Abstract
Dehydration-responsive element-binding protein 1 (DREB1)/C-repeat binding factor (CBF) family plays a key role in plant tolerance against different abiotic stresses. In this study, an orthologous gene of the DWARF AND DELAYED FLOWERING (DDF) members in Arabidopsis, SlDDF2, was identified in tomato plants. The SlDDF2 gene expression was analyzed, and a clear induction in response to ABA treatment, cold, salinity, and drought stresses was observed. Furthermore, two transgenic lines (SlDDF2-IOE#6 and SlDDF2-IOE#9) with stress-inducible overexpression of SlDDF2 under Rd29a promoter were generated. Under stress conditions, the gene expression of SlDDF2 was significantly higher in both transgenic lines. The growth performance, as well as physiological parameters, were evaluated in wild-type and transgenic plants. The transgenic lines showed growth retardation phenotypes and had higher chlorophyll content under stress conditions in plants. However, the relative decrease in growth performance (plant height, leaf number, and leaf area) in stressed transgenic lines was lower than that in stressed wild-type plants, compared with nonstressed conditions. The reduction in the relative water content and water loss rate was also lower in the transgenic lines. Compared with wild-type plants, transgenic lines showed enhanced tolerance to different abiotic stresses including water deficit, salinity, and cold. In conclusion, stress-inducible expression of SlDDF2 can be a useful tool to improve tolerance against multiple abiotic stresses in tomato plants.
1. Introduction
Abiotic stresses, such as cold, drought, high salinity, and extreme heat have adverse effects on plant growth and development. They are considered major constraints for plant production in many areas around the globe. Among them, drought is considered a major limiting factor for the productivity of any given crop. To minimize the negative impact of abiotic stresses on plants, it is necessary to develop new plants that utilize water more efficiently and tolerate such stresses []. In this perspective, a basic strategy is based on the cloning of key regulatory genes and the introduction of their active forms into plants so that they can acquire abiotic stress tolerance phenotypes [].
Under stress, several morphological, physiological, and molecular processes are altered in different organs to improve plant tolerance []. Plants have developed different defense mechanisms against abiotic stresses that involve the interaction among a group of transcription factors and the activation of key effector genes []. Transcription factors control plant responses to different environmental factors through sequence-specific interactions with cis-regulatory DNA elements, which are found in promoter and enhancer regions of their target genes []. Thus, the levels of expression of different abiotic stress-responsive genes are influenced by the manipulation of stress-responsive transcription factors []. Therefore, the manipulation of these transcription factors can improve the tolerance of different plants against different biotic and abiotic stress conditions []. In this perspective, members of dehydration-responsive element-binding protein 1 (DREB1)/C-repeat binding factor (CBF) family, encoding AP2 transcription factors, are known to regulate the expression of several stress-responsive genes by binding to C-repeat/dehydration-responsive cis-element in their promotors, thus enhancing cold, high salinity, and drought tolerance of plants []. In many plants, DREB genes work as the connecting points for multiple plant-response pathways to different stress factors, such as salinity, drought, ABA, and cold pathways [,,]. For instance, the DWARF AND DELAYED FLOWERING (DDF) genes were upregulated in Arabidopsis plant under cold, drought, salinity stress conditions [], while the overexpression of CBF4 gene in Arabidopsis plants improved drought tolerance that was associated with upregulation of several stress-responsive genes and resulted in [].
Tomato (Solanum lycopersicon L.) is one of the most grown vegetable crops all over the world, and it is mainly cultivated under irrigated conditions. Tomato yield is greatly affected in many growing areas by different abiotic stresses including drought, salinity, and low temperatures []. There is a growing need to improve stress tolerance in tomato plants to withstand such adverse conditions. In this study, a new member of DREB transcription factors in tomato plants (named SlDDF2) that is closely related to the DDF gene in Arabidopsis was isolated and characterized. The SlDDF2 gene was expressed in response to a variety of abiotic stimuli, implying a potential role in tolerance against abiotic stresses. Transgenic tomato plants with different levels of inducible overexpression of the SlDDF2 were generated and analyzed for their growth, physiological, biochemical, and gene expression responses to multiple abiotic stress factors. Transgenic tomato plants with inducible overexpression of SlDDF2 showed reduced water loss and improved tolerance against multiple abiotic stresses.
2. Materials and Methods
2.1. Cloning of SlDDF2 Gene and Bioinformatics Analysis
To clone a DDF orthologous gene in tomato plants, bioinformatics and comparative genomics analyses were performed based on a previously published full-length sequence of Arabidopsis DDF1 gene (GenBank accession number: NM_101131 []). a TBLASTN search was performed utilizing DDF1’s amino acid sequence against the annotated ITAG2.3 predicted tomato cDNA sequences database []. The full-length coding sequences of novel and unstudied tomato DNA sequences encoding DDF transcription factors in tomato were retrieved and further analyzed. The predicted coding sequences of a selected tomato DDF gene (SlDDF2; Solyc08g007820) were used to design specific primers (SlDDF2fwd: 5′-ATGAATAACGACTCGAGTTTG-3′ and SlDDF2Rev: 5′-TCAAATACTATAACTCCACA-3′) using the NCBI Primer-BLAST tool to isolate its full-length CDS as described previously by Al-Abdallat et al. [].
