Abstract
The species Brassica rapa includes enormous leafy vegetables with extreme leaf morphological diversity. Leaf traits such as size, shape, weight, and ratio of the leaf blade to the petiole contribute to yield, appearance, and desirability to consumers. These leaf-related traits are controlled by quantitative trait loci (QTLs). The construction of high-density bin maps using low-coverage sequencing is a powerful method for QTL fine-mapping and gene identification. In this study, we performed whole-genome re-sequencing of Wutacai ‘Zhongbaye’ and Chinese cabbage ‘HN53’ and 150 F2 individuals to construct a high-density bin map for QTL mapping of 11 leaf-related traits. The parental lines and F2 population were re-sequenced at 10x and 1x coverage, respectively. A map containing 565 bin markers was constructed based on parental single-nucleotide polymorphisms and a modified sliding window approach. The total map length was 944.6 cM and the average distance of the bins was 1.65 cM. In total, 60 significant QTLs controlling 11 leaf-related traits were detected. We further identified candidate genes responsible for these complex leaf-related traits. These findings suggest that this cost-effective bin-mapping approach is capable of rapid identification of QTLs and candidate genes, and will thus facilitate the dissection of the underlying molecular basis of leaf morphological variations and accelerate the improvement of B. rapa vegetable breeding.
1. Introduction
Brassica rapa (B. rapa, 2n = 20, AA) is an important leafy vegetable crop in Asia, particularly China, Japan, and Korea. B. rapa contains different morphotypes with extreme leaf morphological diversity, including the large curling leaves with knotted leaf surfaces and wide midribs (petioles) of Chinese cabbage, the small round flat leaves with narrow petioles of Wutacai, the smooth leaves with enlarged petioles of Pak choi, and the slender and highly serrated leaves of Mizuna [1,2]. As the leaf is the main edible organ of B. rapa, variations in leaf shape, size, weight, and ratio of the leaf blade to the petiole affect the yield, appearance, and desirability of these vegetables to consumers. Understanding the genetic regulation of these complex leaf-related traits is key to breeding B. rapa vegetable to satisfy consumer preferences.
The genetic regulation of leaf-related traits has been unraveled through quantitative trait locus (QTL) analysis in B. rapa. Genetic linkage maps have been constructed using different molecular markers. For example, simple sequence repeats (SSR) were used to construct genetic linkage maps for the F2, Doubled Haploid (DH), and Recombinant Inbred Lines (RIL) populations to analyze QTLs for agronomic traits such as leaf-head traits, yield, disease resistance, and leaf color in Chinese cabbage [3,4,5,6,7,8]; linkage genetic maps were constructed using restriction fragment length polymorphism (RFLP) markers in different genetic populations of Chinese cabbage such as F2 and RIL for the QTL analysis of traits such as disease resistance and flowering time [5,9,10]; and, genetic linkage maps were constructed using amplified fragment length polymorphism (AFLP) markers for QTL analysis in the F2 population for traits related to root growth, leaf-head formation, pubescence, and disease resistance [11,12,13]. In addition, insertion and deletion (InDel) markers have been used to construct genetic linkage maps [6,14].
A number of QTLs for leaf-related traits have been identified, including leaf size, leaf number, and leaf color. Some well-known candidate genes have been identified based on the analysis of gene functions in trait loci. For example, the genes BrGRF5, BrGA20OX3, BrLNG1, BrKPP2, and BRASSICA RAPA FERREDOXIN-NADP(1)-OXIDOREDUCTASE1, which control leaf shape; the genes BrLNG1 and BrKPP2, which regulate leaf length (LL); BRASSICA RAPA FERREDOXIN-NADP(1)-OXIDOREDUCTASE1, which regulates leaf width (LW); and BrKAN1, BrKAN2, BrREV, BrPNH, and other genes related to the adaxial/abaxial polarity of leaves have been identified [1,15]. In addition, genes including BrSAW1 and BrTCP, which are considered candidate genes involved in leaf-head formation in Chinese cabbage, have been identified [3,16]. Yue et al. also identified the heading candidate genes BrPIN5 and BrSAURs [17]. Although the above traditional molecular markers have been widely used to develop genetic linkage maps for B. rapa [3,10,18], the resulting map density is still too limited for fine-mapping of complex traits of interest.
The wide availability and low cost of next-generation sequencing (NGS) technologies have allowed researchers to sequence the entire genomes of crops within a short period of time. The high-density single-nucleotide polymorphism (SNP) marker map generated from the low-coverage sequencing-based genotyping method has been applied to QTL mapping of agriculturally important traits. Li et al. constructed a linkage map containing 4253 loci using SNP markers and performed QTL analysis and found that a QTL on chromosome A07 was the main cause of stalk color variation in Zicaitai; in addition, several candidate genes associated with stalk color were identified, with BrbHLH49 being the best candidate [19]. Similarly, Liu et al. constructed a genetic linkage map using 5392 SNPs and performed QTL analysis to identify two main QTLs associated with the main flower stalk length in Chinese cabbage [20]. However, low-coverage sequencing results in a relatively large proportion of missing data, a small percentage of SNP genotyping sequencing errors, and false-positive SNPs. Bin markers are more informative for a given population compared with conventional molecular markers, RFLP, SSR, InDel, or single SNP markers, and the sliding window approach can help to remove most of the false-positive SNPs caused by sequencing and mapping errors and can increase the accuracy of SNPs. The bin-mapping strategy based on a sliding-window approach was proven to be powerful for high-density genetic map construction and QTL mapping in crops and vegetables such as maize [21], rice [22], sorghum [23], pepper [24], blueberries [25], carrots [26], and watermelons [27]. However, there is limited published data on bin maps for QTLs in B. rapa. Sun et al. constructed a bin map using the Brassica SNP array for a small DH population (66 lines), which limited the mapping of significant QTLs [28]. Therefore, a large population of 485 F2 plants was used to construct a map with 36 SNPs and 99 InDel markers for the analysis of QTLs linked to rosette leaf and heading traits in Chinese cabbage.
