Abstract
Leaf-feeding phylloxera decreases the photosynthetic activity of a grape plant, leading to decreasing number of fruit buds. In addition, phylloxera larvae emerging from the leaf galls may colonize the roots, negatively affecting the growth of the grape plant. In this study, we evaluated host tolerance of three grapevine hybrid populations obtained from crossing of the same maternal grapevine M. no. 31-77-10 with interspecific hybrids carrying introgressions from Muscadinia and other North American Vitis species against leaf-feeding grape phylloxera. Combining genotyping data of the populations obtained with 12,734 SNPs and their resistance phenotypes evaluated in the laboratory experiment, we performed an association study. As the result of GWAS, nine SNPs with the lowest significant p-values were discovered in the whole sample of 139 hybrids as associated with variation of the scores ‘the percentage of infested leaves’ and ‘intensity of gall formation’. Three of the SNPs on LG 7 were located in the same chromosome interval where a major QTL (RDV6) for root phylloxera resistance was reported from Muscadine background. Two SNPs on LG 8 were detected within the gene, encoding E3 ubiquitin-protein ligase UPL4 involved in apoptosis. SNPs detected on LG 13 and LG 18 may overlap with the previously reported QTLs for phylloxera resistance inherited from V. cinerea.
1. Introduction
Grape phylloxera Daktulosphaira vitifoliae Fitch is one of the main pests of the grape plant that endangers viticulture throughout Europe [1]. Phylloxera was introduced to Europe in the middle of the 19th century with planting material from the United States [2]. Favorable conditions for this invasive pest have developed on European grape varieties, which led to the massive spread of this aphid-like insect in Europe [3]. In Russia, phylloxera was first discovered on the southern coast of the Crimea in 1880, 16 years later the outbreak of the pest was eased. However, since the 1970s, the entire Crimean peninsula—and especially its southern coast—have become zones of continuous distribution of phylloxera.
Over the entire history of the fight against grape phylloxera, more than 300 different methods have been tested. One of the most effective and widespread methods was the use of a grafted vines. However, the long-term (more than a century) usage of phylloxera-tolerant rootstock, changes in the varietal composition of grapes, and climate change have led to the increasing spread of the foliar form of phylloxera in vineyards. The pest adapted and new problems arose, since the already foliar form of phylloxera began to cause economically significant damage to viticulture all over the world [4]. The environmental factors associated with climate change are discussed as a possible reason for the increasing phylloxera infestation of V. vinifera cultivars and interspecific hybrids in Central Europe, as well as changes in vineyard management practices [4,5].
The harmfulness of the leaf-feeding phylloxera in comparison with the root-feeding form is not so obvious, since there is no rapid complete death of the grape plant. However, some previous studies have shown that Vitis vinifera varieties probably decease not because of the phylloxera itself, but mostly from the concomitant development of pathogenic microflora on their roots, causing severe root rot, which leads to the final death of the grape plant [6,7]. It should be taken into account that, starting from the second generation, a part of the larvae of leaf-feeding phylloxera moves to the roots, and the proportion of such larvae increases with each subsequent generation. The negative influence of leaf-feeding grape phylloxera also lies in the fact that the larvae emerging from the leaf galls are spread by the wind over remote distances, settling in other vineyards.
The genetic determinants of phylloxera root resistance were firstly discovered in the genetic background of the interspecific hybrid ‘Börner’ (V. riparia × V. cinerea) [8]. The highly significant QTL was mapped on the ‘lower’ part of linkage group (LG) 13, and later designated as Resistance Daktulosphaira vitifoliae 1 (RDV1). It was assumed that the resistance locus was transmitted from V. cinerea parent, and is expected to operate differently from the resistance determinants inherited from V. riparia. Rootstocks obtained from hybrids with V. riparia have been successfully used as a source of resistance to root phylloxera to safe European viticulture. Another genetic determinant of root phylloxera resistance (RDV2) was mapped on LG 14 in the V. cinerea C2-50 genetic map at position 16.7 cM [9].
Resistance to leaf feeding phylloxera has been reported to be both qualitative and quantitative [10]. Among grape breeding lines of different interspecific backgrounds, various levels of resistances were revealed. For the resistant phenotypes, a reduced number of galls/leaf, incomplete gall formation, and a hypersensitive response was reported [11]. Rather high broad-sense heritability (H = 0.51) for the trait “proportion of leaves with galls” was estimated when conducting greenhouse experiments with artificial infection of rooted grape cuttings with phylloxera eggs. As the result, highly significant QTL for the trait was mapped on LG 14 [10]. The greatest effect on the foliar phylloxera resistance had alleles inherited from MN1264 female parent, whose ancestry has at least six Vitis species, including V. riparia, V. vinifera, V. labrusca, V. rupestris, V. aestivalis, and V. lincecumii. Remarkably, in the same study QTLs for the root form of phylloxera resistance were mapped on other chromosomes (LG 5 and LG 10) using the same mapping population. However, the revealed leaf-specific phylloxera resistance locus on LG 14 overlapped the location of the RDV2 locus, previously identified in the V. cinerea background [9].
Three loci of the resistance to root-specific phylloxera were recently identified in Muscadinia rotundifolia background using the BC1 progeny involving the V. vinifera × M. rotundifolia accession ‘VRH8771′ and the V. vinifera cv. ‘Cabernet-Sauvignon’ [12]. The major QTL (RDV6) was mapped in the lower arm of chromosome 7, which is supposed to be homologous to Muscadinia LG 20. The QTL on LG 7 explained 87% of phenotypic variance for root nodosity number and larval development in a root in vitro experiment, which is expected, given that M. rotundifolia is naturally resistant to grape pathogens and pests. Two other QTLs have been detected on LG3 and LG10 and were officially named as RDV7 and RDV8, respectively [12].