For SlDDF2 cloning, leaves from two-week-old tomato cv. “Money Maker” plants were collected and used for total RNA extraction using the SV Total RNA Isolation System Kit (Promega, Madison, WI, USA), as directed by the manufacturer. Following the manufacturer’s instructions, the extracted RNA was used to synthesize the first-strand cDNA library using the SuperScript® First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) and the oligo T(18) primer. The full-length CDS of SlDDF2 gene was then amplified from the synthesized cDNA using specific pair of primers (SlDDF2Fwd and SlDDF2Rev) in a PCR with a total volume of 25 µL containing 5 μL of cDNA as a template, 2.5 µL of dNTPs (100 µM), 5 μL of 5× PCR buffer, 0.5 µM of each primer and 0.25 µL of 5 U/µL GoTaq DNA polymerase (Promega, Madison, WI, USA). The thermal reaction was conducted using GeneAmp® PCR system 9700 (Applied Biosystems, Carlsbad, CA, USA) under the following conditions: 94 °C for 5 min, followed by 35 cycles at 94 °C for 1 min, 50 °C for 30 s and 72 °C for 2 min and a final extension of 72 °C for 10 min. The amplified CDS fragments were resolved on a horizontal 1% agarose gels stained with ethidium bromide. The PCR products (estimated size 735 bp) were then eluted from the agarose gel using Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA) and then cloned into pGEM®-T Easy Vector System (Promega, Madison, WI, USA). DNA Plasmids containing PCR products were selected, and the DNA products were then sequenced using the M13 reverse and forward sequencing primers by ABI 3730XL machine by Macrogen (Seoul, Korea). The sequenced SlDDF2 cDNA and its deduced amino acids sequences were analyzed by the Vector NTI software (https://www.thermofisher.com/jo/en/home/life-science/cloning/vector-nti-software.html, accessed on 1 July 2021; Invitrogen™, Carlsbad, CA, USA) and further confirmed by a BLAST search (https://solgenomics.net/tools/blast/, accessed on 1 February 2019) for DNA and amino acid sequence homology to further verify the identity of the cloned cDNA.
The Sol Genomics Network [] was used to retrieve and analyze DNA and amino acid sequences, as well as chromosomal position and annotation prediction of several members of DREB-A1 and DREB-A2. Phylogenetic trees were constructed using MEGA version 10 software [] and the amino acids sequences of SlDDf2 and selected ERF subfamily proteins from tomato and Arabidopsis from the DREB-A1 and DREB-A2 groups [] that were retrieved from the Phytozome databases [] were included. The retrieved sequences were aligned using the ClustalW algorithm and the alignment was used to calculate distance matrices for neighbor-joining analyses with the Kimura two-parameter model and Bootstrap analysis with 10,000 replicates was performed to test the robustness of the internal branches, as described previously by Alhindi and Al-Abdallat [].
2.2. Plant Material and Stress Treatments
For gene expression analysis, tomato cv. “Moneymaker” (MM) plants were grown under growth conditions and were subjected to different treatments that included water deficit, salinity, cold, and ABA. Transgenic tomato lines with inducible overexpression of the SlDDF2 gene, on the other hand, were utilized to compare abiotic stress tolerance in MM plants in response to drought, salinity, and cold. The transgenic plants were generated using the binary plasmid pCABIMA1302 harboring the SlDDF2 gene downstream of the stress-inducible Rd29a promoter. For this purpose, the mgfp5 gene was replaced with SlDDF2 CDS at the NcoI and BstEII sites to generate pCAMBIA1302/SlDDF2. Thereafter, the Rd29a promoter (GenBank accession number: AY973635.1) was cloned into pCAMBIA1302/SlDDF2 by replacing the CaMV 35S promoter using EcoRI and NcoI sites to produce pCAMBIA1302/Rd29a::SlDDF2. The constructs were then introduced into tomato cv. “MM” using Agrobacterium-mediated transformation, and two positive plants carrying a single copy of the transgene were identified and selected, as described previously by Al-Abdallat et al. []. Gene expression analysis levels in T2 homozygous selected transgenic lines were analyzed using the quantitative RT–PCR approach under water deficit and control conditions.
