Abstract
Amorphophallus konjac is an economically important horticultural crop, valued for its corms as both a traditional medicine and a food source. The auxin transcription factor (ARF) family plays pivotal roles in auxin signaling and the processes of the morphogenesis of tissues and organs. However, the specific role of the ARF gene family in the regulation of corms remains unknown. In this study, we identified 23 AkARF genes unevenly distributed across 11 chromosomes in A. konjac. Phylogenetic analysis classified these proteins into four distinct clades, with members of the same group sharing conserved gene structures. Expression profiling demonstrated AkARF genes were expressed in a tissue-specific and spatio-temporal manner. Furthermore, lanthanum treatment significantly increased corm biomass and endogenous auxin levels (peak at 20 mg·L−1; p < 0.05). Transcriptome and qRT-PCR analyses revealed coordinated expression of AkARF6/13/14/16/20 with corm biomass accumulation. Pearson’s correlation analysis further confirmed positive correlations of AkARF6/13 with auxin content (p < 0.05). These results suggested the potential regulatory roles of AkARF6/13 in auxin-mediated corm development. This study provides the potential functional role of ARF-mediated growth and development of corms in A. konjac.
1. Introduction
Auxin is involved in many aspects of plant growth and development, which is perceived by auxin receptors such as transport inhibitor response 1 together with Aux/IAA proteins and auxin response factors (ARFs) []. When auxin is low or absent, Aux/IAAs interact with ARF transcription factors and repress gene transcription and auxin responses. In the presence of auxin, Aux/IAA proteins are degraded by ubiquitination, thus releasing the repression on ARFs and allowing transcriptional auxin responses []. ARF is the key transcription factor in auxin response and mediates the auxin signal transduction pathway []. Most ARF proteins are composed of three conserved domains, the B3 DNA binding domain (DBD) in the N-terminal region, the variable middle region (MR), and the C-terminal dimerization domain (CTD) that controls protein–protein interactions [,,,,]. ARFs fall into three deeply conserved evolutionary classes: A, B and C. In the model plant Arabidopsis thaliana, class A, which has an MR rich in glutamine, is constituted as an activator, which includes AtARFs 5–8 and AtARF19. Class C (AtARF10/16/17) and class B (involving the remaining AtARFs) have an MR rich in proline, serine and threonine are considered repressors [,,].
At present, with the development of sequencing technology, a large number of ARF proteins have been identified in a variety of plants, such as Arabidopsis thaliana [], Oryza sativa [], Cucurbita pepo L. [], sweet potato [] and so on. Moreover, the ARF gene family has been demonstrated to be involved in various developmental processes. AtARF2/3/4/5/7/10/16/19 were indicated to regulate root development [,,,], and AtARF1/2/6/8/17 were demonstrated to be involved in floral organ development [,,]. OsARF12/16/19 were involved in auxin-mediated root elongation, the phosphate starvation response and leaf angle change, respectively [,,]. SlARF5/7/8A/8B are involved in the regulation of fruit development in tomato []. In addition, ARFs have been found to play a crucial role in abiotic stress responses in different plant species. AcARF4/5/23a/28a were found to be significantly up-regulated after salt stress induction in kiwifruit []. The transgenic Arabidopsis thaliana lines with the CpARF22 gene enhanced their tolerance to salt stress and drought stress []. The diversity and complexity of the ARF gene family in different species have been extensively studied. However, there is still a lack of research on the function of ARF genes in Amorphophallus konjac.
A. konjac is a perennial herbaceous monocot belonging to the Amorphophallus genus, Araceae family [,], which has been used and cultivated as a traditional medicine and a food source for more than 2000 years in China [,]. Its corms are rich in konjac glucomannan, which has been widely used in agricultural, chemical, medical and industrial industries due to its strong water absorption, high swelling rate, good gelling and film-forming properties [,,,]. Nevertheless, the increasing market demand and rapid cultivation expansion have resulted in the yield reduction associated with monoculture practices and inefficient agricultural management []. Therefore, breeding stable productions is one of the main measures to solve these problems. The ARF family plays pivotal roles in auxin-mediated plant development and organ morphogenesis [,,,,]. To investigate their potential functions in A. konjac corm development, we identified the ARF genes of A. konjac and comprehensively analyzed the basic information of AkARF proteins, gene structure, cis-acting elements, and tissue-specific and spatio-temporal expression patterns. Furthermore, the endogenous auxin contents, corm biomass and differential expression under lanthanum induction have been analyzed to excavate the potential AkARF genes influencing the expansion of konjac corms. Our findings provide the foundation for an in-depth understanding of the function of ARF in corm development and lay the possibility for the cultivation and development of new varieties with high quality and high yield of A. konjac.
2. Materials and Methods
2.1. Identification and Physicochemical Properties of AkARF Gene Family
Genomic data for A. konjac, including the genome sequence file in fasta format and the corresponding gene structure annotation file in GFF3/GTF format, coding sequence (CDS) and amino acid sequence information, were obtained from NCBI under the accession number PRJNA734512 that was published by Gao et al. []. The Hidden Markov Model (HMM) profile of the ARF structural domain (PF06507) was downloaded from the Pfam Protein Family Database (http://pfam.xfam.org/, accessed on 1 May 2025) [], and the candidate AkARF gene (E-value < 1 × 10−10) was identified in ‘A. konjac’ using TBtools II (v2.310) software []. Furthermore, the physicochemical properties of the AkARF proteins, including the molecular weight, isoelectric point, instability index, aliphatic index and hydrophilic index, were predicted using the ProtParam tool on the ExPASy server (https://web.expasy.org/protparam/, accessed on 9 May 2025) []. Subcellular localization of the AkARF was predicted with the online network WoLF PSORT (https://www.genscript.com/wolf-psort.html, accessed on 9 May 2025).
2.2. Phylogenetic Relationships Among AkARF Family Genes
Amino acid sequences of the ARF homologs in Arabidopsis thaliana and Oryza sativa were downloaded from TAIR (https://www.arabidopsis.org/, accessed on 1 May 2025) and Ensembl (https://asia.ensembl.org/index.html, accessed on 1 May 2025), respectively. MEGA 11 software was used to perform multiple sequence alignment of ARF protein sequences of A. konjac, A. thaliana and O. sativa. Subsequently, the phylogenetic tree was constructed by the neighbor-joining method with 1000 bootstrap replicates to assess node support. The dendrogram was further visualized using ChiPlot (https://www.chiplot.online, accessed on 9 May 2025) [,].
2.3. Analysis of Conserved Structural Domains, Motifs, and Gene Structures
MEME online tool (https://meme-suite.org/, accessed on 9 May 2025) was employed to identify conserved motifs in AkARF proteins, with the maximum number of motifs set to 10. All other parameters were maintained at their default values []. Conserved structural domains were predicted via the Conserved Domain Database (CDD) at NCBI (https://www.ncbi.nlm.nih.gov/cdd/, accessed on 9 May 2025) using reverse position-specific BLAST (v3.21) with an E-value cutoff of 1 × 10−3. The gene structure, conserved domains and conserved motif were visualized by TBtools II [].
