Abstract
Long noncoding RNAs (lncRNAs) play important roles in abiotic and biotic stress responses; however, studies on the mechanism of regulation of lncRNA expression are limited in plants. The present study examined the relationship between lncRNA expression level and two active histone modifications (H3K4me3 and H3K36me3) in Brassica rapa. Both histone marks were enriched in the chromatin regions encoding lncRNAs, especially around the transcription start site. The transcription level of long intergenic noncoding RNAs was positively associated with the level of H3K4me3 and H3K36me3, while this association was not observed in natural antisense RNAs (NATs) and intronic noncoding RNAs. As coordinate expression of mRNAs and paired NATs under biotic stress treatment has been identified, the transcriptional relationship between mRNAs and their paired NATs following Fusarium oxysporum f. sp. conglutinans (Foc) inoculation was examined. A positive association of expression levels between mRNAs and their paired NATs following Foc inoculation was observed. This association held for several defense-response-related genes and their NAT pairs. These results suggest that coordinate expression of mRNAs and paired NATs plays a role in the defense response against Foc.
1. Introduction
Brassica rapa L. includes leafy vegetables such as Chinese cabbage (var. pekinensis), pak choi (var. chinensis), and komatsuna (var. perviridis), as well as root vegetables such as turnip (var. rapa) [1]. These vegetables provide nutrition, vitamins, minerals, dietary fiber, and health-promoting substances. Most of the modern cultivars of these vegetables are F1 hybrids [2]. Disease resistance is an important trait in a breeding program of F1 hybrid cultivars [3], and a major disease problem is Fusarium yellows. Fusarium yellows is caused by infection with a soil-borne fungus, Fusarium oxysporum f. sp. conglutinans (Foc) or F. oxysporum f. sp. rapae (For), in B. rapa vegetables, and infection leads to yellowing, wilting, defoliation, stunting, and plant death [3,4,5,6]. A Fusarium yellows resistance gene has been identified in B. rapa [6,7], and a system for DNA marker selection of Fusarium yellows resistance has also been developed [5,6,8]. The transcriptional response to Foc inoculation has also been examined [9], and SA-induced genes involved in systemic acquired resistance (SAR) are considered to play a role in resistance against Foc [9,10].
In all eukaryotic cells, approximately 145–147 bp of DNA wraps the histone octamer of the nucleosome in the chromatin in the nucleus. A histone octamer has two copies of each of four core histone proteins, H2A, H2B, H3, and H4 [11,12,13]. The histone proteins have an N-terminal tail, and modification of amino acid residues in the histone tail can change gene expression. Histone modification includes methylation, acetylation, phosphorylation, ubiquitylation, and sumoylation [14,15,16]. Methylated lysine in histone tails can be found in mono (me1)-, di (me2)-, or tri (me3)-methylated states at lysine 4 (K4), 9 (K9), 27 (K27), and 36 (K36) of histone H3 [14,15,17]. Histone modification has an important role in plant development and response to stresses through regulating chromatin structure and gene expression [18,19]. In plants, tri-methylation of histone H3 lysine 4 (H3K4me3) and H3K36me3 are histone marks that are associated with transcriptional activation, while H3K9me2 and H3K27me3 are associated with transcriptional repression [18,20,21]. Bivalent chromatin states where active and repressive histone marks co-exist have been identified [22,23,24], and this chromatin state may trigger plant stress-responsive gene expression [25,26].
Recent advances in sequencing technology enable us to detect various types of transcripts, not only mRNAs but also noncoding RNAs (ncRNAs), which are defined as RNAs devoid of protein-coding potential [27]. There are two types of ncRNAs, housekeeping ncRNAs and regulatory ncRNAs. Ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) are classified as housekeeping ncRNAs [28,29,30]. Regulatory ncRNAs are classified into two groups based on their size, small RNAs (sRNAs) and long ncRNAs (lncRNAs). sRNAs are usually 18 to 30 nucleotides (nt) in length such as microRNAs (miRNAs) or small interfering RNAs (siRNAs). LncRNAs are longer than 200 nt in length. LncRNAs are classified into three major groups: long intergenic noncoding RNAs (lincRNAs), natural antisense RNAs (NATs) transcribed from the complementary DNA strand of their associated genes, and intronic noncoding RNAs (incRNAs) derived from introns [29,30,31,32,33]. LncRNAs have been identified at the whole genome level in many plant species and are generally expressed at lower levels than mRNAs and have low sequence conservation between species [27,34,35]. Only 10–15% of the lncRNAs in Chinese cabbage had relatively high sequence similarity with lncRNAs of other Brassica species, and less than 2% had similarity with A. thaliana sequences [36]. Transcriptome analyses have shown high diversity of lncRNA expression in different tissues or under abiotic and biotic stresses, suggesting that lncRNAs might play important roles in plant development or abiotic and biotic stress responses. However, the functions of only limited numbers of lncRNAs have been elucidated [37].