For stress treatments, tomato seeds of the selected genotypes were submerged in water for 24 h at 25 °C, and then, they were washed with sterilized water and sown into small pots (10 cm diameter × 10 cm depth) filled with acid-washed sand. The pots were incubated under controlled conditions (at constant temperature (25 °C) and photoperiod of 16 h light–8 h dark, with 250 µmol·m−2·s−1 photon flux density) till full germination. The tomato seedlings were then irrigated daily with a fixed amount of 1× Hoagland solution (Sigma-Aldrich, Gillingham, UK). For water deficit treatment, two-week-old tomato seedlings were exposed to water withholding for 0, 3, 5, and 7 days for gene expression analysis in MM plants and for 7 days for inducible expression analysis in transgenic plants. Furthermore, stress tolerance and wilting behavior in response to water withholding for 10 days were investigated on transgenic and nontransgenic lines, in which 20 plants from each line were used per treatment and the survival percentage was calculated at the end of the dehydration treatment.
To investigate the effect of high salt treatment on SlDDF2 gene expression, the roots of two-week-old MM seedlings were submerged in saline water (300 mM NaCl) for 0, 2, 4, 8, 12, and 24 h. For salinity stress tolerance in transgenic and nontransgenic lines, two-week-old seedlings (20 plants from each line were used per treatment) were irrigated every three days with a fixed volume of 1× Hoagland solution supplemented with 100 mM for 12 days and the wilting behavior was monitored, and survival percentage was calculated at the end of the high salt treatment. For the effect of cold treatment on SlDDF2 gene expression, two-week-old MM seedlings were incubated at 4 °C for 0, 2, 4, 8, 12, and 24 h. For cold stress tolerance in transgenic lines and wild-type plants, two-week-old seedlings (20 plants from each line were used per treatment) were incubated for 24 h at 4 °C, and the wilting behavior was monitored, and the survival percentage was calculated. The gene expression of SlDDF2 was analyzed in response to ABA treatment, two-week-old MM seedlings were sprayed with 100 µM ABA solution, and leaf tissues were collected after 0, 2, 4, 8, 12, and 24 h.
2.3. Growth and Physiological Measurements
Relative water content (RWC) was determined in well-watered and stressed plants (subjected to 10 days of water withholding period), as described in Al-Abdallat et al. [], using fully expanded leaves, and the RWC was calculated according to Barrs and Weatherley []. The water loss rate was measured using fully expanded leaves excised from the well-watered and stressed transgenic lines (subjected to 10 days of water withholding period) that were placed on filter paper for 2 h, as described in Al-Abdallat et al. [], and the water loss rate was measured according to Ristic and Jenks []. Stomatal resistance (s·cm−1) was measured using a fully expanded leaf from well-watered and stressed plants after 10 days of water withholding by using a Porometer device (AP4, Delta-T Devices, Cambridge, UK). The determination of chlorophyll content (Chla, Chlb, and total Chl) was carried out using excised leaves from stress-treated transgenic lines and MM plants following a modified protocol, as described previously by Al-Abdallat et al. []. Finally, growth-related measurements (leaf number, plant height (in cm), and leaf area (cm2) were collected from stress-treated (subjected to 10 days of water withholding period) and well-watered transgenic lines and MM plant. Five replicates were used for all physiological and growth parameters, and the standard error of means was used to compare means.
2.4. Gene Expression Analysis
For quantitative real-time PCR (qRT–PCR) analysis, total RNA was extracted from leaf samples collected from treated plants at indicated time points using the SV Total RNA Isolation System Kit (Promega, Madison, WI, USA). The extracted RNA was used to synthesize the first-strand cDNA library as described above. Specific primers pairs for SlDDF2 expression analysis (SlDDF2Efwd: 5′-ATGAATAACGACTCGAGTTTG-3′ and SlDDF2ERev: 5′-TCAAATACTATAACTCCACA-3′) were used. The qRT–PCR analysis was performed using the stress-inducible Le16 gene (Solyc10g075090, a phospholipid transfer protein from tomato also known as Le16 (Lycopersicon esculentum protein 16) and Solyc03g078400 (encoding actin, a house-keeping gene used as an internal reference control for relative gene expression analysis) as described in Al-Abdallat et al. []. All cDNA samples were analyzed in triplicate, and each replicate was derived from two biological replicates. The relative changes in gene expression were quantified as described in Vandesompele et al. [].
2.5. Statistical Analysis
The data are presented as mean ± SD of three technical replicates from two biological replicates (n = 6) for gene expression analysis and five biological replicates for the growth performance and physiological parameters’ measurements. Student’s t-test was used to determine significant group differences, and means were considered as statistically significant if p < 0.05.