2.4. Analysis of Cis-Acting Elements in AkARF Promoters
The 2000 bp upstream sequence of AkARF gene family members was extracted from the A. konjac Genomics Data as the promoter sequence of AkARF genes, and the cis-regulatory elements in the promoter region were predicted using the online PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 1 May 2025) []. The predicted data were visualized using TBtools II [].
2.5. Chromosome Distribution, Collinearity Analysis and Evolutionary Analysis
The chromosome distribution information of the AkARF gene family was obtained from the A. konjac Genomics Data, and each gene was mapped to the corresponding chromosome using TBtools II software. Meanwhile, the intra-genomic and inter-genomic (Arabidopsis thaliana and Oryza sativa) collinearity relationship, gene duplication events were analyzed and visualized using the TBtools II. Additionally, the non-synonymous substitution rate (Ka), synonymous substitution rate (Ks) and Ka/Ks for each repeated ARF gene were calculated by the same software [].
2.6. Plant Materials and Lanthanum Treatment
One-year-old A. konjac corms used in the experiment were purchased from Qujing City, Yunnan Province, and authenticated by Professor Shuili Zhang from Zhejiang Traditional Chinese Medicine University as A. konjac. The similar-weight corms (approximately 100 g) were selected and planted with a planting density of 100 cm × 100 cm in the plantation of Zhejiang Traditional Chinese Medicine University (30°5′26″ N, 119°53′40″ E) in April 2022. The experimental site was characterized by a subtropical monsoon climate with loam soil (pH 6.5~7.5), and the field management (irrigation, insect and weed control) was implemented according to weather and local agronomic practices.
At the early stages of leaf expansion, when all compound leaf expansion diameters were approximately 20 cm, three concentrations of lanthanum (La) solutions (20, 80 and 160 mg L−1) were evenly sprayed on the leaves of A. konjac (50 mL/plant). The La (III) solutions were prepared with appropriate quantities of LaCl3·7H2O (Sigma-Aldrich Corporation, St. Louis, MO, USA) and distilled water. At the same time, the same amount of distilled water, namely La (III) was 0, which was used as the control (CK) group. After 7 days of La (III) treatment, the A. konjac corms were collected, with three biological replicates, and stored at −80 °C prior to RNA extraction.
2.7. Biomass of A. konjac Corms
The A. konjac corms were harvested after 60 days of La (III) treatment and were dried in an air oven at 80 °C until they reached a constant weight to obtain the biomass with ten biological replicates.
2.8. Detection of Auxin Content
Auxin content of the A. konjac corms was detected using Quantitative Detection Kit (ELISA) of Auxin (Ruixin Biotech Co., Quanzhou, China), with three biological replicates.
2.9. Gene Expression Profiles of AkARFs Based on RNA-Seq
Transcriptomic data for A. konjac were retrieved from the Integrated Medicinal Plantomics platform (IMP; https://www.bic.ac.cn/IMP, accessed on 22 March 2024) []. The expression level of AkARF in different parts and corm developmental periods was obtained by BLASTn alignment with the A. konjac RNA-seq data. Additionally, transcriptome sequencing of A. konjac samples, both untreated and treated with La (III), was conducted by Genepioneer Biotechnologies (Nanjing, China). Differential expression of AkARF genes under La (III) treatment was similarly analyzed via BLASTn alignment. Cluster analysis and heatmap visualization of expression patterns were performed using TBtools II software [].
2.10. RT-qPCR Analysis
The selected AkARF genes (Table S1), with Elongation factor-1 alpha (EF-1α) as the internal reference gene [], were randomly selected, designed with specific primers through PrimerQuest Tool (https://sg.idtdna.com/PrimerQuest/Home/Index, accessed on 10 May 2025), and synthesized at the Biotechnolo-gy Co., Shanghai, China, as shown in Table S1. Full-length cDNA was synthesized using the PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) kit (RR047A, Takara, Dalian, China). According to the procedure outlined, 0.5 μL of cDNA (2.5 ng μL−1), 5 μL of 2 × iTaq™ SYBR® Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), 0.75 μL of upstream and downstream primers, and 3 μL of ddH2O were mixed for the qRT-PCR detection using a CFX96 Real-Time PCR instrument (Bio-Rad). The qRT-PCR procedure was as follows: pre-denaturation at 95 °C for 3 min and 40 cycles of denaturation at 95 °C for 5 s and annealing at 60 °C for 30 s. The relative expression of each gene was calculated using the 2−ΔΔCT protocol. Three biological replicates were performed per sample.
2.11. Statistical Analysis
All the statistical analyses were performed using SPSS 25.0 (IBM, Armonk, NY, USA). The means among various groups were compared by Duncan’s multiple range tests. The data were analyzed and are expressed as the means and standard deviations (SD), and p < 0.05 indicated a significant difference.
3. Results
3.1. Identification of the AkARF Gene Family
A total of 23 putative AkARF genes were identified from A. konjac genome data and verified using the Pfam Database. These genes are named AkARF1 to AkARF23 according to their physical location on the chromosome (Table 1). The results of the bioinformatics analysis showed that the number of amino groups in the AkARF family members ranged from 225 to 1144 aa, and the molecular weight of the protein members is between 24.97 and 128.07 kD. The theoretical isoelectric point (pI) of the AkARF proteins is between 5.35 and 8.25, among which AkARF2/5/9/13/16/19 are alkaline proteins and the other 17 proteins are acidic proteins. The aliphatic index of AkARF is between 63.77 and 82.58. As shown in Table 1, all of the AkARF members are unstable proteins (instability index value > 40) and hydrophilic proteins (Grand Average of Hydropathicity < 0). The analysis of the AkARF proteins’ subcellular localization showed that AkARF2 may be localized to the cytosol, whereas other AkARF protein members are localized and expressed in the nucleus.

Table 1.
Basic analysis information of AkARF family.
The chromosomal localization of the AkARF gene was shown in Figure 1A. A total of 23 genes were mapped unevenly across the 11 chromosomes, of which chromosome 12 has 5 genes; chromosomes 2, 4 and 7 have 3 genes; and chromosomes 5 and 9 have 2 genes, respectively. Chromosomes 1, 3, 8, 10 and 13 each have one gene. Among them, there was a collinearity relationship between eight AkARF gene pairs, which all belong to the segmentally duplicated pairs, and the gene duplication events were mainly concentrated on chromosomes 5, 7 and 12 (Figure 1B). Furthermore, evolutionary selection pressure analysis revealed that all duplicated AkARF gene pairs exhibited Ka/Ks ratios < 1.00 (Table S2), consistent with findings reported for the BpeARF family in Betula pendula []. The results suggest purifying selection has acted on the AkARF gene family, implying functional conservation of these transcription factors.