Chromatin immunoprecipitation sequencing (ChIP-seq) allows us to identify the distribution of the histone modifications. The association between histone modification and expression levels in genes has been analyzed in some plant species including B. rapa [12,26,38,39]. However, only a limited number of studies have examined the relationship between lncRNA expression and histone modifications in plants [40,41]. RNA-sequencing (RNA-seq) has shown coordinate expression of mRNAs and paired NATs under stress treatment [36,42,43], which suggests a role for NATs in stress response. We have identified differentially expressed genes following Foc inoculation [9] and mRNA and NAT pairs [43]. From these datasets, we searched for lncRNAs that are paired with genes whose expression is changed by Foc. We examined both the relationship between lncRNA expression level and active histone modifications (H3K4me3 and H3K36me3) and the transcriptional relationship of mRNAs and their paired NATs following Foc inoculation in B. rapa.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
Fusarium yellows susceptible and resistant inbred lines of Chinese cabbage—RJKB-T24 and RJKB-T23, respectively—were used as plant materials [7]. To examine the transcription of NATs, an additional four inbred lines of Chinese cabbage (RJKB-T39, RJKB-T41, RJKB-T42, and RJKB-T43), four komatsuna commercial cultivars (‘Inasena’ (KANEKO SEEDS Co., Ltd., Maebashi, Japan), ‘Nacchan’ (Nohara Seed Co., Ltd., Kuki, Japan), ‘Nanane’ (Takii & Co., Ltd., Kyoto, Japan), and ‘Wakami’ (Sakata Seed Corporation, Yokohama, Japan)), four commercial pak choi cultivars, (‘Kunyan’ (Sakata Seed Corporation), ‘Natsu-Mikado’ (Sakata Seed Corporation), ‘Niihao-syan’ (Watanabe Noji Co., Ltd., Noda, Japan), and ‘Raiko’ (Sakata Seed Corporation)), and four commercial turnip cultivars (‘Mifune’ (Sakata Seed Corporation), ‘Nitou’ (Sakata Seed Corporation), ‘Shiro-Warabe’ (Takii & Co., Ltd.), and ‘Yukibotan’ (Musashino Seed Co., Ltd., Tokyo, Japan)) were used. After surface sterilization, the seeds were placed on Murashige and Skoog (MS) agar supplemented with 1% (w/v) sucrose and grown under long day (LD) condition (16 h light/8 h dark) at 21 °C. First and second leaves were collected from 14-day-old plants.
2.2. RNA Extraction and Gene Expression Analysis
Total RNAs were isolated from whole plants that were mock and F. oxysporum f. sp. conglutinans (Foc) inoculated (at 24 and 72 h after inoculation (HAI)) by using SV Total RNA Isolation System (Promega Co., Madison, WI, USA). Differentially expressed genes of mock- and Foc-inoculated samples were identified from RNA-seq analysis in RJKB-T24 and RJKB-T23 [9]. For quantitative real-time RT-PCR (qPCR), 500 ng total RNA was used for cDNA synthesis using ReverTra Ace® qPCR RT Master Mix with gDNA Remover (TOYOBO Co., Ltd., Osaka, Japan). Prior to qPCR, single PCR amplification products and the absence of genomic contamination were confirmed by RT-PCR. Using a Light-Cycler 96 (Roche Molecular Systems, Inc., Pleasanton, CA, USA), qPCR was performed using FastStart Essential DNA Green Master (Roche). The qPCR conditions were 95 °C for 10 min followed by 45 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 10 s. Melt temperature analysis (65–97 °C at 0.1 °C/s) was performed for each reaction after amplification cycles to confirm the presence of single amplified product. The relative expression level of all mRNAs and NATs relative to ACTIN (Bractin) was automatically calculated using automatic CQ calling according to the manufacturer’s instructions (Roche) [44]. Data presented are the average and standard error of three biological and experimental replicates and statistically analyzed using Student’s t test, p value < 0.05, 0.01, or 0.001. The primer sets used in this study are listed in Table S1.
Total RNAs from first and second leaves were extracted by SV Total RNA Isolation System (Promega) in RJKB-T24 for RNA-sequencing (RNA-seq) for detection of lncRNAs. A more detailed description of lncRNA identification can be found in [43]. To examine the transcriptional conservation among four different varieties of B. rapa, total RNAs from first and second leaves were extracted by SV Total RNA Isolation System (Promega) in sixteen B. rapa lines. cDNA was synthesized, and RT-PCR using QuickTaq®HS DyeMix was performed. RT-PCR amplified products were electrophoresed using 1.5% agarose gel. PCR conditions were 94 °C for 2 min followed by 30, 35, or 40 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 30 s. The primers used in this study are listed in Table S1.
2.3. DNA Extraction and PCR
The cetyl trimethyl ammonium bromide (CTAB) method was used to isolate genomic DNA [45]. PCR amplification of genomic DNA as template was performed to test whether the coding genomic sequences of NATs were conserved in the sixteen B. rapa lines. PCR amplification of genomic regions corresponding to all six NATs was confirmed by electrophoresis of PCR amplified products using QuickTaq®HS DyeMix (TOYOBO) on 1.5% agarose gel in all sixteen B. rapa lines. PCR conditions were 94 °C for 2 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 30 s, and final extension at 68 °C for 3 min. Primer sequences used in this study are shown in Table S1.
2.4. Detection of Epigenetic States in lncRNA Coding Regions
Chromatin immunoprecipitation sequencing (ChIP-seq) data were previously produced using the same line, tissue, and developmental stages but were harvested independently [26]. We used these data to investigate the epigenetic states (H3K4me3 and H3K36me3) of lncRNA coding regions in B. rapa. Data covering genomic regions encoding for lncRNA in chromosomes A01 to A10 were extracted from the ChIP-seq reads using anti-H3K4me3 (Millipore, 07-473) and H3K36me3 (Abcam, ab9050) antibodies that were mapped to the B. rapa reference genome v.1.5 using Bowtie2 version 2.2.3.
2.5. Sequential ChIP-qPCR
For sequential ChIP analysis, one gram of first and second leaves of RJKB-T24 was used. Anti-H3K4me3 antibodies (Millipore, 07-473) and anti-H3K27me3 antibodies (Millipore, 07-449) were used for the first and second ChIP, respectively. Experiments were performed as described by [26,46]. The average and standard error (s.e.) of three biological and more than three technical replicates are presented. Amplification of target mRNAs/NATs and non H3K4me3 and H3K27me3 marks (Bra011336) were compared by qPCR using immunoprecipitated DNA as a template to calculate the enrichment of bivalent H3K4me3-H3K27me3-marks of the targeted mRNA-NATs pairs (Bra016382-MSTRG.19710 and Bra033594-MSTRG.1355). The difference in the qPCR amplification of the input DNA as a template was used to correct the difference between the primer pairs. The primer sets used in this study are listed in Table S1.
