Abstract
The family Rissoinidae represents a significant component of microgastropod diversity, with a global distribution spanning temperate to tropical zones and encompassing over 300 recorded species. Hainan Island, the largest island in the South China Sea, harbors a rich diversity of mollusks, but the family Rissoinidae remains poorly studied in this region. Here, we report three rissoinid species and one newly recorded species from Hainan Island, providing detailed taxonomic descriptions supported by SEM imaging. For the first time, we provide the mitochondrial genomes of Rissoina cardinalis and Phosinella seguenziana, analyzing their genome structure and nucleotide composition, thereby addressing the existing knowledge gap in Rissoinidae research. A phylogenetic tree of the family Rissoinidae was reconstructed using the COI gene, clarifying the intergeneric relationships within the family. Notably, the genus Rissoina is revealed as a non-monophyletic group, likely due to the limitations of single-gene analyses in providing adequate phylogenetic information.
Key Contribution:
This study reports four Chinese species of the family Rissoinidae, including one newly recorded species in China, and presents the first two mitochondrial genomes within this family.
1. Introduction
Marine microgastropods represent a significant yet underexplored component of gastropod diversity [1]. The family Rissoinidae W. Stimpson, 1865 (Rissooidea) constitutes one of the predominant microgastropod families, encompassing over 300 extant species distributed globally. Members of this family are widely distributed, spanning temperate to tropical regions [2]. These gastropods predominantly inhabit shallow waters, with the highest species diversity observed in the low intertidal and shallow sublittoral zones, where algae, stones, or corals provide refuge. The family Rissoinidae possesses a well-documented fossil record, tracing back to at least the Middle Jurassic period [3]. Research on rissoinids has historically been constrained, predominantly focusing on systematics [2,3,4].
Owing to the pronounced convergence in shell morphology, rissoinids have undergone a complex and evolving classification process. Rissoinids were initially classified within the family Rissoidae, with certain groups designated as the subfamily Rissoininae [5,6]. Taxonomists classified groups with an apophysis on the inner side of the horny operculum as part of the family Rissoinidae, which was later demoted to a subfamily within Rissoidae [7,8,9,10]. Since then, the questions regarding the systematic classification of this group has remained unresolved. Bouchet et al. [11] elevated the subfamily Rissoininae to the rank of family Rissoinidae, a change supported by phylogenetic evidence and widely accepted by scholars. To date, Rissoinidae comprises 37 extant genera and 13 fossil genera [12]. Research efforts have predominantly focused on the Mediterranean, South Pacific, and Atlantic Oceans [13,14,15,16]. However, there is a notable lack of research on the taxonomy and phylogeny of Rissoinidae in the South China Sea.
Hainan Island, the largest island in the South China Sea, harbors a remarkable diversity of mollusks. This study conducted a comprehensive survey of the diversity and taxonomy of Rissoinidae on Hainan Island and reconstructed the molecular phylogeny of the family. Additionally, this study is the first to report and characterize two complete mitochondrial genomes of Rissoinidae. This work offers novel insights into the diversity and evolutionary relationships of Rissoinidae in the South China Sea.
2. Materials and Methods
2.1. Taxon Sampling and Processing
Samples were collected from the intertidal zone of Hainan Island on 20 September 2022 and 10 September 2024. All specimens were preserved in 95% ethanol. Standard measurements were performed using a stereomicroscope equipped with an eyepiece micrometer. DNA extraction was performed on intact specimens using the commercial TIANamp Marine Animals DNA Kit (Tiangen Biotech, Beijing, China). All procedures strictly followed the manufacturer’s protocol. The extracted DNA was maintained at −4°C for short-term preservation. Shells, radulae, and opercula were examined using a scanning electron microscope (SEM) (TESCAN (China) Co., Ltd., Shanghai, China), following the protocols described by Xu et al. [17] and Qi et al. [18]. For SEM studies of the radulae, the surrounding tissues were digested with proteinase K during DNA extraction using the TIANamp Marine Animals DNA Kit. Radulae were precipitated to the bottom of the centrifuge tube after separation and collected using a pipette. Then, the residual material at the bottom of the centrifuge tube was transferred onto a glass slide using a micropipette, and the radula was carefully relocated to a clean slide under a stereomicroscope. Subsequently, the radulae were washed with drops of distilled water or 10% KOH on glass histology slides. The shells, radulae, and opercula were affixed to stubs, coated with a layer of gold via sputtering, and examined using a TESCAN VEGA3 scanning electron microscope (TESCAN (China) Co., Ltd., Shanghai, China).