Recently, we reported the results of evaluating of pathogen resistance in three Crimean grape hybrid populations obtained via crossing the interspecific hybrid “Magarach 31-77-10” as the female parent and Muscadinia rotundifolia × Vitis vinifera BC5 hybrids (2000-305-143 and 2000-305-163) as the male parents [13]. In the study, 139 progenies from three crosses were genotyped using the RADseq method, providing information on 12,734 SNPs polymorphic in all three hybrid populations. Here, we report the results of assessing the resistance of hybrid populations to leaf-feeding phylloxera and the results of studying the association between the detected SNPs and segregation observed in hybrid populations in terms of resistance to foliar phylloxera.
2. Materials and Methods
2.1. Plant Material
Three hybrid populations derived from crosses of the same female M. no. 31-77-10 (Nimrang × Seibel 13-666) with male 2000-305-143, 2000-305-163, and a mixture of pollen (DRX-M5-734, DRX-M5-753, DRX-M5-790) were investigated in the study. Hereafter, the obtained hybrid populations are designed as 3-11, 4-11, and 2-11 respectively. Pedigree of the parental genotypes was described previously [13]. In short, the female parent ‘Magarach no. 31-77-10′ was obtained by crossing the ‘Nimrang’ variety having functionally female type of flower with the ‘Seibel 13666′ variety, which is a complex interspecific hybrid obtained through the use of the species of the Euvitis subgenus: V. riparia, V. berlandieri, V. cinerea, V. aestivalis, V. lincecumii, V. labrusca, and V. rupestris with just 45% of Vitis vinifera genes in the genome [14]. The males 2000-305-143 and 2000-305-163 are progenies of the cross VRH 3082-1-42 × V. vinifera cv. Regent. VRH 3082-1-42 was the BC4 progeny of pseudo-backcrosses tracing back to the famous NC6-15 interspecific hybrid (Muscadinia rotundifolia × Vitis vinifera) [15]. Varieties from the pedigree list of the studied populations (Nimrang, Seibel 13-666, and Regent) were included in the analysis. We also analyzed the variety ‘Dixie’ as a representative of the immune species M. rotundifolia.
DRX-M5 interspecies hybrids were developed at the Institute of Viticulture and Winemaking ‘Vierul’ (Moldova) as F5 progeny from the interspecific crosses of the famous DRX-55 hybrid (F2 from N.C.6-15 open pollinated). In 2011, pollen of three hybrids—DRX-M5-734, DRX-M5-753, and DRX-M5-790—were sent to the ‘Magarach’ Institute and used to pollinate the female parent Magarach no. 31-77-10, resulting in 66 progenies of population 2-11 [13].
The three hybrid populations 3-11, 4-11, and 2-11 (in total 139 plants) were planted as a self-rooted plants in 2013, in the experimental fields at the village Partenit, Yalta District, Crimea (44°34′12.0″ N 34°19′44.1″ E).
2.2. Laboratory Assessment of Resistance to the Leaf Form of Phylloxera
In February 2021, 3–4-eyed, dormant, hardwood cuttings were harvested and packed in damp filter paper and polyethylene to prevent drying out. The cuttings were stored in a refrigerator at a temperature of +4 °C for three months. In the first week of May, the cuttings were placed in water for rooting. The plants were rooted in separate 0.5 L pots in water without using growth regulators. When 3–5 mature leaves were formed on cuttings, a preliminary selection of experimental plants was made according to the uniformity of morphological development for the further infection with the leaf form of phylloxera. The population of the leaf-feeding phylloxera used for inoculation, was collected from an infected vineyard in the Bakhchysarai region of the Crimea, on the rootstock variety Kober 5BB.
The laboratory experiment consisted of a completely randomized design (CRD) with three replications of each genotype: 3 × 43 for population 3–11; 3 × 30 for population 4-11; 3 × 66 for population 2-11.
The experiment was carried out at the beginning of July 2021 with an interval of one week. The leaves were moistened, and 20 eggs of the leaf form of phylloxera were placed on the surface of each of the three upper leaves of the cuttings. Cuttings infected with the eggs of phylloxera were kept in natural light, but not in direct sunlight. The cuttings were sprayed daily with water to maintain moisture.
The resistance to leaf-feeding phylloxera was evaluated in the first, second, and third weeks after infection. First, the percentage of infested leaves (P, %) was calculated as the number of leaves with galls in relation to the total number of leaves in the sample. The second score—intensity of gall formation (R, %)—was estimated as an indicator of the manifestation of the disease (formation of galls) according to the method described below:
First, each leaf of an experimental plant was assessed visually by 1-to-9-point scale according to OIV-461 guideline [16] (Table 1):

Table 1.
Codes OIV-461. Leaf: degree of tolerance to Phylloxera (leaf gall).
Next, intensity of gall formation (R, %) in a particular genotype was calculated by the formula:
where:
a—a score of the scale, according to which the lesion was evaluated in the experiment;
b—the number of affected leaves within the range of this score;
N—the total number of leaves evaluated, pcs;
K—the highest score of the scale;
100—conversion factor.
2.3. SSR and Target SNP Marker Analysis
For SSR marker analysis DNA was extracted from 100 mg of fresh or frozen young grape leaves using a CTAB method (2%) [17]. The amount and purity of the isolated DNA was estimated using a BioPhotometer plus spectrophotometer (Eppendorf, Hamburg, Germany). DNA was diluted to a final concentration of 20 ng/μL.