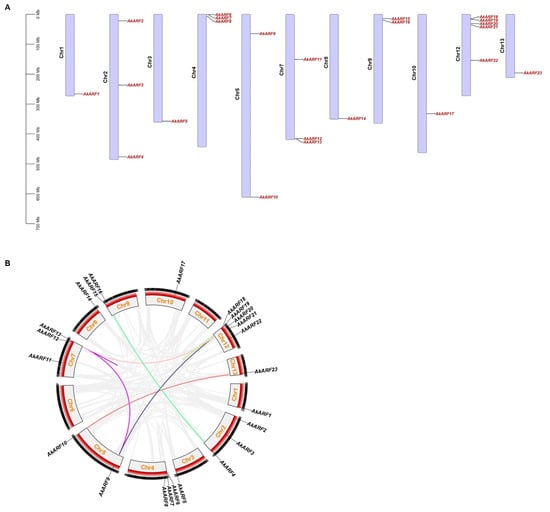
Figure 1.
Chromosome distribution and intro-genomic collinearity analysis of the AkARF gene family. (A) Chromosomal localization of AkARF genes; (B) intro-genomic collinearity analysis. The colored lines represent segmental repeat genes, and the gray line represents all the synteny blocks in the A. konjac genome.
3.2. Phylogenetic Analysis of the AkARF Gene Family
A phylogenetic tree was constructed using full-length ARF protein sequences from A. thaliana, O. sativa and A. konjac (Figure 2A). Phylogenetic analysis classified the ARF genes of A. konjac into four groups, similar to their orthologs in A. thaliana and O. sativa [,]. There are two genes (AkARF4/16) in group I, six genes (AkARF3/5/15/18/19/20) in group II, nine genes (AkARF1/6/7/8/10/14/17/22/23) in group III, and six genes (AkARF2/9/11/12/13/21) in group Ⅳ. AkARFs were in the same branch as OsARFs frequently. Moreover, we performed a collinearity analysis between A. konjac, A. thaliana and O. sativa (Figure 2B). The results showed that the ARFs of A. konjac and O. sativa had a high degree of homology; 14 pairs of orthologous genes were identified between A. konjac and O. sativa, while 3 pairs were found in A. thaliana. These results suggest that AkARF genes may functionally resemble their O. sativa orthologs more closely than those in A. thaliana, potentially reflecting shared evolutionary trajectories in monocot development.
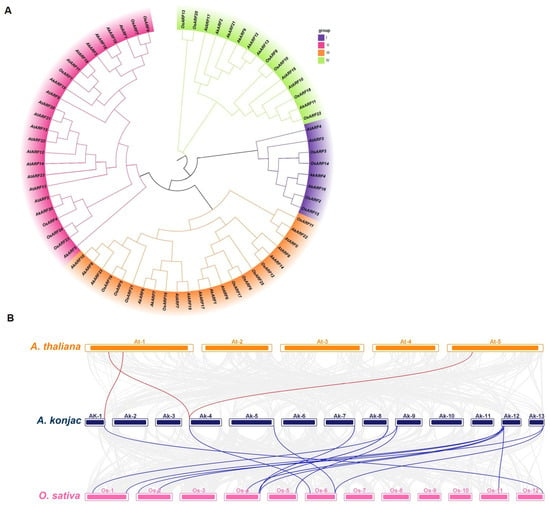
Figure 2.
Phylogenetic relationship and collinearity analysis of ARF between A. thaliana, O. sativa and A. konjac. (A) Phylogenetic tree among the three plant species. All ARF genes were classified into four classes with different colors. (B) Inter-genomic collinearity analysis. Collinear gene pairs are indicated by red lines (A. konjac-A. thaliana) and blue lines (A. konjac-O. sativa), while the gray lines represent homologous blocks in the A. konjac and other plant genomes.
3.3. Gene Structure and Protein Structure Analysis of the AkARF Gene Family
To better understand the crucial insights into gene evolution and function, the DNA and protein sequences of AkARF genes were analyzed. The motif composition analysis revealed that AkARF family proteins contained varying numbers of conserved motifs (ranging from 4 to 10), and all contained motifs 1, 4, 6 and 10. Except for AkARF2/9/19, other AkARF proteins follow the same motif order, which is motif 3, motif 8, motif 2, motif 1, motif 10, motif 4, motif 6 and motif 7 (Figure 3A). This structural homogeneity underscores the functional conservation of the AkARF family. Gene structure analysis showed that motif 5 is associated with the Aux_IAA superfamily domain (Figure 3B). Moreover, the number of exons ranged from 2 to 14 in the AkARF family, with 17 proteins having 10~14 exons. Notably, the AkARF17 encodes the longest protein. The other six proteins have 2~3 exons, and AkARF2 is the shortest protein (Figure 3C).
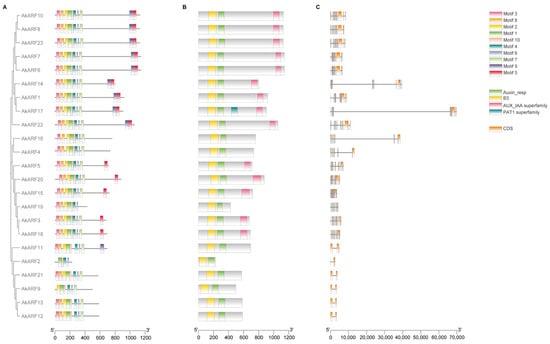
Figure 3.
Gene and protein structure of AkARFs. (A) Protein motifs. Different motifs were indicated by different numbers in various color boxes. (B) Protein conserved structural domains. DBD, MR, and CTD were marked in yellow, green, and pink, respectively. (C) Gene structure. Orange boxes represented CDS regions.
3.4. Regulatory Element Profiling of AkARF Gene Promoters
Cis-acting elements are transcription factor-specific binding sites, which play an important role in regulating genes responsible for the growth, differentiation, and development of plants. The 2.0 kb upstream region of the AkARF genes’ initiation transcription site was submitted to PlantCARE, and 26 response elements were identified (Figure 4, Table S3). Among them, seven hormone response elements include the auxin response element (TGA, AuxRR-core), gibberellin response element (GARE-motif, TATC-box and P-box), salicylic acid response element (TCA-element), methyl jasmone response element (CGTCA-motif and TGACG-motif), and abscisic acid response element (ABRE). It reflected the AkARF gene family’s comprehensive involvement in hormonal signaling pathways. All proteins contained light-responsive elements (Box II, G-box, GATA-motif, Sp1, etc.). Additionally, elements related to stress-related (low-temperature, defense and stress responsiveness) growth and development elements (meristem and endosperm expression, cell cycle regulation, etc.), MYB-binding elements and zein metabolism regulation. The results showed that the AkARF family’s versatile role in plant growth, development, and stress response mechanisms.