2.2. Sequencing, Assembly, and Annotation
Total genomic DNA was subjected to library preparation and 150 bp paired-end sequencing using Illumina’s HiSeq X platform, with all technical procedures conducted at Beijing Novogene Technology Co., Ltd. (Beijing, China). The bioinformatics analysis followed the pipeline described by Qi et al. [19]. The clean sequencing reads were de novo assembled through SPAdes v3.13.0 [20], employing the software’s standard parameter settings. The complete mitochondrial genome was reconstructed using NOVOPlasty 4.3.5 [21], with the previously published COI gene of Rissoina ambigua as the seed. The mitogenomes were annotated through the MITOS WebServer [22]. The complete circular visualization of the mitochondrial genome was constructed by employing CGView, a specialized tool for mitogenome mapping [23]. The AT and GC skew values were computed according to the standard formulae: AT skew = (A − T)/(A + T), and GC skew = (G − C)/(G + C).
2.3. Phylogenetic and Genetic Divergence Analysis
As no mitochondrial genomes of Rissoinidae were available in GenBank, we reconstructed phylogenetic trees for the family based on COI sequences, combining our data with GenBank sequences, which included six Rissoinidae taxa and one Barleeidae species, Ansola angustata (Pilsbry, 1901), as the outgroup (Table 1). COI genes of Rissoina cardinalis Brazier, 1877 and Phosinella seguenziana (Issel, 1869) from the family Rissoinidae were extracted from their mitochondrial genomes. Phylogenetic analyses were conducted using the maximum likelihood (ML) method. ML analyses were performed using IQ-TREE [24] with the nucleotide substitution model K3Pu+F+G4 selected by ModelFinder, along with 1000 ultrafast bootstrap replicates. The pairwise distances of COI within the Rissoinidae clade were calculated using the p-distance in MEGA X [25].

Table 1.
GenBank accession numbers for specimens included in the molecular analyses.
3. Results and Discussion
3.1. Systematics
Rissoinidae W. Stimpson, 1865
Rissoina d’Orbigny, 1841
Type species: Rissoa inca A. d’Orbigny, 1841 from the delta of Peru.
Rissoina cardinalis Brazier, 1877
Costalynia birestes Laseron, 1956
Rissoina (Rissolina) cardinalis Brazier, 1877
Type locality: Papua New Guinea.
Description: Shell—minute (6.2 ± 0.04 mm in height; 1.3 ± 0.02 mm in width), narrowly conical, stout; most specimens are uniformly white, but some exhibit a pale-brown spiral band on the abapical half of the spire whorls and the adapical half of the body whorl (Figure 1A,B). Protoconch of planktotrophic larval type, of 1.5, relatively weakly convex whorls, each being 1.5 times wider than high; protoconch–teleoconch boundary sharply demarcated, with moderately thickened margin (Figure 1G,H). Teleoconch of 7.5 very weakly convex whorls; spire whorls weakly angulate below and above suture; body whorl weakly angulate below suture and strongly contracted near the base; suture weakly undulating, not impressed (Figure 1C,D). Axial ribs of prominent, very narrow, rather distantly spaced, rounded to sharp, weakly opisthocline ribs; interspaces rather deep and slightly wider than axial ribs (Figure 1F). Spiral sculpture is absent on spire whorls, apart from some very weak and irregularly spaced lirae; last whorl with a rather variable number of spiral riblets—usually two or three—above contraction near shell base; body whorl with prominent, narrow, and nodular spiral fold, which is ornamented with fine, inconspicuous lirae (Figure 1F). Aperture D-shaped; inner lip thin, apart from moderately prominent swelling near rather wide, shallow, and short anterior channel; anterior margin of anterior channel moderately curved dorsally; inner side of outer lip thin; prominent and moderately wide labial varix, bearing very prominent spiral ribs on its entire length or restricted on abapical half; outer lip very weakly opisthocline in profile (Figure 1C,D). No umbilicus.
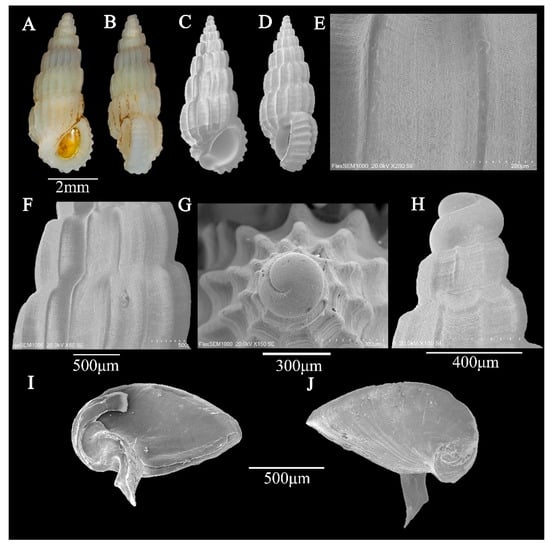
Figure 1.