Three SSR markers VVIQ67 (LG 7), VMC3D7 (LG 10), and VVIN85 (LG 10) co-localized with QTLs for phylloxera resistance [12] were combined in multiplex PCR. The forward primer of each pair was 5′-end labeled with fluorescent dyes (FAM, TAMRA, and R6G) (Syntol, Moscow, Russia). Amplification reactions were carried out using a T100 thermal cycler (Bio-Rad, Hercules, CA, USA) in a total volume of 15 µL with 20 ng of template DNA, 5–10 pmol of each primer and 6 µL 2.5× PCR reaction mixture containing 0.25 mM dNTPs, 3 mM MgCl2 and 3 unit Taq polymerase (Syntol, Moscow, Russia). The cycling program consisted of the following steps: 5 min at 95 °C followed by 35 cycles of 30 s at 95 °C, 30 s at 63 °C, and 35 s at 72 °C and a final extension step of 15 min at 72 °C. The PCR products were evaluated by capillary electrophoresis on an automated Genetic Analyzer ABI Prism 3130 and using GenMapper 4.1 program.
To identify SNPs linked to phylloxera resistance loci [12], we designed three pairs of primers to amplify sequences of a length of 433–475 bp, containing SNPs 3_5494608, 7_13408919, and 7_4261424. The PCR was performed on a T100 amplifier (BIO-RAD, Hercules, CA, USA) in a total reaction volume of 15 μL under the following conditions: (1) 95 °C—5 min; (2) 36 cycles: 95 °C—30 s; 63–64 °C—30 s; 72 °C—35 s; (3) 72 °C—15 min.
The amplicons were purified on columns of the ColGen reagent kit (Syntol, Moscow, Russia). To set up a sequencing reaction a BrilliantDye V.3.1 kit Cycle sequencing kit (NimaGen, The Netherlands) was used according to the manufacturer’s protocol. The sequencing reaction was carried out on a T100 amplifier (BIO-RAD, Hercules, CA, USA).
The products of the sequencing reaction were purified by the ethanol/acetate Na-precipitation method according to the protocol. Then the dried sample was dissolved in 10 μL of formamide and denatured for 5 min at 94 °C. Sequencing was performed on a 4-capillary genetic analyzer ABI 3130 (Applied Biosystems, Waltham, MA, USA) in PDMA-6 polymer (Syntol, Moscow, Russia). Sequencing results were processed using Sequencing Analysis Software V5.3.1. The alignment of reads to the reference genome PN40024 12× (GCF_000003745.3) from the NCBI database and SNP analysis were performed using the Unipro Ugene v.34 [18].
2.4. Genome Wide Association Study (GWAS)
General linear model (GLM) and mixed linear model (MLM) implemented in GAPIT version 3 [19] were used for genome wide association study (GWAS) of 12,734 SNP markers polymorphic within all three hybrid populations under study [13] and the variation of the percentage of infested leaves (P, %) and intensity of gall formation (R, %). Additive effects associated with SNP markers were estimated.
GLM tests the association between a phenotype (Y) and markers (Si) as
where Q is the population structure estimated by PCA (principal components analysis), and e is the residual term.
Y = Si + Q + e,
The association with MLM was estimated as
where K is the kinship matrix, accounting for known relatedness among individuals in the sample.
Y = Si + Q + K + e,
3. Results
3.1. Laboratory Assessment of the Resistance of Hybrid Populations to Leaf Form of Phylloxera
In this study, we evaluated the resistance to the leaf form of phylloxera for three hybrid populations: 3-11 (43 progenies), 4-11 (30 progenies), and 2-11 (66 progenies) in the laboratory experiment. The hybrid populations were obtained from the cross of the same maternal genotype M. no. 31-77-10 with intespecific hybrids carrying Muscadinia introgressions.
When the resistance to leaf-feeding phylloxera was assessed for the populations in the field (2000–2021), the gall formation intensity did not exceed 10% (data not shown). The field evaluating data obtained for the same material over two years varied significantly. This is in line with reports that field tolerance/resistance against grape phylloxera depends on the plant genotype, but also other factors such as insect biotype or environmental conditions [20]. Instead, the controlled artificial inoculation was applied in the laboratory experiment, and segregation in severity of foliar phylloxera infestation was observed between parental genotypes and their hybrid progenies.
First, in the laboratory experiment, we observed that both percentage of infested leaves and intensity of gall formation were the highest in the male parent 2000-305-163, whereas its full sibling 2000-305-143 showed almost complete resistance, as well as female parent M. no. 31-77-10 (Figure 1). Accordingly, segregation of progenies from the cross M. no. 31-77-10 × 2000-305-163 (population 4-11) for the resistance trait significantly differed from that observed for population 3-11 (M. no. 31-77-10 × 2000-305-143). Population 2_11 obtained as a result of pollination of the maternal genotype M. no. 31-77-10 with a mixture of pollen of DRX hybrids segregates significantly in resistance.
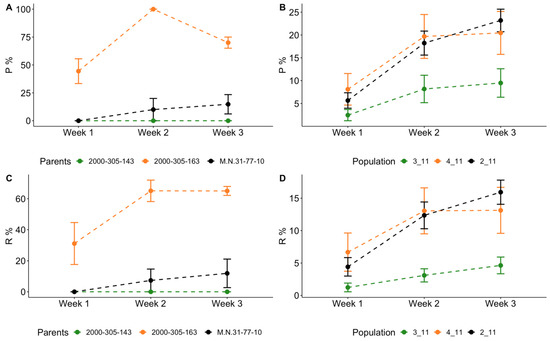
Figure 1.
Assessment of resistance to the leaf form of phylloxera among populations of grape hybrids carrying M. rotundifolia introgressions in a laboratory experiment. Percentage of infested leaves (P, %) scored in first, second, third week after infection by phylloxera eggs: (A) among the parental lines; (B) among the populations of their progenies. Similarly, (C,D) for the intensity of gall formation (R, %).
The resistance to phylloxera of the maternal genotype M. no. 31-77-10 can be assumed, since there are several North American grape varieties in its pedigree for which phylloxera resistance loci have been identified (e.g., V. riparia, V. berlandieri, V. cinerea). However, the contrast resistance level of full siblings 2000-305-143 and 2000-305-163 can be explained by different recombinations of chromosomes of their parents’ (cv.’Regent’ and VRH 3082-1-42), including those carrying introgressions from Muscadinia.