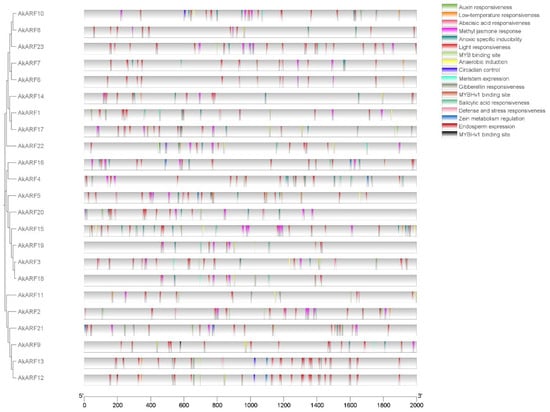
Figure 4.
Analysis of cis-acting elements in the AkARF gene families. Different cis-acting elements are represented by different color blocks.
3.5. Expression Analysis of AkARF Genes in Different Parts and Growth Stages
To explore the spatial and temporal expression patterns, we analyzed the expression of AkARFs at different tissues and stages using RNA-seq data of A. konjac from the IMP Database []. The results showed that AkARF7/8/10/18/20/21/23 were highly expressed in corm, while AkARF1/11/12/13/14/15/16/17/22 were highly expressed in flower. Notably, AkARF15 was only expressed in flower (Figure 5A). Interestingly, AkARF15 co-clustered with known floral regulators AtARF2 and OsARF6/12/17/25 [,] (Figure 2A), indicating its possible involvement in floral development. AkARFs gene families are expressed differently at different stages. 8 genes (AkARF1/2/17/18/15/20/22/23) showed the highest expression in the change head stage (CH) and gradually decreased as they matured. Seven genes (AkARF5/7/10/11/13/16/21) were higher in the corm expansion stage (CE), while AkARF8/12/14 were higher in the maturity stage (MA) (Figure 5B).
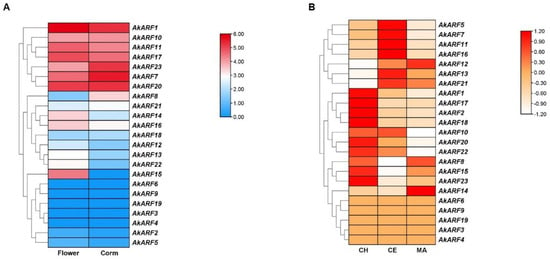
Figure 5.
Transcript profiles (gene expression) of AkARF genes in different parts and different growth stages of corms. (A) Expression of AkARF genes in flower and corm. Red and blue indicate high and low expression levels, respectively. (B) Expression levels of AkARF genes in growth stages of corm. Red and white indicate high and low expression levels, respectively. CH: change head stage, CE: corm expansion stage, MA: maturity stage.
3.6. The Corm Biomass, Auxin Content and AkARF Gene Expression Under La (III) Treatment
The application of La (III) (20–160 mg L−1) significantly promoted corm biomass accumulation (p < 0.05), consistent with our previous report []. Endogenous auxin levels were similarly elevated by La (III) exposure (p < 0.05), with both parameters reaching maximal values at 20 mg L−1 La (III) (Figure 6). Meanwhile, AkARF genes exhibited distinct expression patterns under La (III) exposure, AkARF2/3/4/7/8/15/18 were upregulated in CK but downregulated with La (III) contents increasing. AkARF6/13/14/16/20 were higher at La20, AkARF10/11/12/17/19/23 and AkARF1/5/9/21/22 peaked at La80 and La160, respectively (Figure 7A). RT-qPCR was employed to better understand the expression profiles of AkARF6/13/14/16/20, the result showed that the expression of the selected genes was consistent with those detected by RNA-Seq (Figure 7B). Additionally, Pearson’s correlation analysis showed that AkARF6/13 were correlated positively with endogenous auxin contents (Table S4).
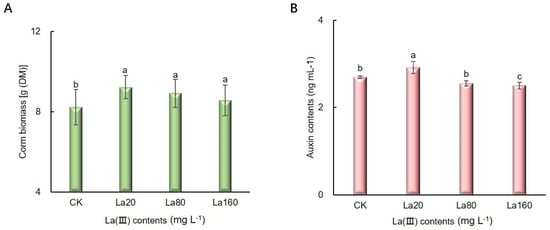
Figure 6.
The corm biomass and endogenous auxin contents of A. konjac under La (III) treatment. (A) corm biomass (mean ± SD, n = 10). (B) Endogenous auxin contents (mean ± SD, n = 3). Bars with different letters indicated significant differences (Duncan’s test, p < 0.05). CK: control group. La20: 20 mg·L−1 La (III) contents. La80: 80 mg·L−1 La (III) contents. La160: 160 mg·L−1 La (III) contents.
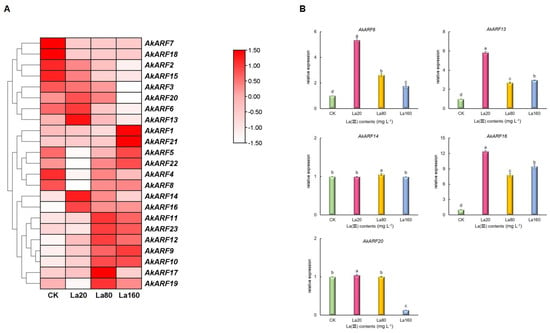
Figure 7.
The expression pattern of AkARF genes under La (III) treatment. (A) Transcript profiles of AkARF genes. Red and white indicate high and low expression levels, respectively. (B) Relative gene expression of AkARF6/13/14/16/20 by RT-qPCR (means ± SD, n = 3). Bars with different letters indicated significant differences (Duncan’s test, p < 0.05). CK: control group. La20: 20 mg·L−1 La (III) contents. La80: 80 mg·L−1 La (III) contents. La160: 160 mg·L−1 La (III) contents.
4. Discussion
Being the first identified phytohormone, auxin functions as a central signaling molecule that modulates transcriptional programs controlling cell division, elongation, differentiation, and organogenesis throughout the plant life cycle [,,,]. Auxin response factors (ARFs), as a class of transcription factors that interacted with Aux/IAAs to respond to auxin, are widely involved in plant growth and development and stress resistance regulation []. The number of ARF family genes varies among different plant species. The ARF gene was identified to have 23 members in A. thaliana [] and 25 members in O. sativa []. However, there is a lack of analysis and functional prediction of A. konjac genome-wide expression profiles. The genomic information of A. konjac chromosome levels published by Gao et al. (2022) [] provides an opportunity to systematically evaluate the function of the ARF gene family in this plant.
In this study, 23 AkARF genes were unevenly distributed on 13 chromosomes; the number of genes was similar to that of Arabidopsis thaliana. Subcellular localization predicts that 22 AkARF gene family members are located in the nucleus, indicating that AkARF plays a major role as a transcription factor in the nucleus. The results are consistent with most ARF family proteins reported in Cucurbita pepo L. [], implying that the subcellular localization of ARF proteins is relatively conservative. According to the phylogenetic analysis, the AkARF genes were divided into four groups, and inter-genomic collinearity analysis showed that ARF of A. konjac were more closely related to that of O. sativa, which indicated that they may encode proteins with the same biological function.