(A–J) Rissoina cardinalis Brazier, 1877. (A,B) shell illustration in color; (C) scanning electron micrographs of frontal view of shell; (D) scanning electron micrographs of lateral view of shell; (E,F) detail of teleoconch surface; (G) scanning electron micrographs of apical view of protoconch; (H) scanning electron micrographs of lateral view of protoconch; (I) inner face of operculum; (J) outer face of operculum.
Operculum: Horny, thick, yellowish, translucent, D-shaped, posteriorly broadly angled, anteriorly rounded; nucleus eccentric, located in the lower left corner; growth line is obvious, arc-shaped, and radiating; there is a strong, curved, grooved nail-like structure growing from the core on the inner side, and the spike-like structure ends in a V shape; a weak ridge runs parallel to the medial edge (Figure 1I,J).
Radula: Radular teeth interlocked moderately in unfolded condition (Figure 2A–E). Central tooth , with long triangular cutting edge, small cusps, and a single pair of basal denticles (Figure 2B); the U-shaped ventral extension area is well developed. Lateral teeth 5 − 6 + 1 + 5 − 6, wide; small cusps at the end, larger primary cusp long and wide; smaller pointed denticles at the sides, 5–6 each inner and outer (Figure 2A). Two rows of marginal teeth, elongated; inner marginal teeth 9 − 12 + 1 + 1 − 2, larger primary cusp, 9–12 cusps on outer, 1–2 on inner; outer marginal teeth 3 − 4 + 1 + 0, only 3–4 cusps on outer (Figure 2C–E).
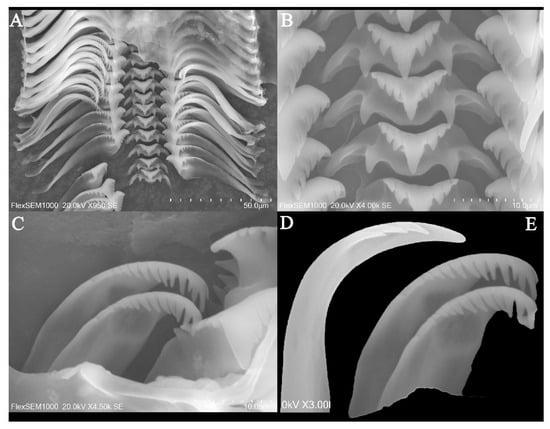
Figure 2.
(A–E) scanning electron micrographs of the radula of Rissoina cardinalis Brazier, 1877. (A) partial photo of radula; (B) central tooth; (C,E) inner marginal teeth; (D) outer marginal teeth.
Distribution: Queensland (Lindeman Island) to Northern Territory (Point Charles) in Australia; southern Papua New Guinea (Katow); and China (Eman Town, Danzhou City, Hainan Island, and Taiwan Province).
Remarks: Rissoina cardinalis Brazier, 1877 strongly resembles Rissoina (Rissolina) angasii Pease, 1871 in shell shape and sculpture, but differs essentially in protoconch structure. R. cardinalis is a planktotrophic larvae type, while R. angasii is a non-planktotrophic larvae type. Furthermore, the outer lip exhibits a prominent and moderately wide labial varix.
Rissoina ambigua (A. Gould, 1849)
Pyramidella ambigua A. Gould, 1849
Rissoina (Rissoina) ambigua (A. Gould, 1849)
Type locality: Clermont Tonnerre Island of Polynesia.
Description: Shell—minute (5.5 ± 0.07 mm in height; 2.3 ± 0.05 mm in width), elongated conical, stout; white with light reddish-brown spiral stripes, 8 whorls (Figure 3A,B). Protoconch of planktotrophic larval type, of 2.5 whorls; of 1 whorl, smooth; of 2–2.5 whorls, weak spiral sculpture; protoconch–teleoconch boundary sharply demarcated (Figure 3H,I). Teleoconch of 6 weakly convex whorls; only axial ribs, interspace slightly wider than axial rib; spiral sculpture absent, apart from some very weak and irregularly spaced lirae (Figure 3C–F). Aperture thin, D-shaped; anterior channel shallow and short, posterior angulated; inner lip thin, smooth, central concave; outer lip thin, outer side of the outer lip without labial varix; axial ribs contract near shell base; no columella (Figure 3G).