3. Results
3.1. Comparison of the H3K4me3 and H3K36me3 States in the lncRNA Coding Region
We identified 1444 long intergenic noncoding RNAs (lincRNAs), 551 natural antisense transcripts (NATs), and 93 intronic noncoding RNAs (incRNAs) in 14-day first and second leaves in RJKB-T24 [43]. We also examined the H3K4me3 and H3K36me3 distribution in 14-day first and second leaves in RJKB-T24 [26]. In the present study, the genomic regions encoding lncRNAs that have H3K4me3 and H3K36me3 marks were examined. In total, 704 of 1173 (60.0%) lincRNAs, 458 of 529 (86.6%) NATs, and 47 of 92 (51.1%) incRNA coding genomic regions had H3K4me3 marks (Figure S1), whereas 574 of 1173 (48.9%) lincRNAs, 380 of 529 (71.8%) NATs, and 38 of 92 (41.3%) incRNA coding genomic regions had H3K36me3 marks (Figure S1). H3K4me3 and H3K36me3 were enriched in the transcribed region of lncRNAs, especially around the transcription start site, similar to the genic regions (Figure 1). The expression level of lincRNAs was higher when the encoding regions had H3K4me3 or H3K36me3 marks than without H3K4me3 or H3K36me3 marks (Figure 2). In NATs and incRNAs, there was no difference of expression level between with and without H3K4me3 or H3K36me3 marks on their encoding regions (Figure 2).
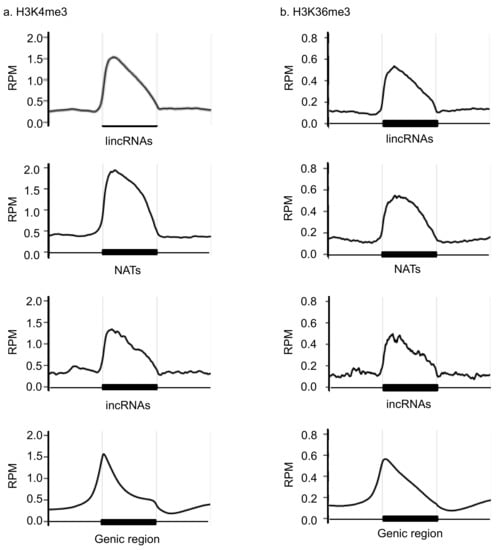
Figure 1.
Enrichment of H3K4me3 (a) and H3K36me3 (b) in the lncRNAs and genic region. The X-axis represents the region including 1 kb upstream and 1 kb downstream. The Y-axis represents the reads per million (RPM).
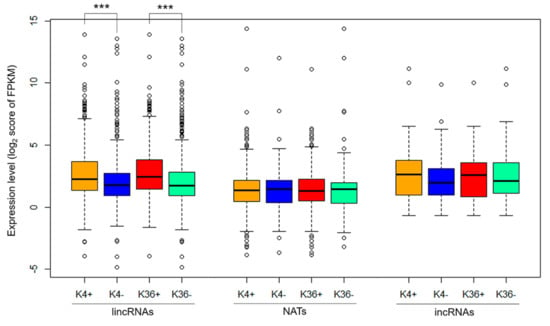
Figure 2.
Boxplots of the expression level of each type of lncRNAs with (+) or without (−) H3K4me3 (K4) and H3K36me3 (K36) on the coding region of RJKB-T24. ***, p < 0.001 (Student’s t-test).
3.2. Characterization of Histone Modification States in Differentially Expressed Genes Following F. oxysporum f. sp. conglutinans (Foc) Inoculation and in Their Overlapping lncRNAs
Previously, we identified differentially expressed genes following 24- or 72-h Fusarium oxysporum f. sp. conglutinans (Foc) inoculation (HAI) in the resistant line RJKB-T23 and the susceptible line RJKB-T24 [9]. In RJKB-T23, 260 and 88 genes showed differential expression at 24 and 72 HAI, respectively, and in RJKB-T24, 253 and 109 genes showed differential expression at 24 and 72 HAI, respectively [9]. We identified overlapped genes between two data sets, DEGs following Foc inoculation (24 and 72 HAI in RJKB-T23 and RJKB-T24) and paired genes overlapping lncRNAs (in RJKB-T24); twelve mRNA and NAT pairs were identified (Figure 3, Figure 4 and Figure 5 and Figure S2).
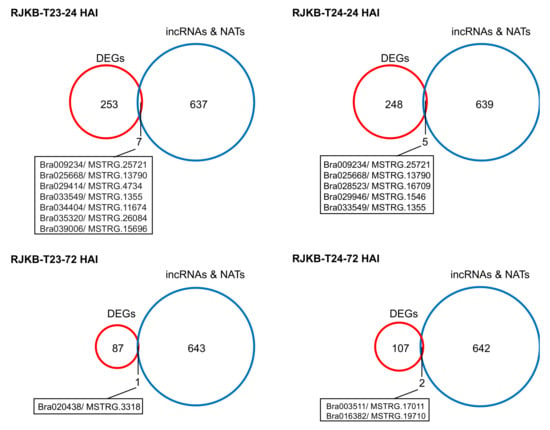
Figure 3.
Venn diagrams between differentially expressed genes (DEGs) following F. oxysporum f. sp. conglutinans (Foc) and paired genes overlapping incRNAs or NATs. HAI of 24 or 72 indicates 24 or 72 h after Foc inoculation, respectively.
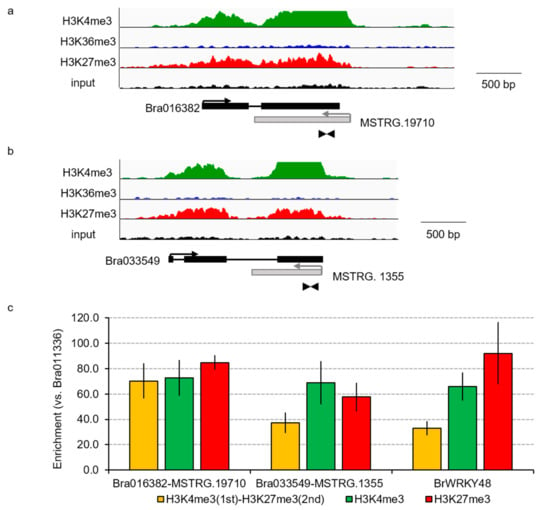
Figure 4.