3.2. Assessment of Parental Genotypes with SSR and SNP Markers Linked to the Phylloxera Resistance Loci in Muscadinia Background
Recently, Rubio et al. [12] reported three SSR and three SNP markers associated with phylloxera root resistance QTLs on chromosomes LG7, LG3, and LG10 of the interspecific hybrid VRH8771 carrying M. rotundifolia introgressions.
To understand better the reason for the difference in tolerance to leaf phylloxera observed between the studied hybrid populations, we evaluated their parental genotypes using these published molecular markers. We also involved in the analysis the variety ‘Dixie’ (M. rotundifolia) and the grapevine varieties from the pedigree list of the studied parental genotypes: Nimrang, Seibel 13-666, and Regent.
Table 2 shows that parental vines M. no. 31-77-10, 2000-305-143, 2000-305-163 do not differ in SNP alleles on LG3, vary in alleles of VMC3D7 and VVIN85 markers on LG10, but none of the alleles were alike to those that could be inherited from Muscadinia background. In contrast, on chromosome LG7, the male parent 2000-305-143 appears to have the VVIQ67 marker allele, similar to the allele found in Muscadinia variety ‘Dixie’.

Table 2.
Polymorphism of parental lines and their ancestors revealed using SNP and SSR markers linked to phylloxera resistance loci identified [12] on chromosomes LG7, LG3, and LG10 respectively.
The VVIQ67 microsatellite marker was mapped in the very lower arm of chromosome LG 7 of the high-density genetic map of a muscadine hybrid VRH8771, and its position was co-locating with the major phylloxera resistance QTL [12]. Taking into account that the Vitis chromosome had been split into the LG 7 (upper arm) and LG 20 (lower arm) in Muscadinia, these authors assumed the location of the putative R factor in the lower arm of chromosome 7, to which the VVIQ67 marker can be linked.
3.3. Genome-Wide Association Analysis (GWAS) of Variability in Resistance to the Leaf Form of Phylloxera in Three Studied Hybrid Populations
In our previous study three hybrid populations 2-11, 3-11, and 4-11 were genotyped using RADseq method, and a common set of 12,734 SNP markers was identified [13]. Here, we use the obtained genotyping data to search for SNPs that may be associated with the variability in resistance to foliar phylloxera observed for hybrid populations. We used two assessed phenotypes as variables: the percentage of infested leaves (P, %) and the intensity of gall formation (R, %) (see Section 2.2).
The classical way to map resistance loci as QTLs on a linkage map developed for a bi-parental population may not be optimal for the studied plant material, since the number of progenies for two hybrid populations (3-11 and 4-11) with known parents was too limited: 43 and 30, respectively. Besides, 66 progenies of population 2-11, in which the mother plant was pollinated with a mixture of pollen from the three paternal genotypes DRX-M5-734, DRX-M5-753, DRX-M5-790, would have to be excluded from such a QTL mapping procedure.
Instead, we decided to conduct an association study (GWAS) for the whole sample of 139 genotypes (43 + 30 + 66) taking into account their relationship and known pedigree. To test the marker–trait associations we used two models that control the population structure of the sample: GLM and MLM. GLM estimates the structure via principal component analysis, using the whole set of SNPs. For MLM we especially prepared the kinship matrix, where relatedness between known full-siblings was estimated as 50%, between half-siblings as 25%. Since the male parents of populations 3-11 and 4-11 are, in fact, full-siblings, those offspring from crosses with the same female M. no. 31-77-10 are related as 37.5%. Progenies of 2-11 populations were split in three non-overlapping clusters on the PCA plot [13]. We assumed that those three clusters may correspond to progenies of three male parents (DRX-M5-734, DRX-M5-753, DRX-M5-790) and consider them within each of the clusters as full-siblings.
The resulting quantile-quantile (QQ) plot showed that mixed linear model perfectly accounted for population structure and does it slightly better than the GLM (Figure 2). However, GLM revealed more SNPs on the upper right section of the graph deviated from the diagonal that are most likely associated with the trait under study.
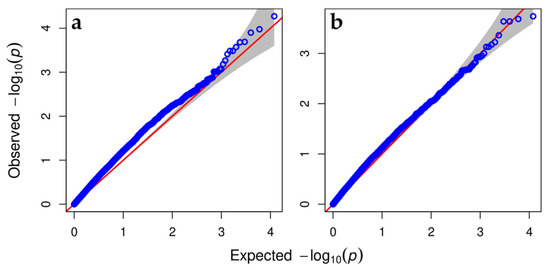
Figure 2.
Quantile–quantile (QQ) plot showing how well the models (a) GLM and (b) MLM, used in GWAS, account for population structure and familial relatedness in a pooled sample of 139 progenies (populations 2-11, 3-11, 4-11) from three crosses. Here, an association of SNPs with the trait ‘intensity of phylloxera gall formation’ (R, %) was analyzed.
The Manhattan plot in Figure 3 summarizes GWAS results for the phenotype ‘intensity of gall formation’ (R, %), suggesting that SNPs associated with variability of the resistance to leaf form of phylloxera observed in the hybrid populations are located on chromosomes LG7, LG8, LG13, and LG18 (Figure 3).
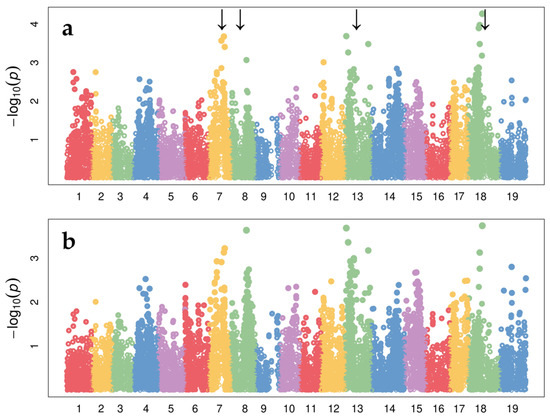
Figure 3.