3. Results and Discussion
3.1. Identification of DDF Orthologous Genes in Tomato
To identify DDF orthologous gene in tomato plants, a TBLASTN search was conducted against the annotated ITAG2.3 predicted tomato cDNA sequences database using the full-length amino acids sequence data of the DDF1 gene from Arabidopsis (GenBank Accession: NM_101131). Using this approach, three tomato orthologous genes were identified: Solyc12g056430, Solyc08g007820, and Solyc08g007830, and the three genes were named SlDDF1, SlDDF2, and SlDDF3, respectively. Phylogenetic analysis using ERF subfamily proteins from tomato and Arabidopsis belonging to groups DREB-A1 and DREB-A2 revealed the identity of SlDDF proteins, which clustered with DDF1 and DDF2 proteins from Arabidopsis (Figure 1). The DDFs are members of the DREB-A1 subfamily of the ERF/AP2 transcription factor family in Arabidopsis, and they are implicated in stress responses and GA biosynthesis regulation [], indicating that SlDDF may have a role in stress tolerance in tomato plants. To investigate this role in tomato plants, the SlDDF2 gene was selected and cloned using specific pair of primers. The SlDDF2 gene was found on the upper arm of chromosome 8, and it was annotated as Solyc08g007820 that encodes ethylene-responsive transcription factor 10 with a single ORF (735 bp with a single exon) intron and a total length of 245 amino acids.
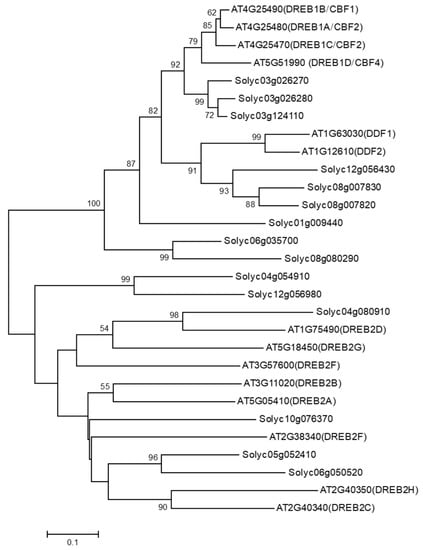
Figure 1.
Phylogenetic analysis of Arabidopsis proteins belonging to the DREB-A1 and DREB-A2 groups and their closest orthologs in tomato plants.
3.2. Expression Analysis of SlDDF2
The expression behavior of SlDDF2 in response to ABA and abiotic stresses was analyzed in MM plants using quantitative real-time PCR. The SlDDF2 expression levels were compared with Lycopersicon esculentum protein 16; Solyc10g075090 (Le16), a stress-inducible gene from tomato encoding a phospholipid transfer protein. The expression of the SlDDF2 gene in MM plants was highly induced after two hours of ABA treatment before returning to the basal expression level of the control (zero time) after 4 h (Figure 2A). This is in general agreement with Li et al. [], who reported that the ABA-induced expression of SlDREB in tomato plants started from 1 to 6 h after treatment before it returned to pre-treatment levels after 12 h. The expression of the Le16 stress-responsive gene, on the other hand, was induced as expected in response to ABA treatment and peaked after 24 h, which is consistent with previous studies [,]. The expression patterns of SlDDF2 and Le16 genes in response to cold treatment (4 °C for 0, 2, 4, 8, 12, and 24 h) were also analyzed in tomato MM plants. The expression of SlDDF2 was induced after 4 h, with a higher level of induction observed at 12 h of cold treatment, while Le16 gene expression was at the highest level after 12 h of cold treatment (Figure 2B). These results are similar to the findings of Zhang et al. [], who found that the expression of the LeCBF1 gene was upregulated upon exposure to low temperature, reaching its highest level after 8 h before returning to its pretreatment levels after 24 h.
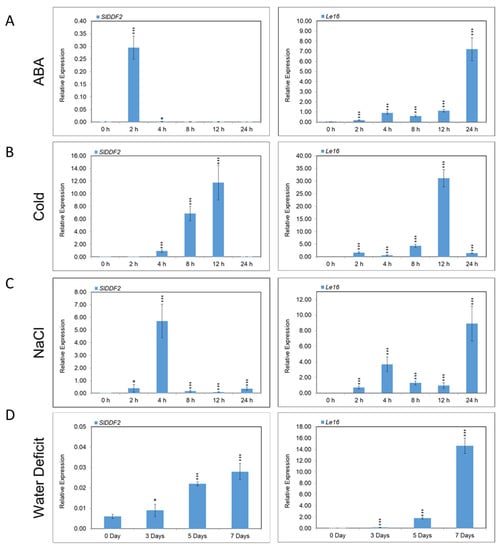
Figure 2.
Relative gene expression analysis of SlDDF2 and Le16 in response to (A) ABA, (B) cold, (C) NaCl, and (D) water deficit treatments. MM plants were compared to untreated plants (control). The stress-responsive Le16 (Solyc10g075090) gene was included as a control. Values are the means ± SD. of six replicates. Relative expressions are significantly different from those at zero time at different levels: * p < 0.05, *** p < 0.001.
The expression of the SlDDF2 gene in MM plants was induced after 2 h in response to 100 mM NaCl treatment, reaching its highest expression at 4 h of incubation (Figure 2D). Le16 gene expression was induced in response to NaCl with time, which is in agreement with Al-Abdallat et al. [], and the highest expression level was observed at 24 h. The upregulation of SlDDF2 in response to high salt stress suggests a potential role in salinity tolerance, which was previously reported by Sakuma et al. [] and Magome et al. [], who found that DDF1 gene expression was induced in Arabidopsis roots under high salinity conditions. These results are also in agreement with Hichri et al. [], who reported the inducible expression of SlDREB2 in tomato plants in response to NaCl treatments.