Gene duplication, including whole-genome duplication, tandem duplication, segmental duplication, proximal duplication, transpositional duplication, and dispersal duplication, with segmental and tandem duplication considered to be the major modes of gene family expansion [,]. In this study, there are seven gene pairs that have a collinearity relationship (AkARF4/16, AkARF18/19, AkARF10/23, AkARF12/21, AkARF12/13, AkARF9/21, and AkARF9/13), and all of them are segmental duplications. So we concluded that segmental duplication is the primary evolutionary mechanism contributing to AkARF’s expansion. The Ka/Ks ratios of collinear gene pairs were less than 1.00, indicating that the gene pairs of AkARF genes have experienced strong purification selection pressure during evolution and limited functional differentiation, and they are highly conserved throughout evolution.
Conservative protein and gene structure studies provided additional evidence for phylogenetic relationships. Most ARF proteins are composed of DBD-MR-CTDs [,]. The AUX_IAA superfamily domain belongs to the CTD, which mediates the interaction between ARF proteins and Aux/IAA proteins in the auxin signaling pathway [,]. Similar to PkorARF [], 17 proteins contain the typical DBD-ARF-CTDs. While all AkARFs of group Ⅳ (AkARF2/9/11/12/13/21) lack the CTD, namely the AUX_IAA superfamily domain (Figure 3B), further research is needed to determine the function of ARF with missing structural domains. Interestingly, motif 5 and the Aux_IAA superfamily domain always appear simultaneously (Figure 3A); it revealed that motif 5 may be associated with the function of the Aux_IAA superfamily domain. Additionally, the analysis of AkARF protein motifs showed that 20 AkARFs have the same eight motifs and order, indicating that the AkARF protein structure and function are relatively conserved. Moreover, phylogenetic analysis of ARF genes, in conjunction with comparative analysis of similar gene structures and conserved motifs within the same group, strongly supports the reliability of group classification.
Cis-acting elements regulate gene expression in plants by binding to transcription factors and controlling the timing and efficiency of gene transcription []. In this study, we identified regulatory elements in the AkARF gene promoter regions, including plant hormone-responsive elements, developmental elements, and stress-responsive elements. Similar to those of the ARF gene family in Dendrobium [], light responsiveness functions are the common element function of all AkARF. Methyl jasmone response elements and abscisic acid response elements were among the most prevalent types. Previous studies have also shown that the overexpression of ARF3 and ARF6 genes in bamboo increases 1H-Indole-3-acetic acid content that regulates lignin biosynthesis []. SlARF5 regulates the set and development of tomato fruit by modulating auxin and gibberellin signaling []. These studies confirmed that ARF transcription factors regulate plant growth and development by binding to auxin-responsive cis-acting elements and integrating multiple hormone signaling pathways. In addition, elements also related to MYB-binding elements (drought-inducibility, flavonoid biosynthetic genes regulation, etc.) and zein metabolism regulation (Figure 4, Table S2). MYB77 interacts with the ARF7 protein and results in a strong reduction in lateral root numbers in Arabidopsis [], which revealed that transcription factors can respond to auxin and regulate the transcriptional expression level of downstream AkARF genes by binding to its upstream promoter.
A comprehensive expression analysis in different organs revealed that many AkARF genes were expressed in a tissue-specific manner, suggesting they have different functions in the growth and development processes of A. konjac. AkARF15 showed the highest expression in the change head stage but gradually decreased as it matured and finally was only expressed in the flower (Figure 5), which implies its potential function in regulating reproductive development. Ellis et al. [] reported that arf2 mutant plants exhibited floral organ abscission and delayed flowering. Meanwhile, this phenomenon was enhanced by nph4 and arf19 mutations, but occurred independently of ethylene and cytokinin response pathways. Phylogenetic analysis showed that AkARF15 and AtARF2 clustered into the same branch; it is supposed that AkARF15 may have a similar function to AtARF2 in floral organs.
Lanthanum, La (III), one of the light rare earth elements [,], has been defined as a beneficial element since its effectiveness in promoting crop yield and improving quality [,]. It exhibits a hormone-like biphasic dose–response in plant growth and development, characterized by low-dose stimulation and high-dose inhibition []. Our preliminary research found that 20~160 mg L−1 La (III) increased the corm yield of A. sinensis []. In this study, to identify the key AkARFs regulating corm expansion, we analyzed AkARF expression patterns under La (III) treatment while monitoring endogenous auxin levels and corm biomass simultaneously. The results demonstrated a consistent pattern for both corm biomass and endogenous auxin content, showing an initial increase followed by a decrease in response to La (III) treatment, with peak values observed at 20 mg L−1 La (III) (Figure 6). Notably, the expression profiles of AkARF6/13/14/16/20 exhibited a similar dose-dependent response, reaching maximum expression levels at the same La (III) concentration (20 mg L−1). This coordinated expression pattern strongly paralleled the corm biomass changes under La (III) treatment. Furthermore, Pearson’s correlation analysis revealed significantly positive correlations between AkARF6/13 expression and endogenous auxin content (p < 0.05), suggesting their potential regulatory roles in auxin-mediated corm development.
The corm serves as a critical storage organ and harvested part of A. konjac, functioning as both a reservoir for water/nutrients during environmental stress and a protective structure for dormant meristems, while also facilitating reproduction []. Similarly to rhizomes and tubers [,], corm growth involves coordinated cellular processes (division/expansion) and physiological changes, including biomass accumulation and carbohydrate metabolism (particularly sucrose/starch conversion) [,]. Auxin plays a pivotal role in tuber metamorphosis and root development, where its concentration directly influences tuberous root thickening []. This aligns with our findings in A. konjac, where auxin levels positively correlate with corm biomass, mirroring previous observations in taro []. ARFs, as the one main auxin response transcription factor, participate in the processes of the morphogenesis of tissues and organs [,]. ARF5/9/17/23 were significantly up-regulated after treatment with 6-BA in LA. Lily ‘Aladdin’ bulbs []. ARF1 expression was significantly increased after potato tuber dormancy breaking from the tuber dormancy to sprouting; it revealed that ARF1 is associated with tuber dormancy and sprouting in potato []. Moreover, adventitious roots are the core structure for nutrient absorption and reproduction of corms. In adventitious root development, AtARF6/8 acted as a positive regulator, while AtARF17 functioned as a negative regulator, with both being post-transcriptionally controlled by miRNAs []. OsARF12 mediates auxin synthesis and transport via the auxin synthesis genes OsYUCCAs and auxin efflux carriers OsPINs and OsPGPs, regulating root elongation positively in rice []. AtARF10/16 are involved in the regulation of the WOX5 transcription and thus contribute to the auxin-promoted distal stem cell differentiation []. Additionally, studies on SlARF4 have demonstrated that ARFs regulate sugar content during tomato fruit development []. An OsARF18-OsARF2-OsSUT1–mediated auxin signaling cascade regulating carbohydrate partitioning between the source and sink tissues in rice reproductive organs was demonstrated by Zhao et al. []. Interestingly, ARF exhibits both autonomous functions and cooperative regulation through inter-family interactions. Although mutations in NPH4/ARF7 and ARF19 did not affect senescence of rosette leaves or floral organs on their own, they enhanced the effects of the arf2 mutation in floral organs []. In summary, ARFs showed functional diversity and complex networks in regulating organ morphogenesis; the specific function and mechanism of AkARF6/13 in corm growth need further verification and exploration.