Bivalent histone modification in the genic regions and their paired NATs. (a,b) Visualization of H3K4me3, H3K36me3, and H3K27me3 peaks by the Integrative Genomic Viewer (IGV) by ChIP-seq. Black and gray boxes represent the exon regions of genes or NATs, respectively. Arrows represent the direction of transcription. Arrowheads represent the position of primer sets for sequential ChIP-qPCR. (c) Sequential ChIP-qPCR analysis in 14-day leaves in RJKB-T24. Bra011336 that does not have H3K4me3 and H3K27me3 is used as the reference gene for qPCR. A previous study using sequential ChIP-qPCR showed that BrWRKY48 has both H3K4me3 and H3K27me3 marks [26]. Values are means ± standard error (s.e.; three biological and technical replicates) of relative H3K4me3, H3K27me3, or H3K4me3/H3K27me3 levels.
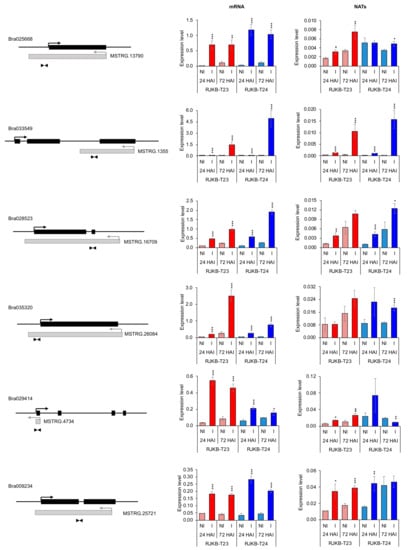
Figure 5.
Expression levels in mRNAs and their paired NATs following F. oxysporum f. sp. conglutinans (Foc) inoculation. Black and gray boxes represent the exon regions of gene or NATs, respectively. Arrows represent the direction of transcription. Arrowheads represent the position of primer sets for qPCR to detect the NAT transcripts. Bar graph represents values that are means ± standard error (s.e.; three biological and technical replicates) of relative Bractin expression levels. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Student’s t-test). NI, non-inoculated; I, inoculated.
The histone modification states (active marks, H3K4me3 and H3K36me3; repressive mark, H3K27me3) of the genomic regions encoding these 12 mRNA and NAT pairs in 14-day leaves in RJKB-T24 were examined (Table 1). Three pairs did not have any histone modifications (Table 1). One pair had only H3K4me3 or H3K27me3 and four pairs had H3K4me3 and H3K36me3 (Table 1). Two pairs had H3K4me3 and H3K27me3, indicating bivalent active and repressive histone modifications (Figure 4, Table 1). One pair had all three histone modifications (Table 1). To examine the simultaneous occupancy of active (H3K4me3) and repressive (H3K27me3) histone modifications, sequential ChIP-qPCR in 14-day leaves in RJKB-T24 was performed in two mRNA and NAT pairs (Bra016382/MSTRG.19710 and Bra033549/MSTRG.1355) that have both H3K4me3 and H3K27me3 marks [26,39]. The genomic region encoding Bra016382/MSTRG.19710 and Bra033549/MSTRG.1355 showed enrichment for the second modification, similar to BrWRKY48 which has been shown to have bivalent active and repressive histone modifications by sequential ChIP-qPCR (Figure 4) [26].

Table 1.
Histone modification states in the selected 12 mRNA and their paired NATs.
3.3. Relationship between mRNA and Their Paired NAT Transcription Following F. oxysporum f. sp. conglutinans (Foc) Inoculation
We selected six of the twelve mRNA and NAT pairs (Bra025668/MSTRG.13790, Bra028523/MSTRG.16709, Bra033549/MSTRG.1355, Bra035320/MSTRG.26084, Bra029414/MSTRG.4734, and Bra009234/MSTRG.25721) and performed qPCR following Foc inoculation at 24 and 72 HAI in RJKB-T23 and RJKB-T24 (Figure 5, Table S2). All six mRNAs showed upregulation following Foc inoculation at 24 and 72 HAI in both RJKB-T23 and RJKB-T24 (Figure 5, Table S2). In NATs, out of a total of 24 comparisons, 16 comparisons showed upregulation following Foc inoculation (Figure 5, Table S2). In MSTRG.1355, the expression levels increased following Foc inoculation at 24 and 72 HAI both in RJKB-T23 and RJKB-T24 (Figure 5, Table S2). Following Foc inoculation, expression was increased in three of the four comparisons in MSTRG.13790, MSTRG.16709, and MSTRG.25721 (Figure 5, Table S2). In MSTRG.4734, the expression levels following Foc inoculation at 24 and 72 HAI were increased in RJKB-T23, while the expression level at 72 HAI was decreased in RJKB-T24 (Figure 5, Table S2). In MSTRG.26084, the expression level increased following Foc inoculation only at 72 HAI in RJKB-T24 (Figure 5, Table S2).
The correlation coefficient between the ratios of the expression change of non-inoculated (NI) and inoculated samples (I) (I/NI ratio) in mRNA and NAT pairs was examined to observe whether the change of NAT expression is associated with the change of expression level of the mRNAs covering NATs. In total, there was a positive correlation in the I/NI ratio between mRNA and paired NATs (r = 0.69, p < 0.001) (Figure 6, Table S3). In each combination of mRNA and NAT pair, Bra033549/MSTRG.1355 (r = 0.94, p > 0.05) and Bra029414/MSTRG.4734 (r = 0.73, p > 0.05), there was a high positive correlation in the I/NI ratio, but not significant (Table S3). When examined in each condition, 72 HAI in RJKB-T23 (r = 0.87, p < 0.05) and RJKB-T24 (r = 0.99, p < 0.001) showed a positive correlation in the I/NI ratio (Table S3).