The expression patterns of the SlDDF2 gene and Le16 were analyzed in MM tomato plants in response to water deficit treatment by water withholding for 3, 5, and 7 days. The expression of the Le16 gene in stressed MM tomato plants was induced under water deficit with time, reaching the highest expression level after seven days (Figure 2D), which is in general agreement with Al-Abdallat et al. []. Similarly, the expression of the SlDDF2 gene was induced in response to water deficit after five days of stress, reaching its highest level after seven days (Figure 2D). Similar results were described by Hichri et al. [], who reported drought-induced expression of SlDREB2 in tomato plants.
3.3. Stress-Inducible Overexpression of SlDDF2 in Tomato
To check if SlDDF2 inducible overexpression can enhance tolerance of tomato against various abiotic stresses, the coding sequence of SlDDF2 was cloned into a binary plasmid under the control of the rd29A, a stress-inducible promoter from Arabidopsis plant, or under the control of the CaMV 35S constitutive promoter. Several independent transgenic tomato lines were generated with transgenic lines carrying the CaMV 35S constitutive promoter showed severe growth retardation phenotypes and did not produce any seeds and, therefore, were discarded from further analysis (data not shown). Severe pleotropic effects of constitutive overexpression of stress-related DREB transcription factors in tomato plants were reported previously []. Similarly, the overexpression of DDF1 in Arabidopsis plants resulted in dwarfism and late-flowering phenotypes []. On the other hand, transgenic lines carrying the rd29A stress-inducible promoter were generated, and two transgenic lines carrying a single insertion event as revealed by real-time PCR analysis were selected for further analysis (SlDDF2-IOE#6 and SlDDF2-IOE#9). To validate the inducible expression of SlDDF2 in response to stress, transgenic lines and MM plants were subjected to water withholding for seven days. The expression of the SlDDF2 gene was significantly higher in both transgenic lines, compared with wild type; however, the expression level was much higher in stressed plants than that in nonstressed plants (Figure 3A). Furthermore, the expression levels in SlDDF2-IOE#9 plants were significantly higher than that of SlDDF2-IOE#6 plants under both treatments. Stress-inducible expression of DREB genes the rd29A stress-inducible promoter was reported previously in different plants species including tomato [,].
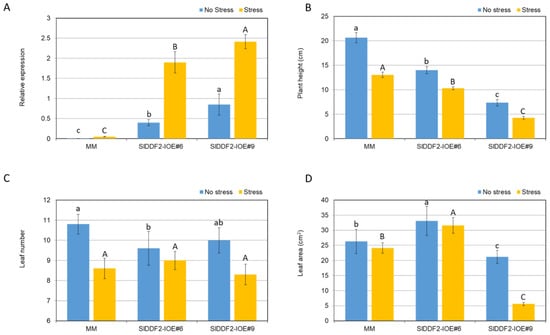
Figure 3.
(A) Relative gene expression analysis of SlDDF2 in MM and SIDDF2-IOE#6 and SIDDF2-IOE#9 transgenic plants in response to water withholding for seven days; values are the means ± SE of six replicates; (B) plant height, (C) leaf number, and (D) leaf area of two-week-old seedlings of SIDDF2-IOE#6 and SIDDF2-IOE#9 transgenic lines and MM plants under normal and water withholding for seven days (stress). Values are the means ± SD. Different lower-case letters indicate a significant difference between transgenic and wild-type plants under nonstressed conditions, and different capital letters indicate a significant difference between transgenic and wild-type plants under stress conditions (p < 0.05).
When compared with the wild-type plants, the two selected transgenic lines (SlDDF2-IOE#6 and SlDDF2-IOE#9) showed growth retardation phenotypes under normal and water deficit conditions, with clear shorter plants and shorter internodes phenotypes (Figure 3B). The wild-type and SlDDF2-IOE#9 plants showed a significant reduction in leaf number mean values under stress conditions when compared with nonstressed plants (Figure 3C). In addition, the SlDDF2-IOE#9 plants showed a significant reduction in leaf area means values under stress conditions when compared with well-watered plants and SlDDF2-IOE#6 plants (Figure 3D). On the contrary to the findings of this study, the use of the stress-inducible rd29A promoter for the overexpression of AtDREB1A in transgenic tomato did not show negative effects on plant growth and development []. However, the observed growth retardation phenotypes in the SlDDF2-IOE lines are similar to previous phenotypes reported in rd29A:DREB1A transgenic tobacco [,] and rd29A:AtCBF3 potato plants, in which growth retardation phenotypes were observed. The differences in growth retardation phenotypes between the two transgenic lines can be explained by the higher expression levels in SlDDF2-IOE#9, as reported previously by Pino et al. []. Additionally, the observed behaviors were comparable to those found in transgenic tomato plants with constitutive overexpression of SlDREB, which has been linked to reduced internode elongation due to lower gibberellin levels [].