To further understand the genetic regulation of the complex leaf-related traits in B. rapa, 150 F2 individuals were developed from Wutacai ‘Zhongbaye’ and Chinese cabbage ‘HN53’ in this study. SNPs from individuals were generated using whole-genome re-sequencing and used to construct a high-density linkage map using a modified sliding window approach. This study confirmed the effectiveness of the bin-mapping approach for genetically mapping 11 leaf-related traits in B. rapa, and further identified candidate genes regulating leaf morphology and development. This research will facilitate understanding of the genetic mechanisms of these complex leaf-related traits and molecular breeding of B. rapa vegetable crops.
2. Materials and Methods
2.1. Plant Materials and Phenotype Evaluation
An F2 population consisting of 150 individuals was obtained from the selfed cross of Wutacai (ssp. narinosa cv ‘Zhongbaye’) as the male parent and Chinese cabbage (ssp. pekinensis cv ‘HN53’) as the female parent. The maternal parent ‘HN53’ is a heading-type Chinese cabbage that has large leaves with knotted leaf surfaces and enlarged short petioles, whereas the paternal parent ‘Zhongbaye’ has relatively small, smooth round leaves that are dark green in color, with narrow petioles. All plants were grown in a field trial at the experimental farm of the Chinese Academy of Agricultural Sciences in Beijing, China. In this study, a total of 150 F2 individuals were randomly selected for the investigation of 11 leaf morphological traits (Table 1). To investigate leaf characteristics, three fully expanded outer leaves were taken from the two parents and 150 F2 individuals and measured. Of these 11 leaf-related traits, leaf weight traits included LWT and PWT, while leaf size traits included LL, LW, PL, LA, and PA. Leaf shape can be represented by the leaf index, including the LI and the total LL divided by the PL (PI). In addition, the PAI (LA/PA) and PLTI (LWT/PWT) are important for evaluating the appearance or attractiveness of vegetables to consumers. To precisely measure LL, LW, LA, PL, and PA, images of the leaves were scanned and analyzed using ImageJ 1.46r software. The average value of three leaf traits was taken as the trait data. Pearson correlation coefficients were calculated to analyze the relationships between traits.

Table 1.
Description of Brassica rapa leaf trait measurements.
2.2. Isolation of Genomic DNA and Re-Sequencing
Sequencing data for the parental lines ‘HN53’ and ‘Zhongbaye’ were obtained from a previous study conducted by this research group, referred to as ‘sample 19’ and ‘sample 130’ [29], which were sequenced to ~10× coverage. Total genomic DNA of 150 F2 individuals was isolated from leaf tissues using a CTAB procedure as previously described [30], but with modifications. The extracted DNA was submitted for re-sequencing on an Illumina HiSeq 2500 using 150-bp paired-end reads under a 350-bp insert size library, generating ~1× coverage data.
2.3. Genotyping and Bin-Map Construction
The raw reads were filtered as described previously [29]. Filtered reads were aligned to the B. rapa reference genome (Chiifu-401-42) version 3.0. A pooled mapping approach was used to call variations in 150 F2 individuals, and high-quality SNPs between parents were used as confident polymorphic loci [31]. Finally, ungenotyped loci were imputed based on linkage disequilibrium (LD) using the k-nearest neighbor (k-NN) algorithm [32].
A slightly modified sliding-window approach was used to identify recombination breakpoints and to construct a bin map of F2 populations [24]. The genotype in each window was called with a window size of 1 Mb and a step size of 400 kb. The ratio of SNPs with ‘HN53’ and ‘Zhongbaye’ genotypes was calculated. When >70% of SNPs had one parental genotype, the window was called homozygous; otherwise, the window was called heterozygous. Adjacent windows with the same genotypes were combined into blocks and recombinant breakpoints were determined based on the physical locations where the genotype of the window changed. Based on the recombination breakpoint position, a physical bin map was constructed as described by Han et al. [24]. Bins were used as markers to construct a high-density genetic map using JoinMap 4.0 software [33].
2.4. QTL Analysis of Morphological Traits
All phenotyping data were used for QTL analysis in MAPQTL 4.0 software [34]. First, the interval mapping procedure was performed to detect major QTLs. A 1000× permutation test was performed for each trait to calculate the LOD threshold corresponding to a genome-wide false discovery rate of 5% (p < 0.05). Markers with LOD scores equal to or exceeding the threshold were used as cofactors in multiple-QTL-model (MQM) mapping. If new QTLs were detected, the linked markers were added to the cofactor list and the MQM analysis was repeated. If the LOD value of a marker dropped below the threshold in the new model, it was removed from the cofactor list and the MQM analysis was rerun. This procedure was repeated until the cofactor list stabilized.
