. J. Korean Soc. Mar. Environ. Saf. 2016, 22, 766–772. [Google Scholar] [CrossRef]
- Baeck, G.; Huh, S.H.; Park, J.M. Diet composition of juvenile Korean piscivorous chub, Opsariichthys uncirostris amurensis in the surf zone of Nakdong river estuary. Korea. J. Korean Soc. Fish. Technol. 2014, 50, 334–341. [Google Scholar] [CrossRef]
- Choi, H.C.; Han, I.S.; Suh, Y.S.; Huh, S.H. Feeding habits of larval Konosirus punctatus from the Nakdong River estuary, Korea. Korean J. Fish. Aquat. Sci. 2015, 48, 752–759. [Google Scholar]
- Huh, S.-H.; Kwak, S.N. Feeding habits of Syngnathus schlegeli in eelgrass (Zostera marina) bed in Kwangyang Bay. Korean J. Fish. Aquat. Sci. 1997, 30, 896–902. [Google Scholar]
- Choi, H.C. Species Composition of Fishes and Feeding Habits of Dominant Species Takifugu niphobles in the Eelgrass Bed of Jangpyeong in Tongyeong, Korea; Gyeongsang National Uniersity: Jinju, Republic of Korea, 2018. [Google Scholar]
- Azzurro, E.; Fanelli, E.; Mostarda, E.; Catra, M.; Andaloro, F. Resource partitioning among early colonizing Siganus luridus and native herbivorous fish in the Mediterranean: An integrated study based on gut-content analysis and stable isotope signatures. J. Mar. Biolog. Assoc. UK 2007, 87, 991–998. [Google Scholar] [CrossRef]
- Hansson, S. Methods of studying fish feeding: A comment. Can. J. Fish. Aquat. Sci. 1998, 55, 2706–2707. [Google Scholar] [CrossRef]
- Nishimoto, A.; Iida, M.; Yokouchi, K.; Fukuda, N.; Yamamoto, T. Eels as natural samplers highlight spatial heterogeneity in energy flow in an estuary. Estuar. Coast. Shelf Sci. 2023, 281, 108215. [Google Scholar] [CrossRef]
- Beals, E.W. Bray-Curtis ordination: An effective strategy for analysis of multivariate ecological data. In Advances in Ecological Research; Elsevier: Amsterdam, The Netherlands, 1984; Volume 14, pp. 1–55. [Google Scholar] [CrossRef]
- Ward, J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Int. J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’hara, R.; Simpson, G.; Solymos, P. Vegan: Community Ecology Package, Version 2.5-6; The Comprehensive R Archive Network: Redmond, WA, USA, 2019.
- Fang, Q.; Huang, S.Q. A directed network analysis of heterospecific pollen transfer in a biodiverse community. Ecology 2013, 94, 1176–1185. [Google Scholar] [CrossRef]
- Levins, R. Evolution in Changing Environments: Some Theoretical Explorations; Princeton University Press: Princeton, NJ, USA, 1968. [Google Scholar]
- Horn, H.S. Measurement of “overlap” in comparative ecological studies. Am. Nat. 1966, 100, 419–424. [Google Scholar] [CrossRef]
- Cortes, E. Standardized diet compositions and trophic levels of sharks. ICES Mar. Sci. Symp. 1999, 56, 707–717. [Google Scholar] [CrossRef]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V. Fishes of the World; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Colloca, F.; Carpentieri, P.; Balestri, E.; Ardizzone, G. Food resource partitioning in a Mediterranean demersal fish assemblage: The effect of body size and niche width. Mar. Biol. 2010, 157, 565–574. [Google Scholar] [CrossRef]
- O’Shea, O.R.; Thums, M.; van Keulen, M.; Kempster, R.M.; Meekan, M.G. Dietary partitioning by five sympatric species of stingray (Dasyatidae) on coral reefs. J. Fish Biol. 2013, 82, 1805–1820. [Google Scholar] [CrossRef] [PubMed]