Under water deficit conditions, the transgenic plants were found to have darker green leaf color, which was associated with increased chlorophyll a, chlorophyll b, and total chlorophyll pigments concentrations in comparison with the wild-type plants (Figure 4). The levels of Chla, Chlb, and total chlorophyll were increased only under water deficit stress conditions compared with the nonstressed groups. These findings are in general agreement with Li et al. [] and Al-Abdallat et al. [], who observed increased total chlorophyll pigments in transgenic tomato plants overexpressing DREB genes, which was attributed previously to reduced GA levels in transgenic lines [].
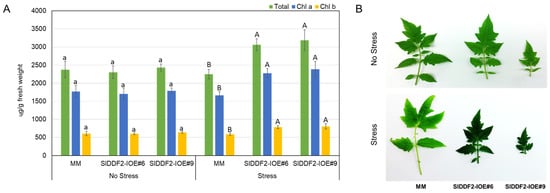
Figure 4.
(A) Total chlorophyll (green), Chlorophyll a (blue), and Chlorophyll b (yellow) contents in MM and SIDDF2-IOE#6 and SIDDF2-IOE#9 transgenic plants in response to water withholding for seven days or no stress conditions; (B) detached leaves from stressed and nonstressed wild-type plants and the two transgenic lines plants. Values are the means ± SD. Different lower-case letters indicate a significant difference between transgenic and wild-type plants under nonstressed conditions, and different capital letters indicate a significant difference between transgenic and wild-type plants under stress conditions (p < 0.05).
The physiological behavior of the transgenic plants under water deficit stress was compared with MM plants. Initially, the relative water content was measured immediately after detaching leaves from each plant. As shown in Figure 5, MM plants exposed to stress conditions showed lower RWC than the transgenic lines, which indicates a better water status in cells of transgenic lines. To investigate the effect of water deficit on water loss rate (g·h−1·g−1 DW), fully expanded wild-type and transgenic-line leaves from both treatments were detached and subjected to dehydration for 2 h. The results showed that the water loss rate from the stressed transgenic lines was lower than that in control and stressed wild-type plants (Figure 5). These results suggest that the drought resistance of the transgenic plants overexpressing the SlDDF2 gene was improved, compared with MM plants. It has been reported that transgenic tomato plants overexpressing DREB genes showed enhanced drought tolerance by maintaining higher water content and reduced water loss rate [].
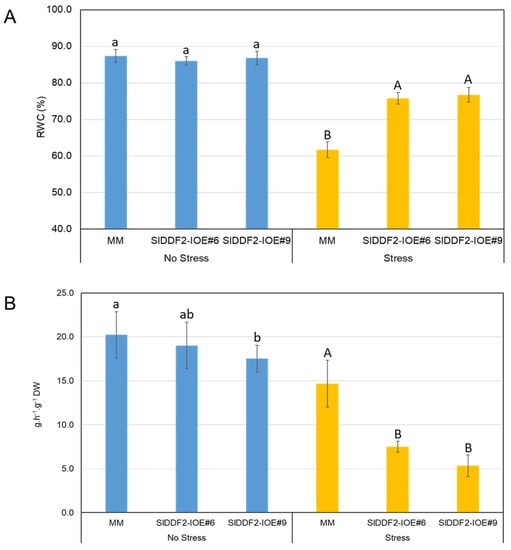
Figure 5.
Relative water content of leaves of MM and SIDDF2-IOE#6 and SIDDF2-IOE#9 transgenic lines after 10 days of (A) water withholding and (B) water loss rate, as measured by decrease in fresh weight after two hours in detached leaves from MM and SIDDF2-IOE#6 and SIDDF2-IOE#9 transgenic plants. Values are the means ± SD. Different lower-case letters indicate a significant difference between transgenic and wild-type plants under nonstressed conditions, and different capital letters indicate a significant difference between transgenic and wild-type plants under stress conditions (p < 0.05).
To analyze the impact of water deficit on physiological responses of SlDDF2 transgenic lines, two-weeks old transgenic and MM (included as control) seedlings were grown under stress conditions for 10 days by water withholding and observed for their growth and wilting behaviors at the end of treatment. Under drought stress conditions, the majority of MM plants were welted (60% survival rate), and an obvious, adverse effect was observed, while transgenic tomato lines showed enhanced tolerance to water deficit stress and showed a delayed wilting behavior and higher survival rate when compared with MM plants (Figure 6B). For salinity stress tolerance, SlDDF2-IOE#9 transgenic plants displayed improved tolerance to high salt stress (survival rate 50%), followed by SlDDF2-IOE#6 plants (survival rate 50%), while MM wild-type plants suffered severely from salinity stress (Figure 6C). For cold stress tolerance, the survival rate of the wild-type plants was 10%, whereas the SlDDF2-IOE transgenic plants showed enhanced tolerance to cold stress and higher survival rate when compared with wild-type plants, with 45% and 75% for SlDDF2-IOE#6 and the SlDDF2-IOE#9, respectively (Figure 6D). These results suggested that the overexpression of the SlDDF2 gene improved drought, cold, and salt stresses tolerance in tomato plants. In line with our results, the overexpression of SlDREB2 enhanced Arabidopsis and tomato tolerance to salinity stress (125 mm NaCl) [], while the stress-inducible overexpression of Arabidopsis CBF1 in transgenic tomato plants improved tolerance against low temperatures, water-deficit, and high salt treatments [].