3. Results
3.1. Variation of Leaf Morphological Traits in the F2 Population
The maternal parent ‘HN53’ has large leaves with short, large petioles, while the paternal parent ‘Zhongbaye’ has relatively small leaves with narrow petioles. The two parental lines showed significant differences in leaf traits (Table 2). A survey of 11 leaf morphological traits in 150 randomly selected F2 individuals revealed that ‘HN53’ had the highest phenotypic values for all traits except the LL/LW ratio (LI). In the F2 population, all 11 leaf-related traits showed a continuous distribution and wide genetic variation (Table 2, Figure 1), indicating that all evaluated traits were quantitatively controlled.

Table 2.
Data for 11 leaf morphological traits in the parents and the F2 population.
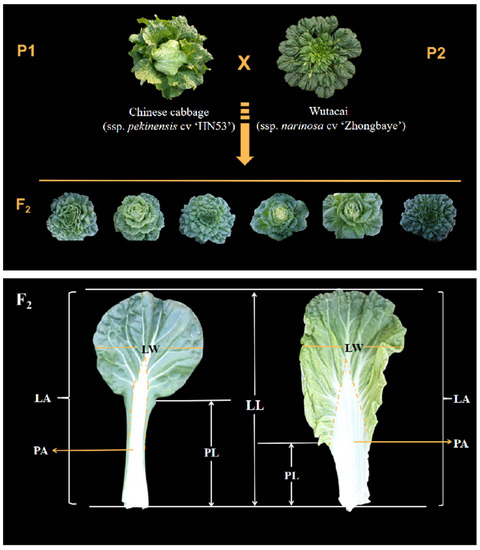
Figure 1.
Morphological traits of heading and non-heading cabbage and their F2 populations are shown on the left. The figure on the right shows two typical plant leaves as examples to describe the different traits measured in this study in relation to leaf morphology. All shapes and their descriptions are listed in Table 1. See Table 1 for trait abbreviations.
Of the 11 leaf-related traits, leaf weight traits included LWT and PWT, and leaf size traits included LL, LW, PL, LA, and PA. Leaf shape can be expressed using the leaf indices LI, PI, PAI, and PLTI. These traits and their phenotypic frequency distribution in the F2 population and their parents are shown in Figure 2.
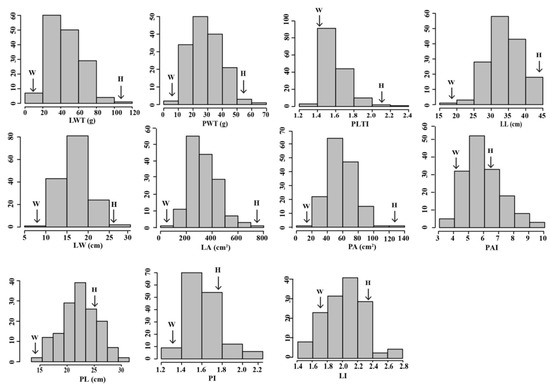
Figure 2.
Frequency distributions of 11 traits in the F2 lines and their parental lines. The vertical axis in each figure represents the number of F2 lines. W and H represent the parental lines ‘Zhongbaye’ and ‘HN53’, respectively. See Table 1 for trait abbreviations.
Most of the traits showed significant positive correlations with other traits (Figure 3). LWT, PWT, LL, LW, LA, and PA were significantly and positively correlated with each other. The highest correlation coefficient was observed between LWT and PWT (0.97). LI was positively correlated with LL and PL, but negatively correlated with the other traits. PLTI was negatively correlated with PL. PI and PAI were negatively correlated with PL. PA had a low correlation with PLTI and LI.
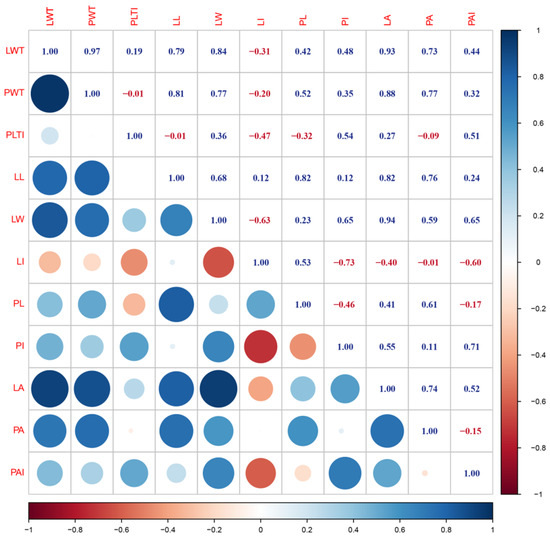
Figure 3.
Correlations between morphological traits evaluated in the F2 population. Blue represents positive correlations and red represents negative correlations. See Table 1 for trait abbreviations.
3.2. Construction of Bin Map Using Low-Coverage Sequencing
One hundred and fifty F2 individuals were sequenced on an Illumina HiSeq 2500, generating a total of 0.3 billion 150-bp paired-end reads. After filtering out low-quality reads, 80 billion re-sequencing data, with an average of 536 Mb per F2 individual, were obtained. This was greater than the one-fold coverage of the B. rapa genome of each F2 individual (Table 3). All filtered reads were used to call SNPs using the pooled mapping method, and SNPs between parental lines were defined as confident polymorphic loci [31]. In addition, the B. rapa reference genome was used for SNP calling and imputation. A total of 637,647 high-quality SNPs were identified from the 150 F2 individuals. After imputation, 636,866 high-quality SNPs with an average density of 2.15 SNPs/kb were obtained and used to construct a bin map (Table 4).