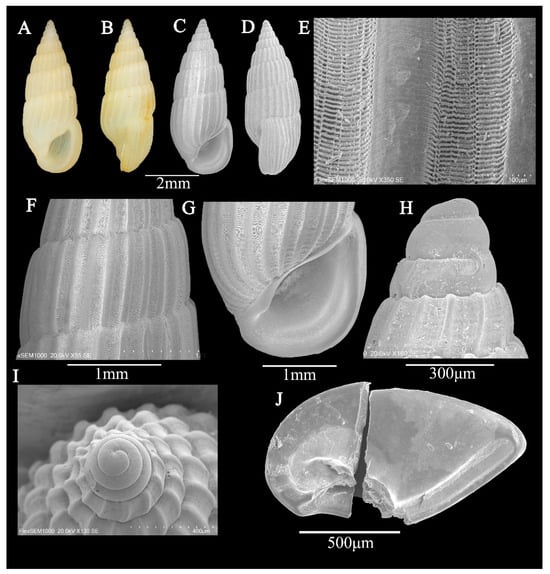
Figure 3.
(A–J) Rissoina ambigua (A. Gould, 1849). (A,B) Shell illustration in color; (C) scanning electron micrographs of frontal view of shell; (D) scanning electron micrographs of lateral view of shell; (E,F) detail of teleoconch surface; (G) detail of aperture; (H) scanning electron micrographs of lateral view of protoconch; (I) scanning electron micrographs of apical view of protoconch; (J) inner face of operculum.
Operculum: thick, solid, D-shaped, posterior angulated; inner edge with groove, nail-like structure growing from the core on the inner side (Figure 3J).
Distribution: Rissoina ambigua is a widely distributed and abundant species primarily inhabiting the tropical Indian and western Pacific Oceans. Its range extends from Tanzania and Mozambique in the Indian Ocean to the Marquesas and Pitcairn Islands in the east. It is also found in the subtropical and tropical waters of China, Japan, and southwestern Australia, as well as in regions such as the Red Sea and Hawaii.
Remarks: The protoconch of R. ambigua suggests its planktotrophic larval development, typical of species inhabiting subtropical and tropical intertidal zones. It was frequently misidentified as the common species Rissoa rosea Deshayes, 1863, which subsequent taxonomic revisions have reassigned to the genus Rissoina. R. ambigua differs from R. rosea via its larger size and greater color variability, confirming that they are distinct species.
R. ambigua bears a strong resemblance to Rissoina stricta (Menke, 1851), a common species in the Panamic faunal province, particularly in teleoconch characteristics. Both are quite variable in size, slenderness, and number and density of the axial ribs, which resulted in many synonyms. They might be synonyms; however, “the protoconch is somewhat more conical in R. stricta … the penes … differ markedly” [26]. Since there is no information on its anatomy, we provisionally regard this single Clipperton specimen as R. stricta [5].
Apataxia Laseron, 1956
Type species: Apataxia erecta Laseron, 1956 = Rissoina miltozona Tomlin, 1915 from Australia.
Apataxia cerithiiformis (Tryon, 1887)
Rissoina balteata Pease, 1870
Rissoina cerithiiformis “Dunker” Tryon, 1887
Apataxia erecta Laseron, 1956
Type locality: Red Sea.
Description: Shell—minute (2.9 ± 0.03 mm in height; 1.2 ± 0.01 mm in width), thick, rather stout, elongated conical, white, translucent, with yellowish or bright-brown spiral band above the suture line, 9 whorls (Figure 4A–C). Protoconch of planktotrophic larval type, yellowish-brown, 3.5 convex, smooth apart from wavy sculptures in the lower part of the last convex, protoconch–teleoconch boundary clear (Figure 4G,H). Teleoconch of 5.5 weakly convex to almost flat whorls; axial ribs prominent, narrow, interspaces wider than axial ribs, microscopic (only observable through SEM) striae in interspaces between axial ribs (Figure 4D,F), with weak ribs; different whorl have different number of ribs; sutures distinct, grooved; the axial ribs of body whorl contract near shell base; without umbilicus. Aperture small, thick; anterior channel shallow and short, posterior angulated; inner lip smooth, central concave; 4 weak ridges on the inner side of the outer lip.
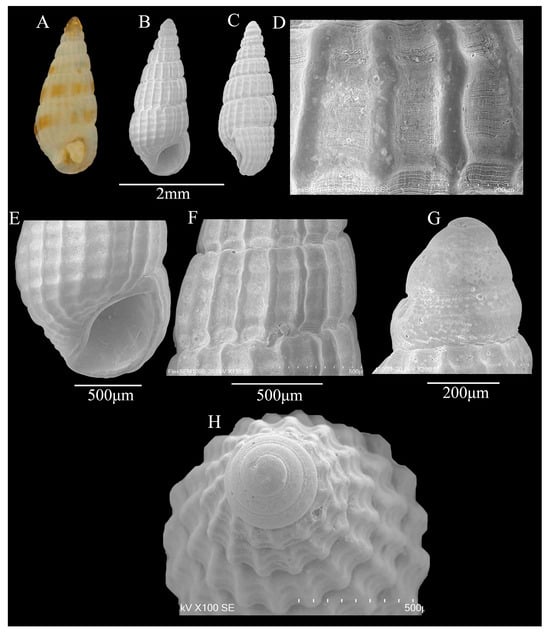
Figure 4.