Figure 6.
Representative MM and SIDDF2-IOE#6 and SIDDF2-IOE#9 transgenic lines grown under (A) normal conditions, (B) water deficit, (C) salinity, and (D) cold stresses (percentages are describing survival rate out of 20 plants per treatment).
4. Conclusions
The SlDDF2 gene was identified in tomato plants, and the phylogenetic analysis clustered it with the DREB1 family, indicating a potential role in abiotic stress tolerance. Furthermore, gene expression analysis of SlDDF2 showed inducible expression patterns in response to multiple abiotic stresses including cold, salinity, and drought. Stress-inducible overexpression of the SlDDF2 gene in tomato plants enhanced tolerance against different abiotic stresses when compared with MM plants, with clear pleotropic effects observed on them. The identified stress-related SlDDF2 gene could be a useful tool for tomato improvement and tolerance under abiotic stress conditions.
Author Contributions
T.A.-D. and A.M.A.-A. conceived and had designed the experiments, analyzed the data, and wrote the manuscript. R.A.-S. helped in molecular work, RNA extraction, relative gene expression, and data analysis. M.A.G., N.E.-A. and H.A.-D. helped in stress experiments, physiological measurements, and data analysis. All authors edited and provided a critical review of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Deanship of Scientific research/The University of Jordan (Grants Number: 1014).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets supporting the results of this article are freely available upon reasonable request from A.M.A.-A.
Acknowledgments
We sincerely thank Jamal Ayad and Shireen Qasrawi, for their technical assistance. We also gratefully acknowledge the financial support of the Deanship of Scientific research/The University of Jordan to A.M.A.-A.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shao, H.B.; Chu, L.Y.; Jaleel, C.A.; Zhao, C.X. Water-deficit stress-induced anatomical changes in higher plants. Comptes Rendus Biol. 2008, 331, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Fujita, M.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Engineering drought tolerance in plants: Discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 2006, 17, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Wahid, A.; Kobayashi, N.S.; Fujita, D.B.; Basra, S.M. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef] [Green Version]
- Qu, A.L.; Ding, Y.F.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Coego, A.; Brizuela, E.; Castillejo, P.; Ruíz, S.; Koncz, C.; del Pozo, J.C.; Piñeiro, M.; Jarillo, J.A.; Paz-Ares, J.; León, J. The TRANSPLANTA Consortium. The TRANSPLANTA Collection of Arabidopsis Lines: A resource for Functional Analysis of Transcription Factors based on their conditional overexpression. Plant J. 2014, 77, 944–953. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.D.; Seo, P.J.; Yoon, H.K.; Park, C.M. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Chen, Y.; Ye, M.; Lu, H.; Wang, D.; Chen, Q. Evolutionary history of the C-repeat binding factor/dehydration-responsive element-binding 1 (CBF/DREB1) protein family in 43 plant species and characterization of CBF/DREB1 proteins in Solanum tuberosum. BMC Evol. Biol. 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Gupta, A.; Soni, D.; Garg, R.; Pathre, U.V.; Nath, P.; Sane, A.P. Ectopic expression of a tomato DREB gene affects several ABA processes and influences plant growth and root architecture in an age-dependent manner. J. Plant Physiol. 2017, 214, 97–107. [Google Scholar] [CrossRef]
- Wang, G.; Xu, X.; Wang, H.; Liu, Q.; Yang, X.; Liao, L.; Cai, G. A tomato transcription factor, SlDREB3 enhances the tolerance to chilling in transgenic tomato. Plant Physiol. Biochem. 2019, 142, 254–262. [Google Scholar] [CrossRef]
- Yang, X.; Lu, M.; Wang, Y.; Wang, Y.; Liu, Z.; Chen, S. Response mechanism of plants to drought stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. Dwarf and delayed flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 2004, 37, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Haake, V.; Cook, D.; Riechmann, J.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhyay, R.K.; Handa, A.K.; Mattoo, A.K. Transcript abundance patterns of 9- and 13-lipoxygenase subfamily gene members in response to abiotic stresses (heat, cold, drought or salt) in tomato (Solanum lycopersicum L.) highlights member-specific dynamics relevant to each stress. Genes 2019, 10, 683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008, 56, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Al-Abdallat, A.M.; Al-Debei, H.S.; Ayad, J.Y.; Hasan, S. Over-expression of SlSHN1 gene improves drought tolerance by increasing cuticular wax accumulation in tomato. Int. J. Mol. Sci. 2014, 15, 19499–19515. [Google Scholar] [CrossRef] [Green Version]