5. Conclusions
In this study, we identified 23 AkARF genes unevenly distributed across 11 chromosomes in A. konjac. Phylogenetic analysis classified these proteins into four distinct clades, and the members in the same group exhibited similar gene structures. Cis-acting elements profiling indicated potential roles for AkARF family genes in plant hormone regulation. Expression analysis revealed tissue-specific and developmental stage-dependent patterns of AkARF. Moreover, lanthanum treatment significantly increased corm biomass and endogenous auxin levels (peak at 20 mg·L−1; p < 0.05). Integrated transcriptomic and qRT-PCR analyses demonstrated that AkARF6/13/14/16/20 exhibited expression patterns synchronized with corm expansion dynamics. Notably, AkARF6/13 showed significantly positive correlations with auxin levels (Pearson’s correlation, p < 0.05). These results can better analyze the function of the ARF gene in corm development and provide a theoretical basis for the genetic breeding of A. konjac.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11060687/s1, Table S1: Primer Sequences for qRT-PCR; Table S2. Ka, Ks and Ka/Ks of AkARF gene pairs; Table S3. Cis-acting elements of AkARF; Table S4. Pearson’s correlation analysis of endogenous auxin contents and the expression of AkARF6/13/14/16/20 under La (III).
Author Contributions
Conceptualization, X.L. and Z.Y.; methodology, X.L.; validation, J.L. and H.H.; resources, H.W.; data curation, X.L.; writing—original draft preparation, X.L.; writing—review and editing, X.L. and Z.Y.; visualization, X.L. and J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Zhejiang Provincial Natural Science Foundation of China (grant numbers ZCLQN25H2803 and LY23H280003) and the National Natural Science Foundation of China (grant number 32000257).
Data Availability Statement
All data generated or analyzed in this study are included in the main text and its Supplementary Materials.
Acknowledgments
We appreciate the experimental support from the Public Platform of Medical Research Center, Academy of Chinese Medical Science, Zhejiang Chinese Medical University.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| ARF | auxin response factor |
| A. konjac | Amorphophallus konjac |
| A. thaliana | Arabidopsis thaliana |
| O. sativa | Oryza sativa |
| La (III) | lanthanum |
References
- Xie, L.; Chen, F.; Du, H.; Zhang, X.; Wang, X.; Yao, G.; Xu, B. Graphene oxide and indole-3-acetic acid cotreatment regulates the root growth of Brassica napus L. via multiple phytohormone pathways. BMC Plant Biol. 2020, 20, 101. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Di, P.; Wang, Y. Genome-Wide Identification and Analysis of the Aux/IAA Gene Family in Panax ginseng: Evidence for the Role of PgIAA02 in Lateral Root Development. Int. J. Mol. Sci. 2024, 25, 3470. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Strader, L.C. AUXIN RESPONSE FACTOR protein accumulation and function. BioEssays 2023, 45, e2300018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.J.; Xue, Y.Y.; Xu, S.; Jin, X.R.; Man, X.C. Identification of ARF genes in Cucurbita pepo L. and analysis of expression patterns, and functional analysis of CpARF22 under drought, salt stress. BMC Genom. 2024, 25, 112. [Google Scholar] [CrossRef]
- Xu, Y.X.; Mao, J.; Chen, W.; Qian, T.T.; Liu, S.C.; Hao, W.J.; Li, C.-F.; Chen, L. Identification and expression profiling of the auxin response factors (ARFs) in the tea plant (Camellia sinensis (L.) O. Kuntze) under various abiotic stresses. Plant Physiol. Biochem. 2016, 98, 46–56. [Google Scholar] [CrossRef]
- Xian, F.; Liu, S.; Huang, J.; Xie, B.; Zhu, L.; Zhang, Q.; Lv, C.; Xu, Y.; Zhang, X.; Hu, J. The OsIAA3-OsARF16-OsBUL1 auxin signaling module regulates grain size in rice. Plant Physiol. 2025, 197, kiaf122. [Google Scholar] [CrossRef]
- Cancé, C.; Martin Arevalillo, R.; Boubekeur, K.; Dumas, R. Auxin response factors are keys to the many auxin doors. New Phytol. 2022, 235, 402–419. [Google Scholar] [CrossRef]
- Pratt, I.S.; Zhang, B. Genome-Wide Identification of ARF Transcription Factor Gene Family and Their Expression Analysis in Sweet Potato. Int. J. Mol. Sci. 2021, 22, 9391. [Google Scholar] [CrossRef]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional Genomic Analysis of theAUXIN RESPONSE FACTOR Gene Family Members in Arabidopsis thaliana: Unique and Overlapping Functions of ARF7 and ARF 19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and Their AUXIN RESPONSE FACTOR Targets Define an Autoregulatory Network Quantitatively Regulating Lateral Root Growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef] [PubMed]
- Schlereth, A.; Möller, B.; Liu, W.; Kientz, M.; Flipse, J.; Rademacher, E.H.; Schmid, M.; Jürgens, G.; Weijers, D. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 2010, 464, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 Regulate Lateral Root Formation via Direct Activation of LBD/ASL Genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Friml, J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2010, 107, 12046–12051. [Google Scholar] [CrossRef]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1andAUXIN RESPONSE FACTOR2regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef]
- Tabata, R.; Ikezaki, M.; Fujibe, T.; Aida, M.; Tian, C.E.; Ueno, Y.; Yamamoto, K.T.; Machida, Y.; Nakamura, K.; Ishiguro, S. Arabidopsis AUXIN RESPONSE FACTOR6 and 8 Regulate Jasmonic Acid Biosynthesis and Floral Organ Development via Repression of Class 1 KNOX Genes. Plant Cell Physiol. 2010, 51, 164–175. [Google Scholar] [CrossRef]