(A–H) Apataxia cerithiiformis (Tryon, 1887). (A) Shell illustration in color; (B) scanning electron micrographs of frontal view of shell; (C) scanning electron micrographs of lateral view of shell; (D,F) detail of teleoconch surface; (E) detail of aperture; (G) scanning electron micrographs of lateral view of protoconch; (H) scanning electron micrographs of apical view of protoconch.
Distribution: A. cerithiiformis is a widely distributed and common Indo-Pacific species, recorded from the Red Sea and Tanzania to China, Japan, Queensland, and the Hawaiian Islands. There are no previous eastern Pacific records [5]. It has a protoconch indicating planktotrophic larval development.
Remarks: Apataxia Laseron, 1956, initially proposed by Laseron (1956), was subsequently classified as a subgenus of Rissoina by Ponder [9]. Faber and Kaiser [5] reclassified Apataxia as a full genus, rather than a subgenus of Rissoina d’Orbigny, 1840. The former differs from Rissoina in that it does not have an incrassate outer lip; instead, the adult aperture is internally reinforced with a series of spiral ribs ending in tooth-like structures. Consequently, the adult aperture is narrower than that of the sub-adult stage. A. cerithiiformis is a widespread and common species occurring throughout the Indo-Pacific region. Rissoina balteata Pease, 1869, is sometimes considered synonymous with A. cerithiiformis. However, no type material is known [27], and the species is described as having a varicose outer lip [5]. This characteristic does not align with the diagnostic features of the genus Apataxia.
Phosinella Mörch, 1876
Type species: Rissoina pulchra (C. B. Adams, 1850) from Jamaica
Phosinella seguenziana (Issel, 1869)
Rissoina seguenziana Issel, 1869
Rissoina tornatilis A.A. Gould, 1861
Type locality: the Gulf of Suez.
Description: Shell—minute (3.7 ± 0.02 mm in height; 1.6 ± 0.04 mm in width), thick, solid, turret-like, with an elongated spire, white or yellowish, translucent, whorls: 7.5 (Figure 5A–D). Protoconch of planktotrophic larval type, yellowish, about 2.5 convex, smooth, protoconch–teleoconch boundary clear (Figure 5I,J). Teleoconch of 5 weakly convex whorls, spire whorls weakly angulate below suture; the axial ribs and spiral ribs on the shell surface are interlaced in a clathrate sculpture, and the intersection points are nodular; fine spiral sculptures in each gird; 3 ribs on 1-3 whorls of teleoconch, 4 ribs on 5 whorl of teleoconch, 4 ribs on body whorl; the axial ribs of body whorl contract near shell base; single weak spiral near the shell base; sutures moderately deep (Figure 5C,D,F–H). Aperture D-shaped; anterior channel shallow and short, posterior angulated; inner lip thin, smooth, central concave; outer lip thick, slopes forward; the outer side of the outer lip with labial varix; no columella (Figure 5E).
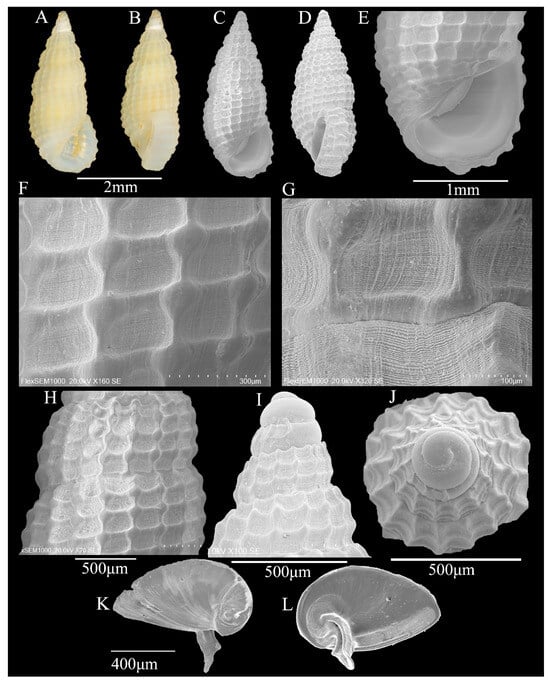
Figure 5.
(A–L) Phosinella seguenziana (Issel, 1869). (A,B) Shell illustration in color; (C) scanning electron micrographs of frontal view of shell; (D) scanning electron micrographs of lateral view of shell; (E) detail of aperture; (F–H) detail of teleoconch surface; (I) scanning electron micrographs of lateral view of protoconch; (J) scanning electron micrographs of apical view of protoconch; (K) outer face of operculum; (L) inner face of operculum.