The Manhattan plot that summarizes GWAS results obtained from GLM (a) and MLM (b) for the trait ‘intensity of phylloxera gall formation’ (R, %). The x-axis is the genomic position of the SNPs, and the y-axis is the negative log base 10 of the p-values. Each chromosome is colored differently. Loci with the strongest associations with the trait on chromosomes 7, 8, 13, and 18 are marked by arrows.
Pearson correlation coefficient calculated for P (%) and R (%) scores was high (0.859, p-value < 2.2 × 10−16), however, the lists of significantly associated SNPs differed slightly for the two traits. We have investigated in detail only the SNPs significantly associated with variation of both resistance phenotypes under analysis (Table 3). Eight out of nine SNPs associated with the resistance to leaf form of phylloxera in our study were found within the coding sequences. For one SNP on LG 13 we referred the nearest transcript match (Table 3).

Table 3.
SNPs polymorphic in three hybrid grape populations and associated with the traits ‘the percentage of infested leaves (P, %) and ‘intensity of phylloxera gall formation’ (R, %) according to GWAS results.
The SNPs on LG7 associated with resistance to foliar phylloxera in our analysis (7_12058709, 7_14355208, 7_15121714) were located nearby the SNP 7_13408919, reported by Rubio et al. [12] as a major QTL for phylloxera larval development.
Figure 4 shows how the intensity of phylloxera gall formation (R, %) depends on alleles of the associated SNPs on LG7. The highly resistant male parent 2000-305-143 was homozygous for all three SNPs: 7_12058709 (A/A), 7_14355208 (C/C), 7_15121714 (T/T). Another male parent 2000-305-163 and female M. no. 31-77-10 were heterozygous: 7_12058709 (G/A), 7_14355208 (T/C), 7_15121714 (T/C). The less foliar phylloxera severity was observed for those plants that inherited the ‘2000-305-143-like’ homozygote genotype (Figure 4). Expectedly, there were about a half (42%) of that kind of progenies in the highly resistant population 3-11 (M. no. 31-77-10 × 2000-305-143), and just a quarter of those (25%) in the most susceptible to foliar phylloxera population 4-11 (M. no. 31-77-10 × 2000-305-163).
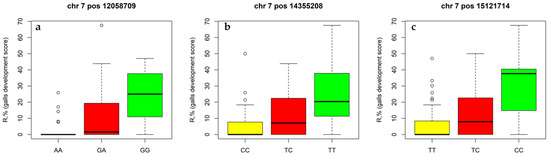
Figure 4.
The box plots showing how the intensity of phylloxera gall formation (R, %, y-axis) varies depending on alleles of associated SNPs revealed on LG7. SNPs: (a)—7_12058709 (G/A), (b)—7_14355208 (T/C), (c)—7_15121714 (T/C).
To our knowledge, there were no reports about polymorphisms on LG 8 in Vitis species linked to phylloxera resistance. In our analysis, though, two linked SNPs 8_ 14223679 and 8_14223697 from the same coding sequence LOC100243094 showed significant association with the resistance to the leaf form of phylloxera assessed by both P, % and R, % scores. The gene encodes E3 ubiquitin-protein ligase UPL4, which is involved in the regulation of the fundamental proteins affecting apoptosis, such as ubiquitin proteasome system (UPS) [21]. Since a hypersensitive response to phylloxera attack usually associated with local necrosis, the possible contribution of the apoptosis related gene to the resistance of a grape plant to phylloxera can be assumed.
SNP 13_21213131 on LG 13 was the only one in Table 2 found outside of the coding sequences. All three parental genotypes of the studied populations were heterozygous (A/G) at this locus, indicating the absence of an obvious source of the resistant allele. However, segregating progenies carrying G/G genotype (11% in 2-11 population, 32% in 3-11, 26% in 4-11) did not show any sign of infection in the laboratory experiment with artificial infection by leaf-feeding phylloxera. RDV1 locus was also mapped on the ‘lower’ region of LG 13 [8]. The possible linkage disequilibrium of the detected SNP 13_21213131 with RDV1 locus requires further investigation.
Two of three SNPs detected on LG 18 (18_10506582 and 18_10506585) are closely linked and belong to the same coding sequence LOC104882551 with unknown function (Table 3). Another SNP 18_12898048 was discovered approximately 2Mb away from the gene. Earlier, a QTL was mapped by Clark et al. on LG 18 (102 cM) in the complex interspecies hybrid’s MN1246 genetic map for two traits of foliar phylloxera resistance: log10 transformation of gall counts and the number of leaves with galls [10]. The traits assessed in our study (the percentage of infested leaves and intensity of gall formation) were consistent with those scores. It is rather difficult to assume whether the SNPs on LG 18 revealed in our study are close to the QTL reported by Clark et al. [10]. On the high-density genetic map of muscadine hybrid VRH8771 the genetic distance 103 cM corresponds to SNP with physical position 18_11890988, whereas on the genetic map of V. vinifera cv. ‘Cabernet-Sauvignon’ the SNP 18_17399516 was mapped in position of 99 cM. As a rough estimate, we propose that the physical distance 10506582–12898048 at LG18 harboring associated SNPs may overlap with the previously reported QTL on LG 18 (102 cM).
4. Discussion
Phylloxera poses a danger to European viticulture, including the Crimean vineyards. In past, many Crimean local grapevine varieties were lost due to the phylloxera epidemic that broke out in Crimea and caused enormous damage to local vines. For example, in 1954 the vineyards in two Crimean districts affected by phylloxera were uprooted on an area of about 140 hectares.