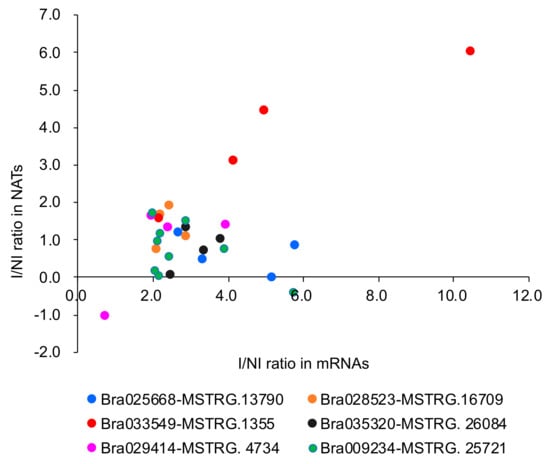
Figure 6.
Scatter plot representing the ratio of the expression levels of mRNAs and their paired NATs following F. oxysporum f. sp. conglutinans (Foc) inoculation (I) compared with non-inoculated (NI).
3.4. Transcriptional Conservation of NATs among B. rapa Varieties
As there is low sequence conservation of lncRNAs between closely related species, SNP number per length in each lncRNA (mutation rate) was examined in RJKB-T23 and RJKB-T24 using previously generated SNP data [47]. Contrary to our expectation, the average mutation rate of the lincRNA and incRNA encoding regions of RJKB-T23 was lower than that of the genic region, and the average mutation rate of the NAT encoding region was similar to that of the genic region in RJKB-T23 (Figure 7). In RJKB-T24, there was no difference in the average of mutation rate of the region encoding lincRNA, incRNA, or NAT compared with that of the genic region (Figure 7).
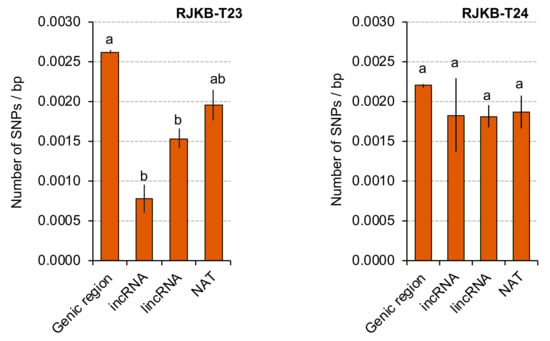
Figure 7.
The average number of SNPs per length (base pair) in region coding lncRNAs of RJKB-T23 and RJKB-T24. The values with the same letter are not significantly different with p < 0.001 by Tukey’s HSD test. Values are means ± standard error (s.e.) of SNP number per base pair.
Next, to examine the transcriptional conservation of NATs, we tested whether six NATs were expressed in 14-day leaves of sixteen B. rapa lines comprising four varieties. All six NATs were expressed in all sixteen lines of B. rapa, though there was a small variation of expression levels among lines (Figure 8).
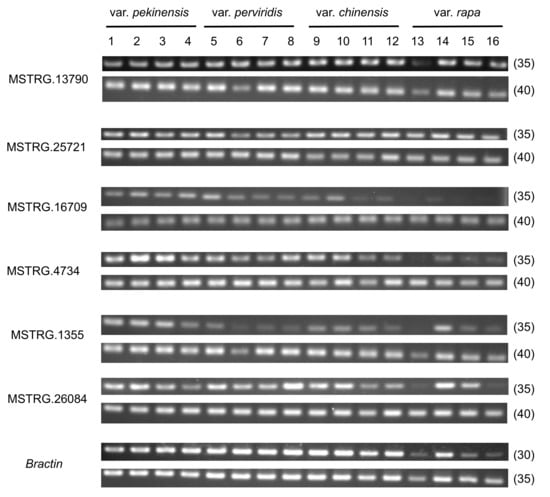
Figure 8.
RT-PCR of NATs in sixteen B. rapa lines. The number in parentheses (30, 35, or 40) represents the number of PCR cycles. The number represents each line as follows: For Chinese cabbage lines (var. pekinensis), 1 = RJKB-T39, 2 = RJKB-T41, 3 = RJKB-T42, 4 = RJKB-T43. For komatsuna lines (var. perviridis), 5 = ‘Inasena’, 6 = ‘Nacchan’, 7 = ‘Nanane’, 8 = ‘Wakami’. For pak choi lines (var. chinensis), 9 = ‘Kunyan’, 10 = ‘Natsu-Mikado’, 11 = ‘Niihao-syan’, 12 = ‘Raiko’. For turnip lines (var. rapa), 13 = ‘Mifune’, 14 = ‘Nitou’, 15 = ‘Shiro-Warabe’, 16 = ‘Yukibotan’.
4. Discussion
LncRNAs are not just transcriptional noise, and thousands of lncRNAs have been identified in plants [27,30,32,34,48,49]. In general, expression levels of lncRNAs are lower than those of mRNAs and are more tissue specific [34,50,51,52,53,54], suggesting a difference in transcription regulation between mRNAs and lncRNAs. However, our knowledge of the mechanisms of the regulation of lncRNA expression is limited. Histone modification plays a role in the regulation of mRNA expression [18,19], but it remains uncertain whether the role of histone modifications in the expression of lncRNAs is the same as that of mRNAs. Previously, we have examined H3K27me3 distribution in the genomic regions encoding lncRNAs in B. rapa [41], and unlike mRNAs, there was no negative association between the presence of H3K27me3 marks and lncRNA expression levels [39,41]. incRNAs with encoding regions having H3K27me3 marks showed higher expression levels than those without H3K27me3 marks; this trend is the opposite of mRNA [41]. There was a lower proportion having H3K27me3 marks in the region encoding lncRNAs than that in mRNAs [41]. In this study, we identified lncRNAs with active histone modification (H3K4me3- and H3K36me3-marks) in their encoding regions and examined the relationship of histone marks to expression level. The genomic regions encoding lincRNAs and incRNAs had H3K4me3 or H3K36me3 marks in a similar proportion as mRNAs, whereas regions encoding for NATs were highly enriched. Like genic regions, both active histone marks were highly enriched around the transcription start site of lincRNAs, incRNAs, and NATs. In lincRNAs, the enrichment of active histone marks either H3K4me3 or H3K36me3 in their encoding regions resulted in a higher expression level like in mRNAs, while this association was not found in incRNAs or NATs. In A. thaliana, H3K4me3 and H3K36me3 marks were positively correlated with the expression levels of lncRNAs, while H3K9me2 and H3K27me3 marks were less correlated [40]. These results suggest that in B. rapa, lncRNAs do not follow the same rules as mRNAs with regard to histone modification states.