Operculum: Horny, thick, yellow, D-shaped; nucleus eccentric, located in the lower left corner; growth line is obvious; there is a strong, curved, grooved nail-like structure growing from the core on the inner side, and the spike-like structure ends in a V shape; a weak ridge runs parallel to the medial edge (Figure 5K,L).
Distribution: Red Sea, Indian Ocean, and tropical Western Pacific as far east as Fiji and northward to China’s Hainan Island and Beibu Gulf.
Remarks: The genus Phosinella differs from Rissoina primarily in its clathrate sculptured shell, while remaining similar in other aspects of shell morphology. Phosinella is predominantly distributed in the tropical Indo-Western Pacific region. P. seguenziana strongly resembles P. media (Schwartz von Mohrenstern, 1860) in shell shape and sculpture, but differs in that it has single weak spiral folds near the shell base instead of two or three prominent folds in P. media. In this study, P. seguenziana is recorded in China for the first time.
3.2. Mitogenome Architecture Genome
The mitogenomes of Rissoina cardinalis (Figure 6, Genbank PQ767088) and Phosinella seguenziana (Figure 7, Genbank PQ767087) are 17321bp and 17221bp, respectively. Each comprises 37 genes, including 13 PCGs, 2 rRNAs genes, 22 tRNAs genes, and a putative control region (CR). The CRs are 605bp and 666bp, respectively. The total lengths of the concatenated 13 PCGs of Rissoina cardinalis and Phosinella seguenziana are 11310bp and 11403bp, respectively, with an average A + T contents of 63.72% and 63.57% (Table 2 and Table 3). ATG is the most frequently used start codon; however, nad4 uses ATA in Rissoina cardinalis (Table 4), while cox3, nad2, and nad4 use ATA and TTA in Phosinella seguenziana (Table 5). The terminal codons of Rissoina cardinalis and Phosinella seguenziana consist of TAG and TAA, respectively. The total lengths of the tRNA sequences in R. cardinalis and P. seguenziana are 1541 bp and 1545 bp, with an A + T contents of 67.16% and 66.34%, respectively. The rrnL and rrnS genes are 1562 bp and 1199 bp in size, with an A + T contents of 68.56% and 69.22% in R. cardinalis, and 1635 bp and 1135 bp, with an A + T contents of 67.62% and 64.84% in P. seguenziana (Table 2 and Table 3), respectively.
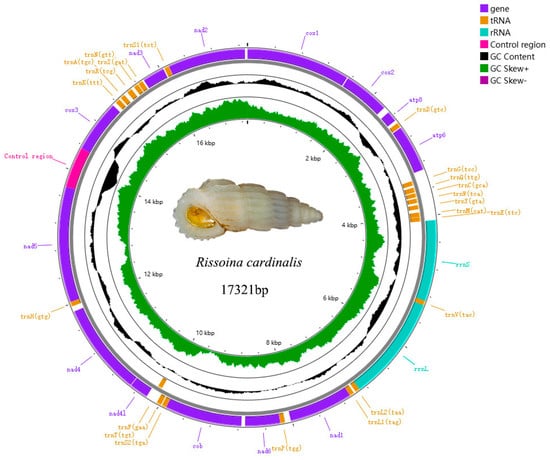
Figure 6.
Map of the complete mitochondrial genome of Rissoina cardinalis Brazier, 1877.
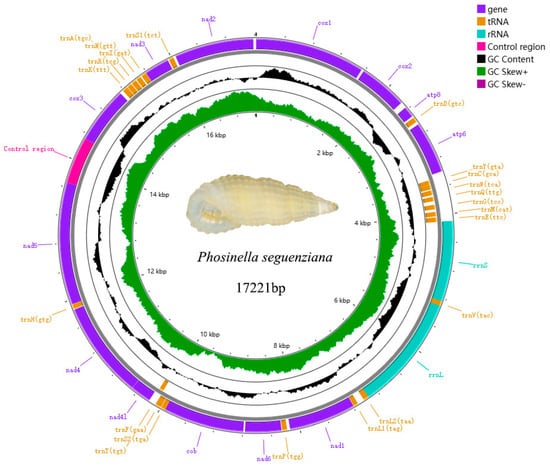
Figure 7.
Map of the complete mitochondrial genome of Phosinella seguenziana (Issel, 1869).

Table 2.
The nucleotide composition of the Rissoina cardinalis mitogenome.

Table 3.
The nucleotide composition of the Phosinella seguenziana mitogenome.

Table 4.
Annotated mitochondrial genome of Rissoina cardinalis.

Table 5.
Annotated mitochondrial genome of Phosinella seguenziana.