Phylloxera is an obligate biotroph of Vitis spp. and can undergo both belowground (root-feeding) and aboveground (leaf-feeding) life stages. Grafting of the susceptible cultivated grapevine (V. vinifera) on top of tolerant Vitis spp. rootstock, is a common practice in viticulture today, as root infestation no longer causes vine death. Remarkably, the tolerant rootstock vines derived from hybrids of American species are often highly susceptible to leaf infestation. As a result, some commercial vineyards have been showing high intensities of leaf galls for many years [22].
Field evaluations carried out at the Institute of Vine and Wine ‘Magarach’ in Yalta (Crimea, Russia) have found that leaves damaged by leaf-feeding phylloxera are usually deformed, curled, and dry out prematurely, which negatively affects the crown architecture of the grape plant, and the photosynthetic activity of the leaves decreases. A decrease in the assimilation surface of the leaf leads to a deterioration in the quality of the current year’s harvest. Besides, the premature defoliation leads to an insufficient accumulation of carbohydrates, decreasing number of fruit buds, which affects the harvest of the next year. A decrease in the frost resistance of damaged plants was also observed. In years with cold winters, a plant weakened from the loss of leaf apparatus can completely die. Besides, the huge amount of phylloxera larvae emerging from the leaf galls colonizes the roots, negatively affecting the growth of the grape plant [23].
Development of phylloxera has been reported to be highly dependent on the environment (e.g., host and abiotic factors) [20]. Plants exposed to drought stress showed more intense phylloxera root infestation compared to watered ones, suggesting that events of water shortage favor the insect’s feeding damage [24]. Thus, the hot and dry conditions of the Crimean Peninsula contribute to the invasion of phylloxera, which causes significant damage to local viticulture. During the field investigations conducted in Crimea it was found that gall formation of leaf phylloxera reduces the number of developed and fruitful shoots by 10–14%, inflorescences by 15–18%, and the total leaf area of the grape plant is reduced by 1.2–2.2 times. As a result, the yield is reduced by 14–44% and the mass concentration of sugars in the juice of berries decreases by 5–14% [23].
For this obvious reason a breeding program is being implemented at the ‘Magarach’ Institute to saturate the genome of the Crimean grapevine varieties with the genes of Muscadinia rotundifolia and other North American Vitis species, providing a source of resistance to the root and leaf forms of phylloxera [25]. Marker-assisted selection could speed up the breeding process considerably.
In this article, we report a pilot study investigating leaf-feeding phylloxera tolerance of Vitis × Muscadinia hybrids, which are segregating for alleles of genes defensive to the pest. To date, seven groups of phylloxera biotypes have been reported, differing by feeding sites, feeding organ (galled tissue), insect development and by their performance on a particular Vitis host [20]. Remarkably, not all the phylloxera biotypes are able to form leaf galls, showing good performance on nodosities and tuberosities on the roots. There are also phylloxera biotypes forming leaf galls on hybrids of V. vinifera × American Vitis species, but not on the ‘pure’ V. vinifera varieties [20]. The leaf phylloxera strains used for inoculation in our experiments may belong to one of these phylloxera biotypes, since they were collected from the rootstock variety Kober 5BB, which was obtained from the crossbreeding of the North American species Vitis berlandieri and Vitis riparia (https://plantgrape.plantnet-project.org (accessed on 12 November 2021)).
Because of the economic importance of root-feeding phylloxera biotypes a variety of methods have been developed to study interactions between root-feeding grape phylloxera and Vitis hosts, while standardized leaf infestation studies are scarce [22]. In our study, we have chosen to focus on leaf infestation rather than on root phylloxeration; therefore, the experiment methodology in some ways differed from those mentioned in review of Forneck et al. [20] for leaf-feeding phylloxera: whole plant assays potted under controlled environmental conditions or clipped leaves of field-grown plants. Instead, we performed a lab-based assay by rooting cuttings in water. When assessing the resistance to foliar phylloxera, the main target is the leaves—the younger they are, the better. Since leaf growth does not depend too much on the method of rooting (in water or in soil), the cuttings were kept in water for a three-week experimental period. Within the time period, new leaves on the plant have time to grow and the maximum intensity of gall formation of the leaf form of phylloxera was achieved. Prolongation of this short-term experiment aiming to assess several generations of the leaf form of phylloxera on the cuttings would hardly be successful, because new leaves do not develop so quickly, but the old ones, completely affected, dry up. Taken this in account, it was unlikely to observe root phylloxeration, thus the cuttings were submerged in water instead of soil to assess severity of foliar phylloxera infestation.
Due to difficulties with obtaining M. rotundifolia × Vitis spp. hybrids, the reports about genetic determinants of tolerance to phylloxera in Muscadinia background are limited. According to the segregation ratios of F1 hybrids backcrossed to V. vinifera it has been suggested that grape phylloxera resistance in the M. rotundifolia is mediated by a semi-dominant locus, which is regulated by three genetic modifiers [26]. Rubio et al. [12] firstly mapped the putative causative loci on a high-density genetic map of the interspecific hybrid VRH8771 ((‘Cabernet-Sauvignon’ × ‘Alicante Bouschet’) × M. rotundifolia cv. NC184–4). In their experiments, NC184-4 was the original source of muscadine resistance genes, while the parental genotypes 2000-305-143 and 2000-305-163, assessed in our study, had an interspecific hybrid NC6-15 in their pedigree. However, our discovery of SNPs associated with leaf phylloxera resistance in the interval 12058709-15121714 of the LG 7 physical map corresponds very well to the previous report on the main QTL for phylloxera larvae development located in the in the lower arm of chromosome 7.