Transcriptional coordination of mRNAs and their paired NATs has been identified in plants [53,54,55,56], and our previous study showed co-upregulation of some mRNA and their paired NATs following four weeks of cold treatment [43]. In particular, BrHSFB2a and its paired NAT showed similar expression patterns following two, four, and six weeks of cold treatment and six weeks of cold treatment followed by seven days under normal growth conditions [43]. In A. thaliana, both HSFB2a and its paired NAT, asHSFB2a, were induced by heat stress [57]. These examples suggest that the co-expression of mRNA-NAT pairs plays a role in abiotic stress. Co-expression of mRNA-NAT pairs has also been identified in biotic stress [42,58]. In A. thaliana, 15 NATs were detected that were responsive to F. oxysporum infection, and one pair showed a co-regulation pattern following F. oxysporum infection [42]. In this study, we examined the association of transcriptional changes between six mRNAs and their paired NATs following Foc inoculation at 24 and 72 HAI in Fusarium yellows resistant and susceptible lines. A strong association between mRNA and their paired NATs was found, especially in Bra033549-MSTRG.1355 and Bra029414-MSTRG.4734. Bra033549 encodes a putative beta-1,3-endoglucanase, which is known to be involved in defense by hydrolyzing the cell walls of fungal pathogens, and the Bra033549-MSTRG.1355 pair was upregulated following Foc inoculation in both resistant and susceptible lines, suggesting that upregulation of this gene may be a basic defense response and may not be specific to Fusarium yellows resistance. The Bra033549-MSTRG.1355 pair had the bivalent active and repressive (H3K4me3- and H3K27me3-marks) histone marks, suggesting a role in rapid response of transcription to pathogen infection. In contrast, the Bra029414-MSTRG.4734 pair was clearly upregulated following Foc inoculation in the resistant line. The A. thaliana ortholog of Bra029414 belongs to the superfamily of 2-oxoglutarate Fe (II)-dependent dioxygenases that is upregulated by pathogen infection or SA treatment [59] and encodes a salicylic acid 5-hydroxylase that fine-tunes SA homeostasis [60]. In B. rapa, Bra029414 was upregulated by salicylic acid treatment and was more induced in the Fusarium yellows resistant line than in the susceptible line [10]. As previous studies have suggested that systemic acquired resistance (SAR) may be important for Fusarium yellows resistance in B. rapa [9,10], Bra029414 is an important gene for Fusarium yellows resistance and may be involved in SA homeostasis following Foc infection in resistant lines. In A. thaliana, knocking down expression of a NAT resulted in a decreased expression level of its paired mRNA [55], suggesting that this NAT regulates the expression of its paired mRNA. In tomato, overexpression of a NAT induced its paired mRNA and enhanced resistance to Phytophthora infestans infection [58]. The effect of knockdown or overexpression of MSTRG.4734 on the expression of Bra029414 should be examined, but MSTRG.4734 may be involved in Fusarium yellows resistance through co-expression with Bra029414.
We showed that the association between histone modifications and lncRNA expression can be different from that of mRNA. We also showed that the expression patterns of mRNAs and their paired NATs were similar following Foc inoculation, suggesting that the transcriptional responses are interdependent. Although sequences of lncRNAs are poorly conserved between species [41], the sequences of lncRNAs were conserved within B. rapa. Thus, transcriptional coordination of mRNAs and their paired NATs following Foc inoculation may be widely found in B. rapa. The role of transcriptional coordination of mRNAs and their paired NATs will be clarified by increasing the number of lines and by studying the expression of mRNA-NAT pairs under different stress conditions.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae8010017/s1. Figure S1: Proportion of each type of lncRNA and mRNA with (+) or without (−) H3K4me3 (A) and H3K36me3 (B) marks on the coding region in RJKB-T24. K4 and K36 represent H3K4me3 and H3K36me3, respectively; Figure S2: Structure of genes and their paired NATs. Black and gray boxes represent the exon regions of the genes and NATs, respectively. Arrows represent the direction of transcription; Table S1: Sequence of primers used in this study; Table S2: Expression levels of six mRNAs and their paired NATs; Table S3: Correlation coefficient of expression levels between mRNAs and their paired NATs.