- Brabrand, A. Food of roach (Rutilus rutilus) and ide (Leusiscus idus): Significance of diet shift for interspecific competition in omnivorous fishes. Oecologia 1985, 66, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, H.R.; Morrison, C.D.; Ackroff, K.; Sclafani, A. Learning of food preferences: Mechanisms and implications for obesity & metabolic diseases. Int. J. Obes. 2021, 45, 2156–2168. [Google Scholar] [CrossRef]
- Kraft, N.J.B.; Godoy, O.; Levine, J.M. Plant functional traits and the multidimensional nature of species coexistence. Proc. Natl. Acad. Sci. USA 2015, 112, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Schoener, T.W. The newest synthesis: Understanding the interplay of evolutionary and ecological dynamics. Science 2011, 331, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jeong, J.M.; Kim, H.J.; Ye, S.J.; Baeck, G.W. Feeding habits of javelin goby Synechogobius hasta on tide flat in Sangnae-ri Suncheon, Korea. Korean J. Fish. Aquat. Sci. 2015, 48, 982–987. [Google Scholar] [CrossRef]
- Hwang, S.W.; Choi, K.H.; Hwang, H.; Kim, C.K.; Lee, T.W. Migration history and habitat use by javelin goby Synechogobius hasta as inferred from otolith Sr:Ca ratios. J. Coast. Res. 2015, 31, 299–304. [Google Scholar] [CrossRef]
- Pimm, S.; Lawton, J.H. On feeding on more than one trophic level. Nature 1978, 275, 542–544. [Google Scholar] [CrossRef]
- Kurita, Y.; Nakajima, J.; Kaneto, J.; Onikura, N. Analysis of the gut contents of the internal exotic fish species Opsariichthys uncirostris uncirostris in the Futatsugawa River, Kyushu island, Japan. J. Fac. Agric. Kyushu Univ. 2008, 53, 429–433. [Google Scholar] [CrossRef]
- Popova, O.; Sytina, L. Food and feeding relations of Eurasian perch (Perca fluviatilis) and pikeperch (Stizostedion lucioperca) in various waters of the USSR. J. Fish. Res. Board Can. 1977, 34, 1559–1570. [Google Scholar] [CrossRef]
- Castillo-Rivera, M. Influence of rainfall pattern in the seasonal variation of fish abundance in a tropical estuary with restricted marine communication. J. Water Resour. Prot. 2013, 5, 29214. [Google Scholar] [CrossRef]
- Guo, K.; Zhao, W.; Wang, S.; Dong, S.L. Study of food web structure and trophic level in the sea ponds of an optimized culture model (jellyfish-shellfish-fish-prawn). Aquac. Int. 2014, 22, 1783–1791. [Google Scholar] [CrossRef]
- Guo, K.; Zhao, W.; Wang, S.; Liu, B.Z.; Zhang, P. Food web structure and trophic levels in a saltwater pond sea cucumber and prawn polyculture system. Acta Oceanol. Sin. 2016, 35, 58–62. [Google Scholar] [CrossRef]
- Davis, A.M.; Blanchette, M.L.; Pusey, B.J.; Jardine, T.D.; Pearson, R.G. Gut content and stable isotope analyses provide complementary understanding of ontogenetic dietary shifts and trophic relationships among fishes in a tropical river. Freshwater Biol. 2012, 57, 2156–2172. [Google Scholar] [CrossRef]
- Essington, T.E.; Beaudreau, A.H.; Wiedenmann, J. Fishing through marine food webs. Proc. Natl. Acad. Sci. USA 2006, 103, 3171–3175. [Google Scholar] [CrossRef]
- Pauly, D.; Christensen, V.; Dalsgaard, J.; Froese, R.; Torres, F. Fishing down marine food webs. Science 1998, 279, 860–863. [Google Scholar] [CrossRef]
- Pinnegar, J.K.; Polunin, N.V.C. Differential fractionation of delta C-13 and delta N-15 among fish tissues: Implications for the study of trophic interactions. Funct. Ecol. 1999, 13, 225–231. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).