Table 3.
Overview of sequence data for 150 individuals in F2 population.

Table 4.
Summary of SNPs detected in the F2 population.
To construct the bin map using the sliding window approach, window lengths from 0.5 to 1 Mb and step sizes from 0.1 to 0.5 Mb were first tested in all chromosomes in five random individuals. A window length of 1 Mb and a step size of 0.4 Mb showed a reasonable number of recombination breakpoints per chromosome. This parameter was therefore used to construct a bin map, and a total of 4291 recombination breakpoints were obtained from the 150 F2 individuals, with an average of 28.6 per F2 individual. The mean number of crossovers per chromosome for the F2 population was 2.8. All SNPs were grouped into 565 bins. The physical lengths of the bin markers ranged from 200 kb to 3.1 Mb, with an average of 449.0 kb and a median of 400 kb. In total, 88.13% of bin markers were less than 400 kb in length (Figure 4). A total of 565 bins were used to construct a genetic linkage map. Two bins (bin_246 and bin_249) were unanchored to the linkage map. The total genetic distance of the bin map was 944.6 cM for all 10 chromosomes, and the mean distance of the bins was 1.65 cM. The bin marker order was in agreement with the Chinese cabbage reference genome sequence V3.0, with a few exceptions.
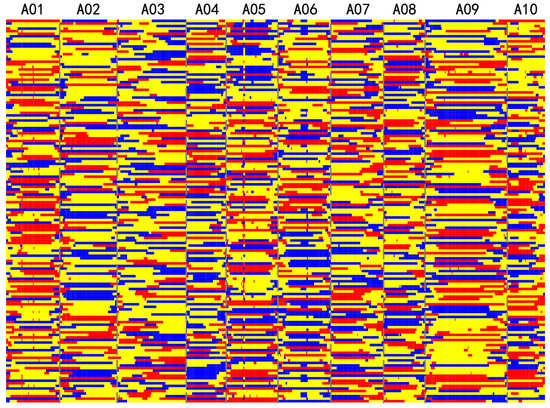
Figure 4.
Recombination bin map of the F2 population. The bin map consists of 565 bin markers. The physical position is based on the ‘Chiifu-401-42’ sequence. Red color indicates the ‘HN53’ genotype, blue color indicates the ‘Zhongbaye’ genotype, and yellow color indicates heterozygotes.
3.3. QTL Analysis
QTL analysis was performed using MAPQTL 4.0 software for MQM. In total, 60 QTLs for 11 traits were identified that satisfied the LOD threshold of more than 3.5 in the F2 mapping population (Table 5, Figure 5). Eighty percent of these QTLs had positive additive effects. seven QTLs for LA traits showed positive additive effects, especially LA 4, which explained 18.9% of the genetic variation. LA 7 showed high dominance effects compared with other QTLs, except for seven QTLs for LA traits and LWT6, where dominance effects were insignificant (Table 5). For each trait, three to nine QTLs were detected, and these QTLs were unevenly distributed across the 10 chromosomes. Of these QTLs, 41 were linked with leaf-related traits, including LL, LW, LWT, LA, PL, PWT, and PA. A total of nine QTLs for LL were detected on eight chromosomes. The percentage of phenotypic variation explained by individual QTLs ranged from 3% to 11.6%. Six QTLs were detected for LW, of which LW4 showed the highest LOD value. Six QTLs and seven QTLs were detected for LWT and LA, respectively, accounting for 5.6%–18.4% and 4.8%–18.9%, respectively. Three QTLs for PL, six QTLs for PWT, and four QTLs for PA were detected. Of the QTLs for petiole-related traits, PL3, PWT4, and PA3 showed comparatively higher LOD values and were located on chromosome A06. The leaf index traits—namely LI, PI, PAI, and PLTI—can represent leaf shape and attractiveness to consumers and were also used for QTL analysis.

Table 5.
QTLs for 11 traits identified using high-density SNP bin map.
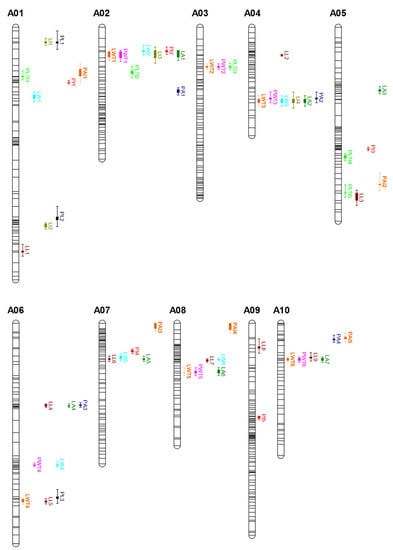
Figure 5.
Distribution of the 11 leaf-related traits on the linkage map. The gray bars represent the 10 chromosomes on the linkage map and the colored bars represent potential quantitative trait loci (QTLs) for the 11 leaf-related traits. Bar length indicates the two-LOD confidence interval of potential QTLs. See Table 1 for trait abbreviations.
A total of four QTLs were detected for LI and explained 7.2%–11.7% of the phenotypic variation. Five QTLs were identified for PI, PAI, and PLTI and explained 5.8%–18.8%, 5.6%–9.8%, and 6.3%–12.6% of the phenotypic variation, respectively.