In addition to the resistance loci transmitted to the studied hybrid populations from Muscadinia background, other resistance loci introgressed from cv Regent genotype or maternal genotype M. no. 31-77-10 could be expected. Both are complex interspecific hybrids with North American phylloxera-resistant species in their pedigree. Therefore, it is not surprising that the associated SNPs found in our study on LG 13 or LG 18 chromosomes may overlap with the previously reported QTLs, where a higher level of resistance was provided by alleles inherited from various North American Vitis species.
In the present study, nine SNPs were found associated with leaf-feeding phylloxera tolerance. Most of them cannot be directly related to defense mechanisms, although the detected polymorphism in the ubiquitin protein ligase E3 gene (LG 8) might be involved in the hypersensitive response to the phylloxera attack. Genes belonging to at least six functional categories were proposed as up- or downregulated during the hypersensitive response (HR) in ‘Börner’ compared to a phylloxera sensitive variety ‘Riesling’ [27]. Among them there were phytoalexins, ethylene-associated gene products, cell wall proteins and transcription factors. Besides, jasmonic and salicylic acid signaling pathways were recently shown to be essential for the activation of host defense responses when grape phylloxera infests vines by the formation of organoid root galls [28].
Among the associated SNPs detected on LG 7 were those in genes that play an important role in the biological process of the cell cycle, such as proteins associated with the cell division cycle and chromatin modification-related proteins. Recently, it was hypothesized that affected vines respond to phylloxeration with a compensatory strategy consisting of enhanced root growth, leaf respiration, and photosynthetic activity [29]. A possible link can be assumed between the compensatory mechanisms and polymorphism of genes responsible for cell division and growth processes in leaf galled vines. However, it is possible that SNPs associated with phylloxera defense genes in our study are linked to causative genes or are in LD with them. Fine mapping of the phylloxera defensive loci using genomic resources developed for Vitis vinifera and Muscadinia rotundifolia may be an effective approach to uncovering the genetic mechanism of tolerance of vines to this destructive pest.
Author Contributions
Conceptualization, V.V. (Vladimir Volynkin) and E.P.; Data curation, I.V. and S.G.; Formal analysis, E.M. and E.L.; Funding acquisition, V.L.; Investigation, E.M., D.K., V.V. (Vitalii Volodin), and G.S.; Methodology, I.V., S.G., E.M. and E.G.; Project administration V.V. (Vladimir Volkov); Resources, I.V. and E.L.; Software, K.L. and E.G.; Supervision, S.G. and V.R.; Validation, I.V.; Visualization, K.L. and E.G.; Writing—original draft, V.V. (Vladimir Volynkin) and E.P.; Writing—review and editing, E.P. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by Russian Science Foundation (project no. 20-16-00060).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in this article; SNP database used in this article was published earlier (https://doi.org/10.3390/plants10061215).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- EPPO Global Database. Available online: https://gd.eppo.int/taxon/VITEVI/categorization (accessed on 12 November 2021).
- Ollat, N.; Cookson, S.J.; Lauvergeat, V.; Marguerit, E.; Barrieu, F.; Gambetta, G.; Goutouly, J.-P.; Tandonnet, J.-P.; Vivin, P.; Delrot, S. Grapevine Roots: The Dark Side. Acta Hortic. 2017, 1188, 213–226. [Google Scholar] [CrossRef]
- Skinkis, P.; Walton, V.M.; Kaiser, C. Grape Phylloxera Biology and Management in the Pacific Northwest. Oregon State University, OSU Extension Catalog 2009. Available online: https://catalog.extension.oregonstate.edu/sites/catalog/files/project/pdf/ec1463.pdf (accessed on 12 November 2021).
- Forneck, A.; Mammerler, R.; Tello, J.; Breuer, M.; Müller, J.; Fahrentrapp, J. First European Leaf-Feeding Grape Phylloxera (Daktulosphaira Vitifoliae Fitch) Survey in Swiss and German Commercial Vineyards. Eur. J. Plant Pathol. 2019, 154, 1029–1039. [Google Scholar] [CrossRef]
- Edwards, J.; Norng, S.; Powell, K.S.; Granett, J. Relationships between grape phylloxera abundance, fungal interactions and grapevine decline. Acta Hortic. 2007, 733, 151–157. [Google Scholar] [CrossRef]
- Usatov, V.; Kireyeva, L.; Volynkin, V.; Klimenko, V.; Oleinikov, N. Evaluation of Interspecific Populations of Grapevine in Breeding for Complex Resistance to Fungal Diseases and Phylloxera. Vitis-J. Grapevine Res. 1990, S, 278–294. [Google Scholar]
- Armengol, J.; Gramaje, D. Soilborne Fungal Pathogens Affecting Grapevine Rootstocks: Current Status and Future Prospects. Acta Hortic. 2016, 1136, 235–328. [Google Scholar] [CrossRef]
- Zhang, J.; Hausmann, L.; Eibach, R.; Welter, L.J.; Töpfer, R.; Zyprian, E.M. A Framework Map from Grapevine V3125 (Vitis Vinifera ‘Schiava Grossa’ × ‘Riesling’) × Rootstock Cultivar ‘Börner’ (Vitis Riparia × Vitis Cinerea) to Localize Genetic Determinants of Phylloxera Root Resistance. Theor. Appl. Genet. 2009, 119, 1039–1051. [Google Scholar] [CrossRef]