- Yang, J.; Tian, L.; Sun, M.X.; Huang, X.Y.; Zhu, J.; Guan, Y.F.; Jia, Q.S.; Yang, Z.N. AUXIN RESPONSE FACTOR17 Is Essential for Pollen Wall Pattern Formation in Arabidopsis. Plant Physiol. 2013, 162, 720–731. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, S.; Shen, C.; Zhang, S.; Chen, Y.; Xu, Y.; Liu, Y.; Wu, Y.; Jiang, D. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 2011, 193, 109–120. [Google Scholar] [CrossRef]
- Shen, C.; Wang, S.; Zhang, S.; Xu, Y.; Qian, Q.; Qi, Y.; Jiang, D.A. OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 2013, 36, 607–620. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, S.; Xu, Y.; Yu, C.; Shen, C.; Qian, Q.; Geisler, M.; Jiang, D.A.; Qi, Y. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2014, 38, 638–654. [Google Scholar] [CrossRef]
- Hu, J.; Li, X.; Sun, T.P. Four class A AUXIN RESPONSE FACTORs promote tomato fruit growth despite suppressing fruit set. Nat. Plants 2023, 9, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Xu, M.; Zhang, J.; Wang, Y.; Lei, Y.; Li, Q. Genome-wide identification of auxin response factor (ARF) family in kiwifruit (Actinidia chinensis) and analysis of their inducible involvements in abiotic stresses. Physiol. Mol. Biol. Plants 2021, 27, 1261–1276. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, R.D.; Reddy, C.K.; Xu, B. Health-promoting effects of konjac glucomannan and its practical applications: A critical review. Int. J. Biol. Macromol. 2019, 126, 273–281. [Google Scholar] [CrossRef]
- Xue, Z.; Huang, F.; Liu, J.; Ke, Y.; Wei, H.; Gao, P.; Qi, Y.; Yu, L. A high trans-zeatin nucleoside concentration in corms may promote the multileaf growth of Amorphophallus muelleri. Front. Plant Sci. 2022, 13, 964003. [Google Scholar] [CrossRef] [PubMed]
- Pouchon, C.; Gauthier, J.; Pitteloud, C.; Claudel, C.; Alvarez, N. Phylogenomic study of Amorphophallus (Alismatales; Araceae): When plastid DNA gene sequences help to resolve the backbone subgeneric delineation. J. Syst. Evol. 2023, 61, 64–79. [Google Scholar] [CrossRef]
- Shan, Y.; Li, J.; Zhang, X.; Yu, J. The complete mitochondrial genome of Amorphophallus albus and development of molecular markers for five Amorphophallus species based on mitochondrial DNA. Front. Plant Sci. 2023, 14, 1180417. [Google Scholar] [CrossRef]
- Basak, S.; Singhal, R.S. Composite hydrogels fabricated from konjac glucomannan and gellan gum: Rheological characterization and their potential application in sustainable agriculture. Carbohydr. Polym. 2024, 336, 122091. [Google Scholar] [CrossRef]
- Hong, J.; Shi, Y.; Chen, J.; Mi, M.; Ren, Q.; Zhang, Y.; Shen, M.; Bu, J.; Kang, Y. Konjac glucomannan attenuate high-fat diet-fed obesity through enhancing β-adrenergic-mediated thermogenesis in inguinal white adipose tissue in mice. Glycoconj. J. 2023, 40, 575–586. [Google Scholar] [CrossRef]
- Bu, N.; Zhou, N.; Cao, G.; Mu, R.; Pang, J.; Ma, C.; Wang, L. Konjac glucomannan/carboxymethyl chitosan film embedding gliadin/casein nanoparticles for grape preservation. Int. J. Biol. Macromol. 2023, 249, 126131. [Google Scholar] [CrossRef]
- Zhang, Y.; Aldamarany, W.A.S.; Deng, L.; Zhong, G. Carbohydrate supplementation retains intestinal barrier and ameliorates bacterial translocation in an antibiotic-induced mouse model. Food Funct. 2023, 14, 8186–8200. [Google Scholar] [CrossRef]
- Gao, Z.; Liu, F.; Lan, H.; Jian, T.; Cao, L.; Deng, M.; Wang, L.; Lan, M.; Li, J. Response and mechanisms of Amorphophallus konjac agronomic traits and disease occurrence after biochar application. Sci. Hortic. 2024, 338, 113657. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, Y.; Feng, C.; Chu, H.; Feng, C.; Wang, H.; Wu, L.; Yin, S.; Liu, C.; Chen, H.; et al. A chromosome-level genome assembly of Amorphophallus konjac provides insights into konjac glucomannan biosynthesis. Comput. Struct. Biotechnol. J. 2022, 20, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Gasteiger, E. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S.; Battistuzzi, F.U. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, T.; Yang, M.; Cui, G.; Tang, J.; Shen, Y.; Liu, J.; Yuan, Y.; Guo, J.; Huang, L. IMP: Bridging the gap for medicinal plant genomics. Nucleic Acids Res. 2024, 52, D1347–D1354. [Google Scholar] [CrossRef]
- Wang, K.; Niu, Y.; Wang, Q.; Liu, H.; Jin, Y.; Zhang, S. Cloning and evaluation of reference genes for quantitative real-time PCR analysis in Amorphophallus. PeerJ. 2017, 5, e3260. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Jin, X.; Lv, S.; Long, S.; Liu, Y.; Chen, L.; Lin, L. Genome-Wide Identification and Expression Analysis of Auxin Response Factor. (ARF) Gene Family in Betula pendula. Horticulturae 2024, 10, 27. [Google Scholar] [CrossRef]
- Zhao, Z.X.; Yin, X.X.; Li, S.; Peng, Y.T.; Yan, X.L.; Chen, C.; Hassan, B.; Zhou, S.X.; Pu, M.; Zhao, J.H.; et al. miR167d-ARFs Module Regulates Flower Opening and Stigma Size in Rice. Rice 2022, 15, 40. [Google Scholar] [CrossRef]
- Li, X.X.; Yu, B.; Dong, Y.Y.; Wang, L.S.; Zhang, S.L.; Shangguan, H.Y.; He, Z.H.; Luo, X.M.; Lai, P.F. Lanthanum chloride enhances the photosynthetic characteristics and increases konjac glucomannan contents in Amorphophallus sinensis Belval. Photosynthetica 2020, 58, 165–173. [Google Scholar] [CrossRef]
- Alonso, M.M.P. In-Depth Analysis of Jasmonate-Mediated Indole-3-Acetic Acid (iaa) Biosynthesis and its Implication in Plant Development. Ph.D. Thesis, Universidad Politécnica De Madrid, Madrid, Spain, 2017. Available online: https://oa.upm.es/47133/ (accessed on 15 May 2025).