3.4. Co-Localization of QTLs and Candidate Gene Prediction
Most chromosomes contained QTLs for several leaf-related traits in apparent co-localization, except for chromosome A09 (Figure 5). Three QTLs for LWT overlapped with QTLs for PWT, LWT1 and PWT1 were co-located on bin_65 of A02, LWT2 and PWT2 were co-located on bin_121 of A03, and LWT3 and PWT3 were co-located on bin_214−215 of A04, suggesting that the entire LWT was significantly correlated with PWT. Six QTLs for six traits—LWT, PWT, LI, LW, LA, and PI—were co-localized on bin_64−65 of A02. Seven QTLs were detected on A04, and six of the seven QTLs for LWT, LA, PWT, LW, LI, and PA traits were co-located in the 36.3−42.3 cM range on bin_214−215. Previous correlation analyses found that LWT, LA, PWT, LW, PA, and PI traits were positively correlated with each other, but negatively correlated with LI. These results indicate that the co-location of QTLs for leaf-related traits may account for the strong correlations among these traits, and these traits are possibly controlled by a common region. A previous QTL study identified leaf-related traits (rosette leaf length and PL) and head-related traits on LG A06 [28,35]. In the present study, LL, LA, LWT, and PA were highly correlated with each other, and their QTLs were co-located on A06.
Candidate genes associated with the investigated traits were identified using physical locations on the B. rapa reference genome corresponding to the bin markers. As a result, a total of 3170 candidate genes were identified. The functional annotations included transcription factors, kinases, hormonal pathways, and photosystem components. Some of the candidate genes play important roles in controlling leaf shape, leaf size, and leaf morphology through cell proliferation, cell expansion, hormone signaling pathways, or adaxial–abaxial patterning (Table 6). These included some cell-proliferation-related and cell-expansion-related genes, for example, GRFs, WOXs, TCPs, and BAMs genes; auxin signaling pathway genes, for example, ARFs, IAAs, and SAURs genes; leaf abaxial-adaxial polarity genes, for example, ARF, PGY; and ribosome-encoding genes (RPSs and RPLs) (Table 6). Among these genes, BrGRF3 (BraA04g025910.3C), a homolog gene of Arabidopsis GRF3, is located near the peak signal on the co-localized QTL (LWT, LA, PWT, LW, LI, and PA traits) region of chromosome A04. Previous studies have shown that growth-regulating factors (GRFs) are plant-specific proteins that play important roles in regulating leaf size. GRFs regulate organ size development through the promotion and/or maintenance of cell proliferation activity [36,37,38]. Furthermore, previous QTL analysis identified the BrGRF5 for LL and LI traits in B. rapa [1]. In the present study, it was found that the BrGRF3 (BraA04g025910.3C) allele of ‘Zhongbaye’ contained three nonsynonymous substitutions and one premature termination compared with that of ‘HN53’. In addition, BrARF5 gene (BraA06g015500.3C), the gene homologous to Arabidopsis ARF5, was located on co-localized QTL (LL, LA, and PA) regions of A06. This may be a candidate gene because it not only controls leaf adaxial–abaxial asymmetry but also influences organ development [39]. Sequence analysis showed that the BrARF5 gene of ‘Zhongbaye’ contained four nonsynonymous substitutions and one alternative splicing site compared with that of ‘HN53’.

Table 6.
60 QTLs localized to candidate genes that may be associated with leaf traits.
4. Discussion
The bin-map strategy was demonstrated to be efficient for the fast identification of QTLs, with high resolution in cereal crops [21,22,23]. However, limited studies on QTL bin maps have been conducted in the B. rapa vegetable.
In this study, we aimed to investigate the genetic regulation mechanisms underlying complex leaf-related traits in cabbage using a bin map of B. rapa constructed using a sliding window approach. The window length was set at 1 Mb instead of fixing the SNP number per window. Our analysis of the F2 population revealed a total of 4291 recombination breakpoints, with an average of 28 recombination breakpoints per individual. This suggested that about two to three recombination events occurred per chromosome. The constructed bin map contained 565 bin markers spanning a total genetic distance of 944.6 cM, and the average distance between the bins was 1.65 cM, suggesting that the bin map could detect differences in recombination frequency between F2 plants and that a QTL could be narrowed down to a small interval harboring dozens of genes. We identified 60 QTLs for 11 leaf-related traits distributed across all 10 chromosomes of B. rapa, indicating that leaf-related traits are complex quantitative traits controlled by many genes. These results are consistent with previous findings [1,28].
The co-localization of QTLs for different leaf traits is of great interest in understanding the genetic regulation mechanisms underlying complex leaf-related traits in cabbage vegetables. In this study, we observed clear co-localization of QTLs for several leaf traits in some chromosomal intervals (Figure 5). For example, three QTLs for LWT overlapped with QTLs for PWT on A02, A03, and A04. Six QTLs were detected and co-located in the bin interval 214–215 on chromosome 4, including QTLs for the LWT, LA, PWT, LW, LI, and PA traits. Obviously, the co-localization of QTLs indicated that leaf size was correlated with leaf weight and shape. This was in accordance with the fact that most of these traits were significantly correlated with each other. Several studies have reported similar results for leaf-related traits in B. rapa. Sun et al. found that QTLs for leaf size traits (LL, LW, and PL), leaf trichome trait, and leafy head traits (head height, head weight, and heading degree) are co-localized, in accordance with the correlation between these traits [28]. QTLs for rosette leaf length and rosette petiole length were co-localized on LG A06 in both investigated populations, suggesting that LL was correlated with PL. Interestingly, the rosette leaf width QTLs were all co-located with the heading degree QTL, revealing a relationship between the rosette leaf and leafy heads, in which the rosette leaf provides nutrients for leafy head formation. These results also indicated that the co-localized QTL regions are more likely to carry important genes that regulate more than one leaf trait.