- Smith, H.M.; Clarke, C.W.; Smith, B.P.; Carmody, B.M.; Thomas, M.R.; Clingeleffer, P.R.; Powell, K.S. Genetic identification of SNP markers linked to a new grape phylloxera resistant locus in Vitis cinerea for marker-assisted selection. BMC Plant Biol. 2018, 18, 360. [Google Scholar] [CrossRef]
- Clark, M.D.; Teh, S.L.; Burkness, E.; Moreira, L.; Watson, G.; Yin, L.; Hutchison, W.D.; Luby, J.J. Quantitative Trait Loci Identified for Foliar Phylloxera Resistance in a Hybrid Grape Population: Foliar Phylloxera Resistance. Aust. J. Grape Wine Res. 2018, 24, 292–300. [Google Scholar] [CrossRef]
- Powell, K.S. Grape phylloxera: An overview. In Root Feeders: An Ecosystem Approach; Johnson, S.N., Murray, P.J., Eds.; CABI: Wallingford, CT, USA, 2008; pp. 96–114. ISBN 978-1-84593-461-3. [Google Scholar]
- Rubio, B.; Lalanne-Tisné, G.; Voisin, R.; Tandonnet, J.-P.; Portier, U.; Van Ghelder, C.; Lafargue, M.; Petit, J.-P.; Donnart, M.; Joubard, B.; et al. Characterization of Genetic Determinants of the Resistance to Phylloxera, Daktulosphaira Vitifoliae, and the Dagger Nematode Xiphinema Index from Muscadine Background. BMC Plant Biol. 2020, 20, 213. [Google Scholar] [CrossRef]
- Volynkin, V.; Vasylyk, I.; Volodin, V.; Grigoreva, E.; Karzhaev, D.; Lushchay, E.; Ulianich, P.; Volkov, V.; Risovannaya, V.; Blinova, S.; et al. The Assessment of Agrobiological and Disease Resistance Traits of Grapevine Hybrid Populations (Vitis Vinifera L. × Muscadinia Rotundifolia Michx.) in the Climatic Conditions of Crimea. Plants 2021, 10, 1215. [Google Scholar] [CrossRef]
- Volynkin, V.; Gorislavets, S.; Volodin, V.; Vasylyk, I.; Lushchay, E.; Likhovskoi, V.; Potokina, E. Immunogenic Breeding Program. Stage I-Phytopathological Screening of the Grape Gene Pool. E3S Web Conf. 2021, 254, 03003. [Google Scholar] [CrossRef]
- Bouquet, A. Introduction Dans l’espèce Vitis Vinifera L. d’un Caractère de Résistance à l’oidium (Uncinula Necator Schw. Burr.) Issu de l’espèce Muscadinia Rotundifolia (Michx.) Small. Vignevini 1986, 12, 141–146. [Google Scholar]
- Codes des Caracteres Descriptifs des Varietes et Especes de Vitis OIV 2009 Website of Organisation Internationale de la Vigne et du vin from. Available online: http://www.oiv.int/fr (accessed on 12 November 2021).
- Angelini, E.; Clair, D.; Borgo, M.; Bertaccini, A.; Boudon-Padieu, E. Flavescence Dorée in France and Italy-Occurrence of Closely Related Phytoplasma Isolates and Their near Relationships to Palatinate Grapevine Yellows and an Alder Yellows Phytoplasma. Vitis 2001, 40, 79–86. [Google Scholar]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Forneck, A.; Powell, K.S.; Walker, M.A. Scientific Opinion: Improving the Definition of Grape Phylloxera Biotypes and Standardizing Biotype Screening Protocols. Am. J. Enol. Vitic. 2016, 67, 371–376. [Google Scholar] [CrossRef][Green Version]
- Sharma, A.; Trivedi, A.K. Regulation of apoptosis by E3 ubiquitin ligases in ubiquitin proteasome system. Cell Biol. Int. 2020, 44, 721–734. [Google Scholar] [CrossRef]
- Wilmink, J.; Breuer, M.; Forneck, A. Effect of Temperature on Host Plant-specific Leaf- and Root-feeding Performances: A Comparison of Grape Phylloxera Biotypes C and G. Entomol. Exp. Appl. 2021, 169, 1113–1125. [Google Scholar] [CrossRef]
- Stranishevskaya, E.; Misiak, A. The Effects of Leaf Phylloxera on the Fruitfulness Parameters, Yield and Quality of the Grape Plant Viticulture and Winemaking. Vitic. Winemak. 2010, 40, 53–55. [Google Scholar]
- Savi, T.; García González, A.; Herrera, J.C.; Forneck, A. Gas Exchange, Biomass and Non-Structural Carbohydrates Dynamics in Vines under Combined Drought and Biotic Stress. BMC Plant Biol. 2019, 19, 408. [Google Scholar] [CrossRef]
- Volynkin, V.; Levchenko, S.; Vasylyk, I.; Likhovskoi, V. Analysis of F2-F6 generations from hybridization with Vitis rotundifolia at the Institute Magarach. Acta Hortic. 2020, 1289, 269–274. [Google Scholar] [CrossRef]
- Bouquet, A. Etude de la résistance au phylloxera radicicole des hybrides Vitis vinifera× Muscadinia rotundifolia. Vitis 1983, 32, 311–323. [Google Scholar]
- Blank, L.; Wolf, T.; Eimert, K.; Schröder, M.-B. Differential Gene Expression during Hypersensitive Response in Phylloxera -Resistant Rootstock ‘Börner’ Using Custom Oligonucleotide Arrays. J. Plant Interact. 2009, 4, 261–269. [Google Scholar] [CrossRef]
- Eitle, M.W.; Griesser, M.; Vankova, R.; Dobrev, P.; Aberer, S.; Forneck, A. Grape Phylloxera (D. Vitifoliae) Manipulates SA/JA Concentrations and Signalling Pathways in Root Galls of Vitis Spp. Plant Physiol. Biochem. 2019, 144, 85–91. [Google Scholar] [CrossRef]
- Eitle, M.W.; Cargnoni, M.; Acar, A.; Crespo Martinez, S.; Failla, O.; Kaul, H.-P.; Griesser, M.; Forneck, A. Phylloxeration Effects on the Sink Activity and Assimilation Rate in Phylloxera (Daktulosphaira Vitifoliae Fitch) Infested Grapevines (Vitis spp.). Acta Hortic. 2017, 1188, 291–298. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).