Author Contributions
Conceptualization, K.O. and R.F.; methodology, validation, and formal analysis, M.A.A., H.M., N.M. and S.T.; writing—original draft preparation, M.A.A., H.M. and R.F.; writing—review and editing, T.T.-Y., M.S., E.S.D., R.F. and K.O.; funding acquisition, N.M. and R.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Grant-in-Aid for Japan Society for the Promotion of Science (JSPS) Research Fellow to N.M. (18J20027) and grants from the Project of the Bio-oriented Technology Research Advancement Institution (Research program on Development of Innovative Technology) to R.F. (30029C).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Tomoko Kusumi and Yumiko Arai for their technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lv, H.; Miyaji, N.; Osabe, K.; Akter, A.; Mehraj, H.; Shea, D.J.; Fujimoto, R. The importance of genetic and epigenetic research in the Brassica vegetables in the face of climate change. In Genomic Designing of Climate-Smart Vegetable Crops; Kole, C., Ed.; Springer: Cham, Switzerland, 2020; pp. 161–255. [Google Scholar]
- Fujimoto, R.; Uezono, K.; Ishikura, S.; Osabe, K.; Peacock, W.J.; Dennis, E.S. Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed. Sci. 2018, 68, 145–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehraj, H.; Akter, A.; Miyaji, N.; Miyazaki, J.; Shea, D.J.; Fujimoto, R.; Doullah, M.A.U. Genetics of clubroot and Fusarium wilt disease resistance in Brassica vegetables: The application of marker assisted breeding for disease resistance. Plants 2020, 9, 726. [Google Scholar] [CrossRef] [PubMed]
- Enya, J.; Togawa, M.; Takeuchi, T.; Yoshida, S.; Tsushima, S.; Arie, T.; Sakai, T. Biological and phylogenetic characterization of Fusarium oxysporum complex, which causes yellows on Brassica spp., and proposal of F. oxysporum f. sp rapae, a novel forma specialis pathogenic on B. rapa in Japan. Phytopathology 2008, 98, 475–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akter, M.A.; Mehraj, H.; Itabashi, T.; Shindo, T.; Osaka, M.; Akter, A.; Miyaji, N.; Chiba, N.; Miyazaki, J.; Fujimoto, R. Breeding for disease resistance in Brassica vegetables using DNA marker selection. In Brassica Breeding and Biotechnology; Islam, A.K.M.A., Hossain, M.A., Islam, A.K.M.M., Eds.; IntechOpen: London, UK, 2021. [Google Scholar]
- Miyaji, N.; Akter, M.A.; Suzukamo, C.; Mehraj, H.; Shindo, T.; Itabashi, T.; Okazaki, K.; Shimizu, M.; Kaji, M.; Katsumata, M.; et al. Development of a new DNA marker for Fusarium yellows resistance in Brassica rapa vegetables. Plants 2021, 10, 1082. [Google Scholar] [CrossRef]
- Shimizu, M.; Fujimoto, R.; Ying, H.; Pu, Z.J.; Ebe, Y.; Kawanabe, T.; Saeki, N.; Taylor, J.M.; Kaji, M.; Dennis, E.S.; et al. Identification of candidate genes for Fusarium yellows resistance in Chinese cabbage by differential expression analysis. Plant Mol. Biol. 2014, 85, 247–257. [Google Scholar] [CrossRef]
- Kawamura, K.; Kawanabe, T.; Shimizu, M.; Okazaki, K.; Kaji, M.; Dennis, E.S.; Osabe, K.; Fujimoto, R. Genetic characterization of inbred lines of Chinese cabbage by DNA markers; towards the application of DNA markers to breeding of F1 hybrid cultivars. Data Brief 2016, 6, 229–237. [Google Scholar] [CrossRef] [Green Version]
- Miyaji, N.; Shimizu, M.; Miyazaki, J.; Osabe, K.; Sato, M.; Ebe, Y.; Takada, S.; Kaji, M.; Dennis, E.S.; Fujimoto, R.; et al. Comparison of transcriptome profiles by Fusarium oxysporum inoculation between Fusarium yellows resistant and susceptible lines in Brassica rapa L. Plant Cell Rep. 2017, 36, 1841–1854. [Google Scholar] [CrossRef] [Green Version]
- Miyaji, N.; Shimizu, M.; Takasaki-Yasuda, T.; Dennis, E.S.; Fujimoto, R. The transcriptional response to salicylic acid plays a role in Fusarium yellows resistance in Brassica rapa L. Plant Cell Rep. 2021, 40, 605–619. [Google Scholar] [CrossRef]
- Itabashi, E.; Osabe, K.; Fujimoto, R.; Kakizaki, T. Epigenetic regulation of agronomical traits in Brassicaceae. Plant Cell Rep. 2018, 37, 87–101. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H. Multifaceted chromatin structure and transcription changes in plant stress response. Int. J. Mol. Sci. 2021, 22, 2013. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Histone variants at a glance. J. Cell Sci. 2021, 134, jcs244749. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannister, A.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Zhan, Z.; Jiang, D. Histone modifications and their regulatory roles in plant development and environmental memory. J. Genet. Genom. 2019, 46, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Demetriadou, C.; Koufaris, C.; Kirmizis, A. Histone N-alpha terminal modifications: Genome regulation at the tip of the tail. Epigenetics Chromatin 2020, 13, 29. [Google Scholar] [CrossRef]
- Kim, J.M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef] [Green Version]
- Meyer, P. Epigenetic variation and environmental change. J. Exp. Bot. 2015, 66, 3541–3548. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, R.; Sasaki, T.; Ishikawa, R.; Osabe, K.; Kawanabe, T.; Dennis, E.S. Molecular mechanisms of epigenetic variation in plants. Int. J. Mol. Sci. 2012, 13, 9900–9922. [Google Scholar] [CrossRef] [Green Version]
- Quadrana, L.; Colot, V. Plant transgenerational epigenetics. Annu. Rev. Genet. 2016, 50, 467–491. [Google Scholar] [CrossRef]
- Sequeira-Mendes, J.; Aragüez, I.; Peiró, R.; Mendez-Giraldez, R.; Zhang, X.; Jacobsen, S.E.; Bastolla, U.; Gutierrez, C. The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 2014, 26, 2351–2366. [Google Scholar] [CrossRef] [Green Version]
- Qian, S.; Lv, X.; Scheid, R.N.; Lu, L.; Yang, Z.; Chen, W.; Liu, R.; Boersma, M.D.; Denu, J.M.; Zhong, X.; et al. Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL. Nat. Commun. 2018, 9, 2425. [Google Scholar] [CrossRef]