The leaf and leaf head are the main edible parts of Chinese cabbage, and their quality significantly impacts the commercial value of the vegetable. Therefore, identifying genes associated with leaf shape, leaf head, and leaf abaxial-adaxial polarity could potentially improve the quality of Chinese cabbage. In this study, we analyzed the 60 QTLs linked with leaf-related traits and identified a total of 3170 candidate genes that may be involved in the regulation of these traits. Among these were some genes involved in the regulation of leaf size, leaf shape, and leaf morphology through the regulation of cell proliferation and cell expansion [36,39,40,41]. BrGRF5 and BrTCP4 are reported to be associated with leaf size and leaf heading in B. rapa [42]. In this study, BrGRF3, BrTCP3, BrWOX5, and BrBAMs were found in the QTL regions. Genes involved in the auxin signaling pathway play important roles in the curling of Chinese cabbage leaves, and IAAs are involved in leaf curling of Chinese cabbage [43]. Here we found that nine auxin signaling pathway genes (BrARF16, BrARF5, BrARF10, BrIAA9, BrIAA26, BrSAU42, BrSAU45, BrSAU46, and BrSAU53) were located in the QTL regions. In addition, leaf adaxial–abaxial polarity genes are reported to have important roles in regulating leaf curvature and leaf heading in B. rapa [29,44,45,46]. Increasing evidence shows that ribosome-encoding genes such as PGYs, RPSs, and RPLs are involved in the adaxial-abaxial patterning and development of leaves [47,48,49]. Here, some ribosome-encoding genes such as PGY1, RPL12, and RPS5B were located in the QTL regions. These genes may be important candidate genes for regulating leaf-related traits in B. rapa.
Further fine mapping of these QTLs should be conducted in order to narrow down the candidate genes, as the efficiency of QTL mapping depends largely on maker density and population size. Therefore, larger populations of F2, BC1S, or other early-generation crosses combined with a high throughput genotyping method is a cost-effective option.
5. Conclusions
In conclusion, a high-density genetic bin map containing 565 bin markers was constructed. The total map length was 944.6 cM, and the average distance of the bins was 1.65 cM. A total of 60 significant QTLs controlling 11 leaf-related traits were identified. This study demonstrated the use of whole genome re-sequencing and high-density genetic bin mapping in the identification of QTLs and candidate genes in an F2 population of B. rapa. For rapid identification of reliable QTLs and candidate genes associated with complex traits in B. rapa, reducing the QTL intervals using larger population sizes and constructing high-density marker maps in early-generation populations will be a cost-effective option in future. Our findings will help to accelerate the improvement of B. rapa vegetable crops.
Author Contributions
J.L., X.W. and J.W. designed the experiment; Z.L. and F.L. prepared the materials, performed the experiments, and analyzed the data; H.C. and X.C. helped analyze the data; H.W. helped revise the manuscript; J.L. and F.L. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32072594), the Agricultural Science and Technology Innovation Program (ASTIP), and the Central Public-interest Scientific Institution Basal Research Fund (Y2022PT23).
Data Availability Statement
The SNP data can be downloaded using this link: http://39.100.233.196:82/download_genome/datasets/150_F2_reseq/SNP%20dataset.xlsx (accessed on 5 February 2023).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xiao, D.; Wang, H.; Basnet, R.K.; Zhao, J.; Lin, K.; Hou, X.; Bonnema, G. Genetic dissection of leaf development in Brassica rapa using a genetical genomics approach. Plant Physiol. 2014, 164, 1309–1325. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.J.; Wang, X.; Deng, B.; Lou, P.; Wu, J.; Sun, R.; Xu, Z.; Vroman, J.; Koornneef, M.; Bonnema, G. Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor. Appl. Genet. 2005, 110, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Yang, X.; Wang, H.; Qiu, N.; Chen, Y.; Wang, F.; Zhang, Y.; Li, H.; Li, J.; Gao, J. Construction of an Intragenic SSR-Based Linkage Map and QTL Mapping for Agronomic Traits in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Horticulturae 2022, 8, 165. [Google Scholar] [CrossRef]
- Kapoor, R.; Banga, S.S.; Banga, S.K. A microsatellite (SSR) based linkage map of Brassica rapa. New Biotechnol. 2009, 26, 239–243. [Google Scholar] [CrossRef]