- Yu, Y.; Tang, W.; Lin, W.; Li, W.; Zhou, X.; Li, Y.; Chen, R.; Zheng, R.; Qin, G.; Cao, W.; et al. ABLs and TMKs are co-receptors for extracellular auxin. Cell 2023, 186, 5457–5471. [Google Scholar] [CrossRef]
- Qian, S.; Zhang, Q.; Li, S.; Shi, R.; He, X.; Zi, S.; Liu, T. Arbuscular mycorrhiza and plant growth promoting endophytes facilitates accumulation of saponin under moderate drought stress. Chin. Herb. Med. 2024, 16, 214–226. [Google Scholar] [CrossRef]
- Feng, L.; Chen, H.; Zhao, J.; Liu, D.; Wei, Z.; Li, Y.; Yang, B.; He, Y.; Zhang, M.; Hou, D.; et al. Auxin induces lateral root formation in Bupleurum: A heme oxygenase dependent approach. Chin. Herb. Med. 2023, 15, 57–62. [Google Scholar] [CrossRef]
- Pernisová, M.; Vernoux, T. Auxin Does the SAMba: Auxin Signaling in the Shoot Apical Meristem. Cold Spring Harb. Perspect. Biol. 2021, 13, a039925. [Google Scholar] [CrossRef]
- Cheng, T.; Li, M.; Zhao, C.; Wang, T.; Zheng, X.; Yang, L.; Diao, Y.; Yang, S.; Hu, Z. Genome-wide identification, expression profile and selection analysis of the CPK gene family in Nelumbo nucifera. BMC Genomics 2025, 26, 461. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, W.; Shen, H.; Yang, L. Genome-wide identification and expression analysis of ARF gene family in embryonic development of Korean pine (Pinus koraiensis). BMC Plant Biol. 2024, 24, 267. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, W.; Ke, S.; Chen, D.; Wang, L.; Zheng, Q.; Huang, Y.; Liu, Z.J.; Yin, W.; Lan, S. Genome-Wide Identification of the ARF Gene Family in Three Dendrobium Species and Its Expression Pattern Analysis in D. nobile Flower. Horticulturae 2024, 10, 568. [Google Scholar] [CrossRef]
- Wang, S.; Hagen, G.; Guilfoyle, T.J. ARF-Aux/IAA interactions through domain III/IV are not strictly required for auxin-responsive gene expression. Plant Signal Behav. 2013, 8, e24526. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, Y.; Cai, C.; Zhu, Q. Auxin response factors fine-tune lignin biosynthesis in response to mechanical bending in bamboo. New Phytol. 2023, 241, 1161–1176. [Google Scholar] [CrossRef]
- Liu, N.; Dong, L.; Deng, X.; Liu, D.; Liu, Y.; Li, M.; Hu, Y.; Yan, Y. Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L. BMC Plant Biol. 2018, 18, 336. [Google Scholar] [CrossRef]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis Transcription Factor MYB77 Modulates Auxin Signal Transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef]
- He, C.; Feng, Y.; Deng, Y.; Lin, L.; Cheng, S. A systematic review and meta-analysis on the root effects and toxic mechanisms of rare earth elements. Chemosphere 2024, 363, 142951. [Google Scholar] [CrossRef]
- Trejo-Téllez, L.I.; Gómez-Merino, F.C. Editorial: Beneficial elements: Novel players in plant biology for innovative crop production, volume II. Front. Plant Sci. 2023, 14, 1303462. [Google Scholar] [CrossRef]
- Syrvatka, V.; Rabets, A.; Gromyko, O.; Luzhetskyy, A.; Fedorenko, V. Scandium–microorganism interactions in new biotechnologies. Trends Biotechnol. 2022, 40, 1088–1101. [Google Scholar] [CrossRef]
- Khosa, J.; Bellinazzo, F.; Kamenetsky Goldstein, R.; Macknight, R.; Immink, R.G.H. PHOSPHATIDYLETHANOLAMINE-BINDING PROTEINS: The conductors of dual reproduction in plants with vegetative storage organs. J. Exp. Bot. 2021, 72, 2845–2856. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, N.; Wen, Y.; Si, H.; Wang, D. Identification of differentially expressed genes in potato associated with tuber dormancy release. Mol. Biol. Rep. 2012, 39, 11277–11287. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.X.; Cheng, L.; Li, S.; Yin, J.; Li, L.; Chen, X. Genome-Wide Analysis of Differentially Expressed Genes Relevant to Rhizome Formation in Lotus Root (Nelumbo nucifera Gaertn). PLoS ONE. 2013, 8, e67116. [Google Scholar] [CrossRef]
- Miao, Y.; Zhu, Z.; Guo, Q.; Yang, X.; Liu, L.; Sun, Y.; Wang, C. Dynamic changes in carbohydrate metabolism and endogenous hormones during Tulipa edulis stolon development into a new bulb. J. Plant Biol. 2016, 59, 121–132. [Google Scholar] [CrossRef]
- Pallotti, C.; Renau-Morata, B.; Cardone, L.; Nebauer, S.G.; Albiñana Palacios, M.; Rivas-Sendra, A.; Seguí-Simarro, J.M.; Molina, R.V. Understanding the Saffron Corm Development—Insights into Histological and Metabolic Aspects. Plants 2024, 13, 1125. [Google Scholar] [CrossRef]
- Zhang, E.; Shen, W.; Jiang, W.; Li, W.; Wan, X.; Yu, X.; Xiong, F. Research progress on the bulb expansion and starch enrichment in taro (Colocasia esculenta (L). Schott). PeerJ. 2023, 11, e15400. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, B.; Liu, X.; Shan, N.; Sun, J.; Zhang, H.; Huang, Y.; Xiao, Y.; Zhou, Q. Uncovering the mechanism preliminarily of formation and development of taro corm in vitro by morphological physiology and transcriptomic analysis. Sci. Hortic. 2022, 291, 110575. [Google Scholar] [CrossRef]
- Sun, Q.; Zhang, B.; Xiang, L.; Wang, Y.; Chan, Z. Auxin Receptor TRANSPORT INHIBITOR RESPONSE1 Promotes Tulip Plant Growth and Bulb Swelling. J. Plant Growth Regul. 2024, 43, 4691–4703. [Google Scholar] [CrossRef]
- Zhang, K.; Lyu, T.; Lyu, Y. Transcriptional Insights into Lily Stem Bulblet Formation: Hormonal Regulation, Sugar Metabolism, and Transcriptional Networks in LA Lily ‘Aladdin’. Horticulturae 2024, 10, 171. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mongelard, G.; Floková, K.; Pacurar, D.I.; Novák, O.; Staswick, P.; Kowalczyk, M.; Pacurar, M.; Demailly, H.; Geiss, G.; et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef]
- Sagar, M.; Chervin, C.; Mila, I.; Hao, Y.; Roustan, J.P.; Benichou, M.; Gibon, Y.; Biais, B.; Maury, P.; Latché, A.; et al. SlARF4, an Auxin Response Factor Involved in the Control of Sugar Metabolism during Tomato Fruit Development. Plant Physiol. 2013, 161, 1362–1374. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, C.; Yu, X.; Tian, Y.; Wang, W.; Zhang, Y.; Bai, W.; Yang, N.; Zhang, T.; Zheng, H.; et al. Auxin regulates source-sink carbohydrate partitioning and reproductive organ development in rice. Proc. Natl. Acad. Sci. USA 2022, 119, e2121671119. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).