3.3. Phylogenetic and Genetic Diversity Analysis
The phylogenetic analysis of the family Rissoinidae, including most Rissoinidae species with COI in the NCBI, inferred the phylogenetic relationships of rissoinids (Figure 8). Pandalosia forms a strongly supported sister group with Stosicia. Rissoina does not form a monophyletic group. However, Phosinella and Rissoina sp. cluster together to form a branch, which in turn forms an unsupported sister group with Rissoina ambigua. Although Rissoina cardinalis represents the outermost branch within the family Rissoinidae, it is unsupported. The non-monophyly of Rissoina is further corroborated by Francesco et al. [28] and may be attributed to the limitations of single-gene analyses in providing a sufficient phylogenetic signal. Pairwise genetic distances among species of Rissoinidae, derived from the COI gene, are presented in Table 6. Pairwise genetic distances within the family Rissoinidae range from 2.9% (Rissoina sp._PNG1679 vs. Rissoina sp._USNM_1466845) to 33.4% (Rissoina cardinalis vs. Pandalosia subulata). Genetic distances between genera range from 16.1% (Phosinella seguenziana vs. Rissoina ambigua) to 33.4% (Pandalosia subulata vs. Rissoina cardinalis). Genetic distances within the genus Rissoina range from 2.9% to 22.9%.
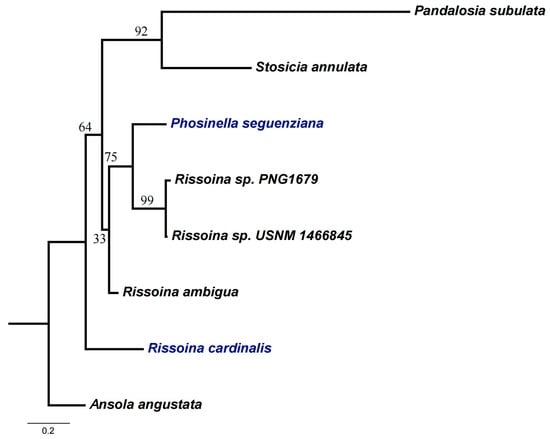
Figure 8.
Summary tree from maximum likelihood analysis of COI sequences. Taxa highlighted in blue represent specimens newly sequenced in this study. Ansola angustata (Barleeiidae) was selected as the outgroup.

Table 6.
Pairwise p-distance among species of Rissoinidae (%).
4. Conclusions
Based on SEM examination, we report four species of Rissoinidae from Hainan Island, China, revealing detailed morphological characteristics of the shells, protoconchs, and opercula, among which Phosinella seguenziana represents a new record for China. The phylogenetic relationships of Rissoinidae were reconstructed using the COI gene, revealing that the genus Rissoina is not monophyletic, likely due to the limitations of single-gene analyses in providing sufficient phylogenetic signals. This study presents the first two complete mitochondrial genomes of Rissoinidae, offering foundational data for future research to resolve the phylogenetic relationships within the family.
Author Contributions
Funding acquisition, L.K. and Z.M.; resources, L.K.; writing—original draft, L.Q.; writing—review and editing, L.K. and Z.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Central Public-Interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, grant number CAFS (NO. 2024TS01); the Key R&D Program of Shandong Province, China (2022TZXD002); the Shandong Provincial Natural Science Foundation (ZR2023MD008); the Qingdao Natural Science Foundation (23-2-1-166-zyyd-jch); the Key Laboratory of Efficient Utilization and Processing of Marine Fishery Resources of Hainan Province (KLEU-2024-10); and the National Key R&D Program of China (2024YFD2401804).
Institutional Review Board Statement
All samples used in this study were obtained from dead shells or alcohol-preserved specimens, not from live animals. Therefore, ethical approval is not required.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data used in this study are available upon request from the corresponding author.