- Blanco, E.; González-Ramírez, M.; Alcaine-Colet, A.; Aranda, S.; Croce, L.D. The bivalent genome: Characterization, structure, and regulation. Trends Genet. 2020, 36, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zhang, W.; Marand, A.P.; Zhu, B.; Buell, C.R.; Jiang, J. Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato. Genome Biol. 2019, 20, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehraj, H.; Takahashi, S.; Miyaji, N.; Akter, A.; Suzuki, Y.; Seki, M.; Dennis, E.S.; Fujimoto, R. Characterization of histone H3 lysine 4 and 36 tri-methylation in Brassica rapa L. Front. Plant Sci. 2021, 12, 659634. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liu, K.; Yu, R.; Zhou, B.; Huang, P.; Cao, Z.; Zhou, Y.; Wang, J. From “Dark Matter” to “Star”: Insight into the regulation mechanisms of plant functional long non-coding RNAs. Front. Plant Sci. 2021, 12, 650926. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.J.; Penny, D. The RNA infrastructure: Dark matter of the eukaryotic cell? Trends Genet. 2009, 25, 120–128. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [Green Version]
- Ariel, F.; Romero-Barrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Rai, M.I.; Alam, M.; Lightfoot, D.A.; Gurha, P.; Afzal, A.J. Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, Q.H.; Kaufmann, K. Long non-coding RNAs in plants: Emerging modulators of gene activity in development and stress responses. Planta 2020, 252, 92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Guo, H.; Hu, W.; Ji, W. The emerging role of long non-coding RNAs in plant defense against fungal stress. Int. J. Mol. Sci. 2020, 21, 2659. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Hu, J.; Wu, T.; Yang, Q.; Feng, X.; Lin, H.; Feng, S.; Cui, C.; Yu, Y.; Zhou, R.; et al. Comparative analysis of long noncoding RNAs in angiosperms and characterization of long noncoding RNAs in response to heat stress in Chinese cabbage. Hortic. Res. 2021, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liu, S.; Qi, H.; Cai, H.; Xu, M. Research progress on plant long non-coding RNA. Plants 2020, 9, 408. [Google Scholar] [CrossRef] [Green Version]
- Asensi-Fabado, M.A.; Amtmann, A.; Perrella, G. Plant responses to abiotic stress: The chromatin context of transcriptional regulation. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 106–122. [Google Scholar] [CrossRef] [Green Version]
- Akter, A.; Takahashi, S.; Deng, W.; Shea, D.J.; Itabashi, E.; Shimizu, M.; Miyaji, N.; Osabe, K.; Nishida, N.; Suzuki, Y.; et al. The histone modification H3 lysine 27 tri-methylation has conserved gene regulatory roles in the triplicated genome of Brassica rapa L. DNA Res. 2019, 26, 433–443. [Google Scholar] [CrossRef]
- Hung, F.Y.; Chen, C.; Yen, M.R.; Hsieh, J.W.A.; Chenlong, L.; Shih, Y.H.; Chen, F.F.; Chen, P.Y.; Cui, Y.; Wu, K. The expression of long non-coding RNAs is associated with H3Ac and H3K4me2 changes regulated by the HDA6-LDL1/2 histone modification complex in Arabidopsis. NAR Genom. Bioinfor. 2020, 2, lqaa066. [Google Scholar] [CrossRef]
- Mehraj, H.; Shea, D.J.; Takahashi, S.; Miyaji, N.; Akter, A.; Seki, M.; Dennis, E.S.; Fujimoto, R.; Osabe, K. Genome-wide analysis of long noncoding RNAs, 24-nt siRNAs, DNA methylation and H3K27me3 marks in Brassica rapa. PLoS ONE 2021, 16, e0242530. [Google Scholar]
- Zhu, Q.H.; Stephen, S.; Taylor, J.; Helliwell, C.A.; Wang, M.B. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 2014, 201, 574–584. [Google Scholar] [CrossRef]
- Shea, D.J.; Nishida, N.; Takada, S.; Itabashi, E.; Takahashi, S.; Akter, A.; Miyaji, N.; Osabe, K.; Mehraj, H.; Shimizu, M.; et al. Long noncoding RNAs in Brassica rapa L. following vernalization. Sci. Rep. 2019, 9, 9302. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, R.; Sasaki, T.; Nishio, T. Characterization of DNA methyltransferase genes in Brassica rapa. Genes Genet. Syst. 2006, 81, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finnegan, E.J.; Bond, D.M.; Buzas, D.M.; Goodrich, J.; Helliwell, C.A.; Tamada, Y.; Yun, J.Y.; Amasino, R.M.; Dennis, E.S. Polycomb proteins regulate the quantitative induction of VERNALIZATION INSENSITIVE 3 in response to low temperatures. Plant J. 2011, 65, 382–391. [Google Scholar] [CrossRef]
- Shea, D.J.; Shimizu, M.; Itabashi, E.; Miyaji, N.; Miyazaki, J.; Osabe, K.; Kaji, M.; Okazaki, K.; Fujimoto, R. Genome re-sequencing, SNP analysis, and genetic mapping of the parental lines of a commercial F1 hybrid cultivar of Chinese cabbage. Breed. Sci. 2018, 68, 375–380. [Google Scholar] [CrossRef]
- Liu, X.; Hao, L.; Li, D.; Zhu, L.; Hu, S. Long non-coding RNAs and their biological roles in plants. Genom. Proteom. Bioinform. 2015, 13, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.L.; Chekanova, J.A. Long noncoding RNAs in plants. Adv. Exp. Med. Biol. 2017, 1008, 133–154. [Google Scholar]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.C.; Kean, M.J.; Xu, J.; Chua, N.H. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014, 24, 444–453. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Niu, Q.W.; Wu, H.W.; Liu, J.; Ye, J.; Yu, N.; Chua, N.H. Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 2015, 84, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Dong, H.; Zhou, D.; Li, M.; Liu, Y.; Zhang, F.; Feng, Y.; Yu, D.; Lin, S.; Cao, J. Systematic identification of long non-coding RNAs during pollen development and fertilization in Brassica rapa. Plant J. 2018, 96, 203–222. [Google Scholar] [CrossRef] [Green Version]
- Deforges, J.; Reis, R.S.; Jacquet, P.; Sheppard, S.; Gadekar, V.P.; Hart-Smith, G.; Tanzer, A.; Hofacker, I.L.; Iseli, C.; Xenarios, I.; et al. Control of cognate sense mRNA translation by cis-Natural Antisense RNAs. Plant Physiol. 2019, 180, 305–322. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant noncoding RNAs: Hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, J.; Lian, B.; Gu, H.; Li, Y.; Qi, Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018, 9, 5056. [Google Scholar] [CrossRef] [Green Version]
- Reis, R.S.; Poirier, Y. Making sense of the natural antisense transcript puzzle. Trends Plant Sci. 2021, 26, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, M.; Gross-Hardt, R.; Schöffl, F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 2014, 85, 541–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Luan, Y.; Jiang, N.; Bao, H.; Meng, J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017, 89, 577–589. [Google Scholar] [CrossRef] [Green Version]
- Van Damme, M.; Huibers, R.P.; Elberse, J.; den Ackerveken, G.V. Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 2008, 54, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.; Liu, C.J.; Zhang, K. S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).