- Mueller, L.A.; Solow, T.H.; Taylor, N.; Skwarecki, B.; Buels, R.; Binns, J.; Lin, C.; Wright, M.H.; Ahrens, R.; Wang, Y.; et al. The SOL genomics network: A comparative resource for Solanaceae biology and beyond. Plant Physiol. 2005, 138, 1310–1317. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms, molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Alhindi, T.; Al-Abdallat, A.M. Genome-wide identification and analysis of the MADS-box gene family in American beautyberry (Callicarpa americana). Plants 2021, 10, 1805. [Google Scholar] [CrossRef] [PubMed]
- Al-Abdallat, A.M.; Ali-Sheikh-Omar, M.A.; Alnemer, L.M. Overexpression of two ATNAC3-related genes improves drought and salt tolerance in tomato (Solanum lycopersicum L.). Plant Cell Tissue Organ Cult. 2015, 120, 989–1001. [Google Scholar] [CrossRef]
- Barrs, H.D.; Weatherley, P.E. A re-examination of the relative turgidity techniques for estimating water deficits in leaves. Aust. J. Biol. Sci. 1962, 15, 413–428. [Google Scholar] [CrossRef] [Green Version]
- Ristic, Z.; Jenks, M. Leaf cuticle and water loss in maize lines differing in dehydration avoidance. J. Plant Physiol. 2002, 159, 645–651. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Sima, W.; Ouyang, B.; Wang, T.; Ziaf, K.; Luo, Z.; Liu, L.; Li, H.; Chen, M.; Huang, Y.; et al. Tomato SlDREB gene restricts leaf expansion and internode elongation by downregulating key genes for gibberellin biosynthesis. J. Exp. Bot. 2012, 63, 6407–6420. [Google Scholar] [CrossRef]
- Kahn, T.L.; Fender, S.E.; Bray, E.A.; O’Connell, M.A. Characterization of expression of drought- and abscisic acid-regulated tomato genes in the drought-resistant species Lycopersicon pennellii. Plant Physiol. 1993, 103, 597–605. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.; Moses, M.; Plant, Á.; Bray, E.A. Multiple mechanisms control the expression of abscisic acid (ABA)-requiring genes in tomato plants exposed to soil water deficit. Plant Cell Environ. 2002, 22, 989–998. [Google Scholar] [CrossRef]
- Zhang, X.; Fowler, S.G.; Cheng, H.; Lou, Y.; Rhee, S.Y.; Stockinger, E.J.; Thomashow, M.F. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 2004, 39, 905–919. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Hichri, I.; Muhovski, Y.; Clippe, A.; Žižková, E.; Dobrev, P.I.; Motyka, V.; Lutts, S. SlDREB2, a tomato dehydration-responsive element-binding 2 transcription factor, mediates salt stress tolerance in tomato and Arabidopsis. Plant Cell Environ. 2016, 39, 62–79. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.H.; Lee, J.T.; Charng, Y.Y.; Chan, M.T. Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 2002, 130, 618–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rai, G.K.; Rai, N.P.; Rathaur, S.; Kumar, S.; Major, S. Expression of rd29A:AtDREB1A/CBF3 in tomato alleviates drought-induced oxidative stress by regulating key enzymatic and non-enzymatic antioxidants. Plant Physiol. Biochem. 2013, 69, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Prasad, V.; Yang, P.T.; Wu, J.; David Ho, T.H.; Charng, Y.Y.; Chan, M.T. Expression of Arabidopsis CBF1 regulated by an ABA/stress inducible promoter in transgenic tomato confers stress tolerance without affecting yield. Plant Cell Environ. 2003, 26, 1181–1190. [Google Scholar] [CrossRef]
- Kasuga, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K.A. Combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 2004, 45, 346–350. [Google Scholar] [CrossRef] [Green Version]
- Pino, M.T.; Skinner, J.S.; Park, E.J.; Jeknić, Z.; Hayes, P.M.; Thomashow, M.F.; Chen, T.H.H. Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol. J. 2007, 5, 591–604. [Google Scholar] [CrossRef]
- Nir, I.; Moshelion, M.; Weiss, D. GAMT1 promotes drought tolerance. Plant Cell Environ. 2014, 37, 113–123. [Google Scholar] [CrossRef]
- Satish, L.; Rathinapriya, P.; Muthuramalingam, P.; Pandian, S.; Ceasar, S.A.; Ramesh, M. Overexpression of Erianthus arundinaceus DREB2 transcription factor ameliorates the salinity and drought tolerance in Eleusine coracana cultivars. Biol. Life Sci. Forum 2021, 4, 8. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).