Acknowledgments
Special thanks to Chunsheng Liu and Yi Yang of Hainan University for their generous support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bouchet, P.; Lozouet, P.; Maestrati, P.; Heros, V. Assessing the magnitude of species richness in tropical marine environments: Exceptionally high numbers of molluscs at a New Caledonia site. Biol. J. Linn. Soc. 2002, 75, 421–436. [Google Scholar]
- Sleurs, W.J.M. A review of the present-day Australian species of the gastropod subgenus Rissoina (Rissolina) (Rissooidea: Rissoinidae) with descriptions of two new species. Basteria 2023, 87, 37–76. [Google Scholar]
- Faber, M.J.; Gori, S. Infralittoral Rissoinidae (Gastropoda, Rissooidea) of Maledives with the introduction of a new subfamily and one replacement name, the description of three new species, and a note on the identity of Rissoa rosea Deshayes, 1863. Basteria 2016, 80, 95–112. [Google Scholar]
- Laseron, C.F. The Families Rissoinidae and Rissoidae (Mollusca) from the Solanderian and Dampierian Zoogeographical Provinces. Mar. Freshw. Res. 1956, 7, 384–484. [Google Scholar]
- Faber, M.; Kaiser, K.L. The Rissoinidae of Île Clipperton in the tropical eastern Pacific (Mollusca: Gastropoda). Misc. Malacol. 2015, 7, 19–23. [Google Scholar]
- Stimpson, W. Researches upon the Hydrobiinae and allied forms: Chiefly made upon materials in the Museum of the Smithsonian Institution. Ann. Mag. Nat. Hist. 1865, 17, 393–395. [Google Scholar]
- Coan, E. A proposed revision of the rissoacean families Rissoidae, Rissoinidae, and Cingulopsidae (Mollusca: Gastropoda). Veliger 1964, 6, 164–171. [Google Scholar]
- Ponder, W.F. The classification of the Rissoidae and Orbitestellidae with descriptions of some new taxa. Trans. R. Soc. N. Z. 1967, 9, 193–224. [Google Scholar]
- Ponder, W.F. A review of the genera of the Rissoidae (Mollusca: Mesogastropoda: Rissoacea). In Records of the Australian Museum; Australian Museum: Darlinghurst, Australia, 1985; Volume 4, (Supplement), pp. 1–221. [Google Scholar]
- Bouchet, P.; Rocroi, J.P. Classification and nomenclator of gastropod families. In Malacologia; Conchbooks: Harxheim, Germany, 2005; Volume 47, pp. 1–397. [Google Scholar]
- Bouchet, P.; Rocroi, J.P.; Hausdorf, B.; Kaim, A.; Kano, Y.; Nützel, A.; Parkhaev, P.; Schrödl, M.; Strong, E.E. Revised classification, nomenclator and typification of gastropod and monoplacophoran families. Malacologia 2017, 61, 1–526. [Google Scholar]
- MolluscaBase (Ed.) Electronic File. 2024. Available online: http://www.molluscabase.org (accessed on 21 July 2024).
- Sleurs, W.J.M.; Preece, R.C. The Rissoininae (Gastropoda: Rissoidae) of the Pitcairn Islands, with the description of two new species. J. Conchol. 1994, 35, 67–82. [Google Scholar]
- Sleurs, W.J.M. Rissoina ponderi n. sp. (Caenogastropoda: Rissoinidae) a new endemic species from New South Wales and a comparison with the related species Rissoina elegantula Angas, 1880. Molluscan Res. 2022, 42, 221–228. [Google Scholar]
- Kaim, A. Gradual evolution of the Early Cretaceous marine gastropod Rissoina lineage in central Poland. Acta Palaeontol. Pol. 2002, 47, 667–672. [Google Scholar]
- Ozturk, B. New alien Molluscs in the Mediterranean Sea. Cah. Biol. Mar. 2014, 56, 205–212. [Google Scholar]
- Xu, B.; Qi, L.; Kong, L.; Li, Q. Description of Alvania wangi Xu, Qi & Kong, sp. nov. (Mollusca, Gastropoda, Littorinimorpha, Rissoidae) from the East China Sea. ZooKeys 2022, 1110, 201–217. [Google Scholar]
- Qi, L.; Xu, B.; Kong, L.; Li, Q. Checklist of the micromolluscs in the intertidal zone of the Yellow Sea and Bohai Sea, China. Biodivers. Data J. 2023, 11, e105444. [Google Scholar]
- Qi, L.; Kong, L.; Li, Q. Redescription of Stenothyra glabra A. Adam, 1861 (Truncatelloidea, Stenothyridae), with the first complete mitochondrial genome in the family Stenothyridae. ZooKeys 2020, 991, 69–83. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.; Nikolen, S.; Pham, S.; Prjibelski, A.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar]
- Nguyen, L.T.; Schmidt, H.A.; Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [PubMed]
- Sleurs, W.J.M. A zoogeographical analysis of the rissoinine fauna of the eastern Pacific with special reference to a comparison with the Caribbean fauna and with a checklist of the eastern Pacific Rissoininae, Stimpson 1865. Ann. Soc. R. Zool. Belg. 1989, 119, 155–164. [Google Scholar]
- Johnson, R.I. Types of shelled Indo-Pacific mollusks described by W.H. Pease. Bull. Mus. Comp. Zool. 1994, 154, 1–61. [Google Scholar]
- Francesco, C.; Winston, F.P.; Frank, K.; Tsuyoshi, T.; Yasunori, K. A molecular phylogeny of Rissoidae (Caenogastropoda: Rissooidea) allows testing the diagnostic utility of morphological traits. Zool. J. Linn. Soc. 2017, 179, 23–40. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).