- Suwabe, K.; Tsukazaki, H.; Iketani, H.; Hatakeyama, K.; Kondo, M.; Fujimura, M.; Nunome, T.; Fukuoka, H.; Hirai, M.; Matsumoto, S. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: The genetic origin of clubroot resistance. Genetics 2006, 173, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, S.; Liu, B.; Wang, H.; Deng, J.; Liao, Y.; Wang, Q.; Cheng, F.; Wang, X.; Wu, J. A sequence-based genetic linkage map as a reference for Brassica rapa pseudochromosome assembly. BMC Genom. 2011, 12, 239. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Xing, J.; Liu, Z.; Feng, H. Mapping quantitative trait loci for yield-related traits in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Euphytica 2013, 193, 221–234. [Google Scholar] [CrossRef]
- Choi, S.R.; Yu, X.; Dhandapani, V.; Li, X.; Wang, Z.; Lee, S.Y.; Heon Oh, S.; Pang, W.; Ramchiary, N.; Hong, C.; et al. Integrated analysis of leaf morphological and color traits in different populations of Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 2017, 130, 1617–1634. [Google Scholar] [CrossRef]
- Kim, J.S.; Chung, T.Y.; King, G.J.; Jin, M.; Yang, T.J.; Jin, Y.M.; Kim, H.I.; Park, B.S. A sequence-tagged linkage map of Brassica rapa. Genetics 2006, 174, 29–39. [Google Scholar] [CrossRef]
- Panigrahi, J.; Patnaik, A.; Kole, P.; Koleb, C. Addition of restriction fragment length polymorphism markers to the genetic linkage map of Brassica rapa L. (syn. campestris). Z. Naturforsch C J. Biosci. 2009, 64, 882–890. [Google Scholar] [CrossRef]
- Lu, G.; Cao, J.; Yu, X.; Xiang, X.; Chen, H. Mapping QTLs for root morphological traits in Brassica rapa L. based on AFLP and RAPD markers. J. Appl. Genet. 2008, 49, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Soengas, P.; Hand, P.; Vicente, J.G.; Pole, J.M.; Pink, D.A. Identification of quantitative trait loci for resistance to Xanthomonas campestris pv. campestris in Brassica rapa. Theor. Appl. Genet. 2007, 114, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Kubo, N.; Saito, M.; Tsukazaki, H.; Kondo, T.; Matsumoto, S.; Hirai, M. Detection of quantitative trait loci controlling morphological traits in Brassica rapa L. Breed. Sci. 2010, 60, 164–171. [Google Scholar] [CrossRef]
- Bagheri, H.; El-Soda, M.; van Oorschot, I.; Hanhart, C.; Bonnema, G.; Jansen-van den Bosch, T.; Mank, R.; Keurentjes, J.J.; Meng, L.; Wu, J. Genetic analysis of morphological traits in a new, versatile, rapid-cycling Brassica rapa recombinant inbred line population. Front. Plant Sci. 2012, 3, 183. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Kitashiba, H.; Inaba, K.; Nishio, T. A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 2009, 16, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, H.; Zhong, W.; Bai, J.; Liu, P.; He, Y. QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS ONE 2013, 8, e76059. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Su, T.; Xin, X.; Li, P.; Wang, W.; Yu, Y.; Zhang, D.; Zhao, X.; Wang, J.; Sun, L. The Adaxial/Abaxial Patterning of Auxin and Auxin Gene in Leaf Veins Functions in Leafy Head Formation of Chinese Cabbage. Front. Plant Sci. 2022, 13, 918112. [Google Scholar] [CrossRef]
- Li, X.; Ramchiary, N.; Choi, S.R.; Van Nguyen, D.; Hossain, M.J.; Yang, H.K.; Lim, Y.P. Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Genome 2010, 53, 939–947. [Google Scholar] [CrossRef]
- Li, G.H.; Chen, H.C.; Liu, J.L.; Luo, W.L.; Xie, D.S.; Luo, S.B.; Wu, T.Q.; Akram, W.; Zhong, Y.J. A high-density genetic map developed by specific-locus amplified fragment (SLAF) sequencing and identification of a locus controlling anthocyanin pigmentation in stalk of Zicaitai (Brassica rapa L. ssp. chinensis var. purpurea). BMC Genom. 2019, 20, 343. [Google Scholar] [CrossRef]
- Liu, S.; Wang, R.; Zhang, Z.; Li, Q.; Wang, L.; Wang, Y.; Zhao, Z. High-resolution mapping of quantitative trait loci con-trolling main floral stalk length in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 2019, 20, 437. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, B.; Dong, X.; Liu, H.; Ren, L.; Chen, J.; Hauck, A.; Song, W.; Lai, J. An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F(2) maize population. BMC Genom. 2014, 15, 433. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Feng, Q.; Qian, Q.; Zhao, Q.; Wang, L.; Wang, A.; Guan, J.; Fan, D.; Weng, Q.; Huang, T. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009, 19, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Zou, G.; Zhai, G.; Feng, Q.; Yan, S.; Wang, A.; Zhao, Q.; Shao, J.; Zhang, Z.; Zou, J.; Han, B. Identification of QTLs for eight agronomically important traits using an ultra-high-density map based on SNPs generated from high-throughput sequencing in sorghum under contrasting photoperiods. J. Exp. Bot 2012, 63, 5451–5462. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Jeong, H.J.; Yang, H.B.; Kang, S.M.; Kwon, J.K.; Kim, S.; Choi, D.; Kang, B.C. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 2016, 23, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Ogden, E.L.; Bostan, H.; Sargent, D.J.; Ward, J.; Gilbert, J.; Iorizzo, M.; Rowland, L.J. High-Density Linkage Map Construction and QTL Identification in a Diploid Blueberry Mapping Population. Front. Plant Sci. 2021, 12, 692628. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.; Sun, T.; Liu, X.; Li, C.; Li, M.; Wang, X.; Ren, H.; Zhao, Z.; Zhuang, F. Detection of Chromosomal Segments Introgressed from Wild Species of Carrot into Cultivars: Quantitative Trait Loci Mapping for Morphological Features in Backcross Inbred Lines. Plants 2022, 11, 391. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Ray, R.; Li, P.; Xu, J.; Zhang, M.; Liu, G.; Yao, X.; Kilian, A.; Yang, X. Construction of a high-density DArTseq SNP-based genetic map and identification of genomic regions with segregation distortion in a genetic population derived from a cross between feral and cultivated-type watermelon. Mol. Genet. Genom. 2015, 290, 1457–1470. [Google Scholar] [CrossRef]
- Sun, X.; Luo, S.; Luo, L.; Wang, X.; Chen, X.; Lu, Y.; Shen, S.; Zhao, J.; Bonnema, G. Genetic Analysis of Chinese Cabbage Reveals Correlation Between Rosette Leaf and Leafy Head Variation. Front. Plant Sci. 2018, 9, 1455. [Google Scholar] [CrossRef]
- Cheng, F.; Sun, R.; Hou, X.; Zheng, H.; Zhang, F.; Zhang, Y.; Liu, B.; Liang, J.; Zhuang, M.; Liu, Y. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016, 48, 1218–1224. [Google Scholar] [CrossRef]
- Rizzo, D.; Da Lio, D.; Bartolini, L.; Francia, C.; Aronadio, A.; Luchi, N.; Campigli, S.; Marchi, G.; Rossi, E. DNA Extraction Methods to Obtain High DNA Quality from Different Plant Tissues. Methods Mol. Biol. 2022, 2536, 91–101. [Google Scholar]
- Fu, L.; Cai, C.; Cui, Y.; Wu, J.; Liang, J.; Cheng, F.; Wang, X. Pooled mapping: An efficient method of calling variations for population samples with low-depth resequencing data. Mol. Breed. 2016, 36, 48. [Google Scholar] [CrossRef]
- Larose, D.T.; Larose, C.D. Discovering Knowledge in Data: An Introduction to Data Mining; Wiley-Interscience: New York, NY, USA, 2004. [Google Scholar]
- Ooijen, J. JoinMap 4.0: Software for the Calculation of Genetic Linkage Maps in Experimental Population; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Van Ooijen, J.W.; Boer, M.P.; Jansen, R.C.; Maliepaard, C.A. MapQTL 4.0: Software for the Calculation of QTL Positions on Genetic Maps (User Manual); Plant Research International: Wageningen, The Netherlands, 2000. [Google Scholar]
- Ge, Y.; Ramchiary, N.; Wang, T.; Liang, C.; Wang, N.; Wang, Z.; Choi, S.R.; Lim, Y.P.; Piao, Z. Mapping quantitative trait loci for leaf and heading-related traits in Chinese cabbage (Brassica rapa L. ssp. pekinesis). Hortic. Environ. Biotechnol. 2011, 52, 494–501. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, B.H. Growth-regulating factor4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 2006, 49, 463–468. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef]
- Krogan, N.T.; Berleth, T. A dominant mutation reveals asymmetry in MP/ARF5 function along the adaxial-abaxial axis of shoot lateral organs. Plant Signal. Behav. 2012, 7, 940–943. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Beltramino, M.; Ercoli, M.F.; Debernardi, J.M.; Goldy, C.; Rojas, A.M.L.; Nota, F.; Alvarez, M.E.; Vercruyssen, L.; Inze, D.; Palatnik, J.F. Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci. Rep. 2018, 8, 13447. [Google Scholar] [CrossRef]
- Mao, Y.; Wu, F.; Yu, X.; Bai, J.; Zhong, W.; He, Y. MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 2014, 164, 710–720. [Google Scholar] [CrossRef]
- He, Y.K.; Xue, W.X.; Sun, Y.D.; Yu, X.H.; Liu, P.L. Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell Res. 2000, 10, 151–160. [Google Scholar] [CrossRef]
- Guo, X.; Liang, J.; Lin, R.; Zhang, L.; Wu, J.; Wang, X. Series-Spatial Transcriptome Profiling of Leafy Head Reveals the Key Transition Leaves for Head Formation in Chinese Cabbage. Front. Plant Sci. 2021, 12, 787826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, J.; Cai, X.; Chen, H.; Wu, J.; Lin, R.; Cheng, F.; Wang, X. Divergence of three BRX homoeologs in Brassica rapa and its effect on leaf morphology. Hortic. Res. 2021, 8, 68. [Google Scholar] [CrossRef]
- Liang, J.; Liu, B.; Wu, J.; Cheng, F.; Wang, X. Genetic Variation and Divergence of Genes Involved in Leaf Adaxial-Abaxial Polarity Establishment in Brassica rapa. Front. Plant Sci. 2016, 7, 94. [Google Scholar] [CrossRef] [PubMed]
- Pinon, V.; Etchells, J.P.; Rossignol, P.; Collier, S.A.; Arroyo, J.M.; Martienssen, R.A.; Byrne, M.E. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 2008, 135, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Ling, Q.; Wang, H.; Huang, H. Ribosomal proteins promote leaf adaxial identity. Development 2008, 135, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liang, J.; Lin, R.; Zhang, L.; Zhang, Z.; Wu, J.; Wang, X. Single-cell transcriptome reveals differentiation between adaxial and abaxial mesophyll cells in Brassica rapa. Plant Biotechnol. J. 2022, 20, 2233–2235. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).