Molecular Responses to Drought and Waterlogging Stresses of Kiwifruit (Actinidia chinensis var. deliciosa) Potted Vines
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatments
2.2. Plant Physiological Measurements
2.3. RNA Isolation and Sequencing
3. Results
3.1. Physiological Response to Drought and Waterlogging
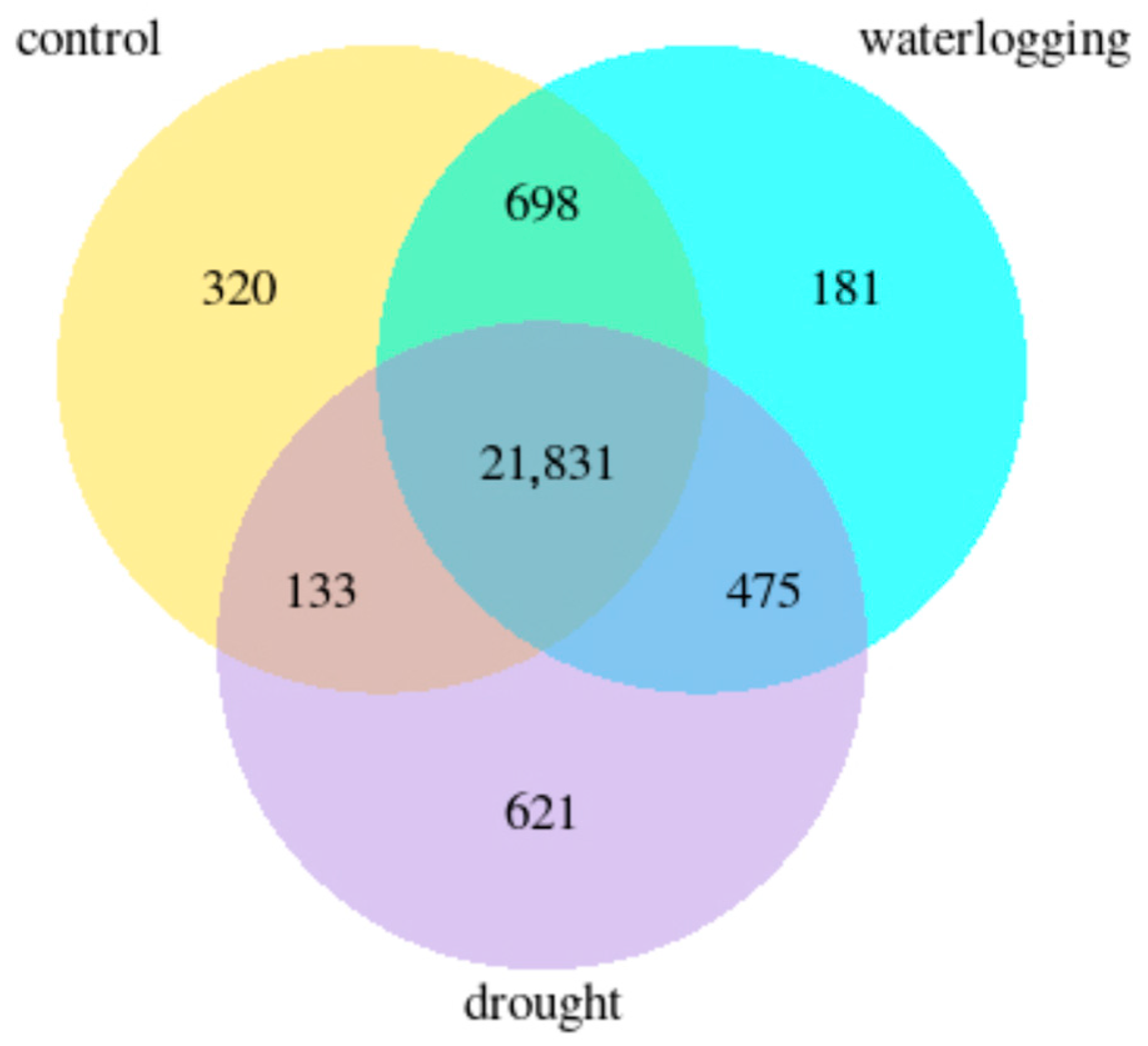
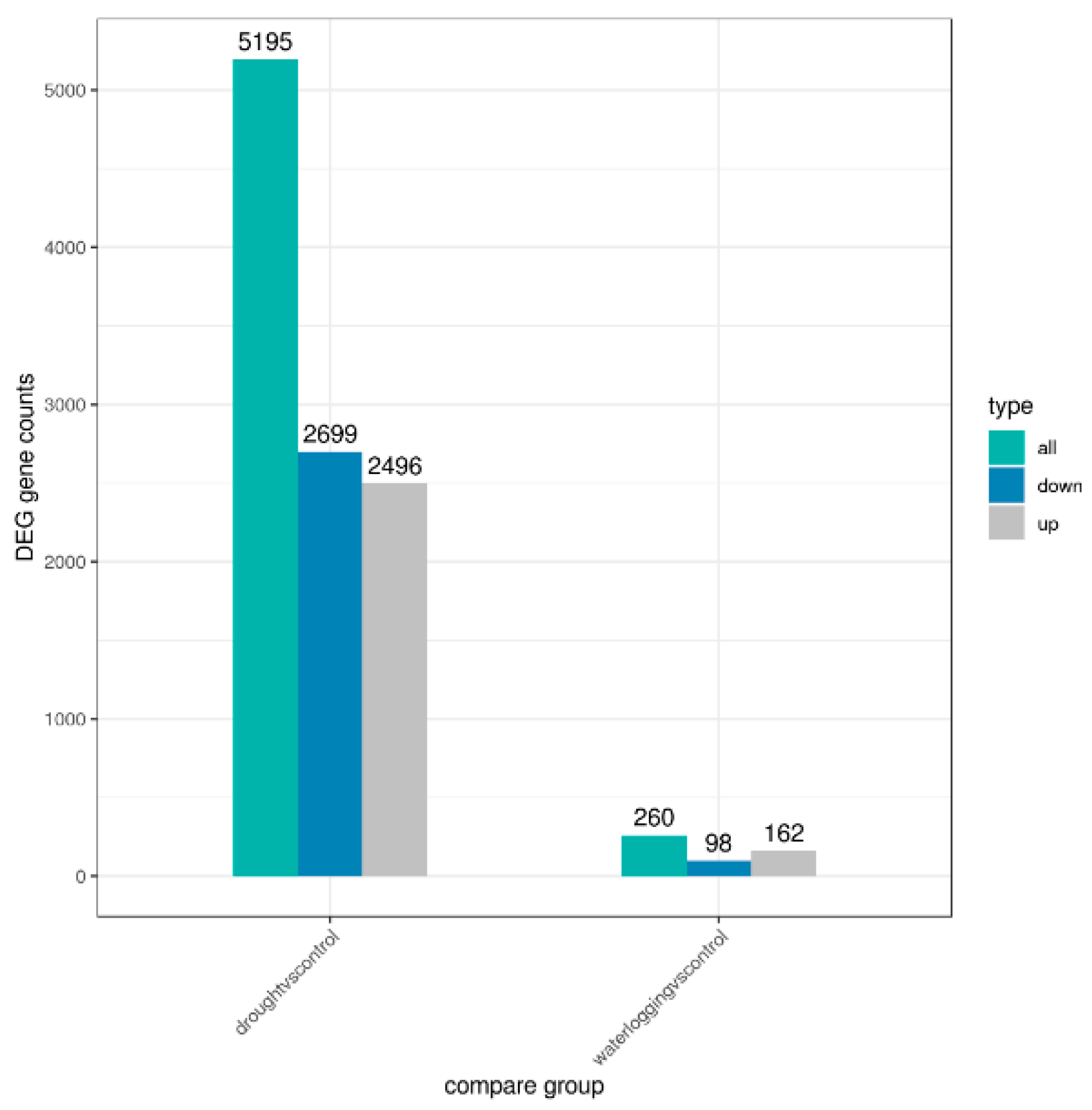
3.2. Overview of Sequencing Data and Statistical Summary of DEGs
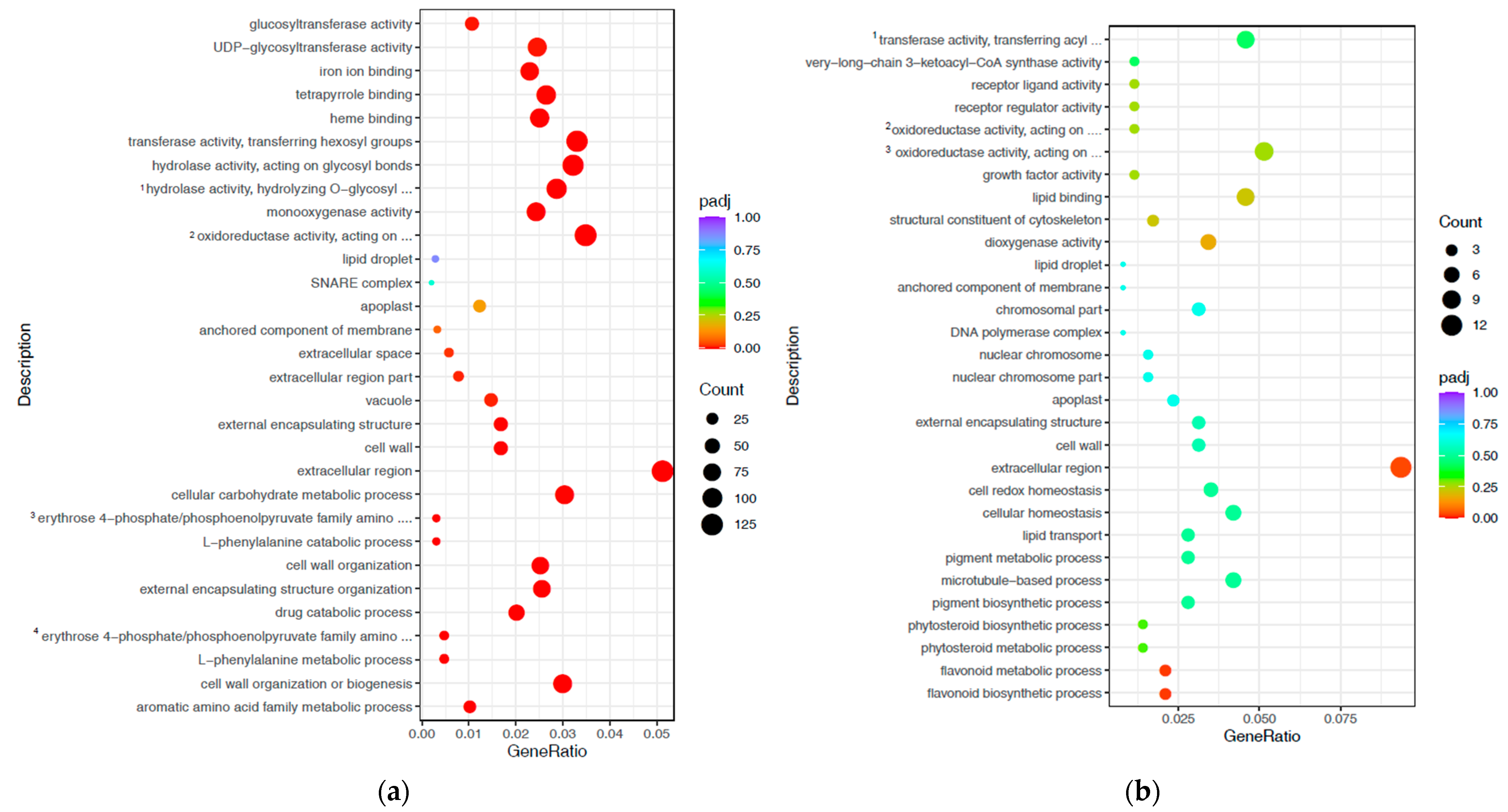
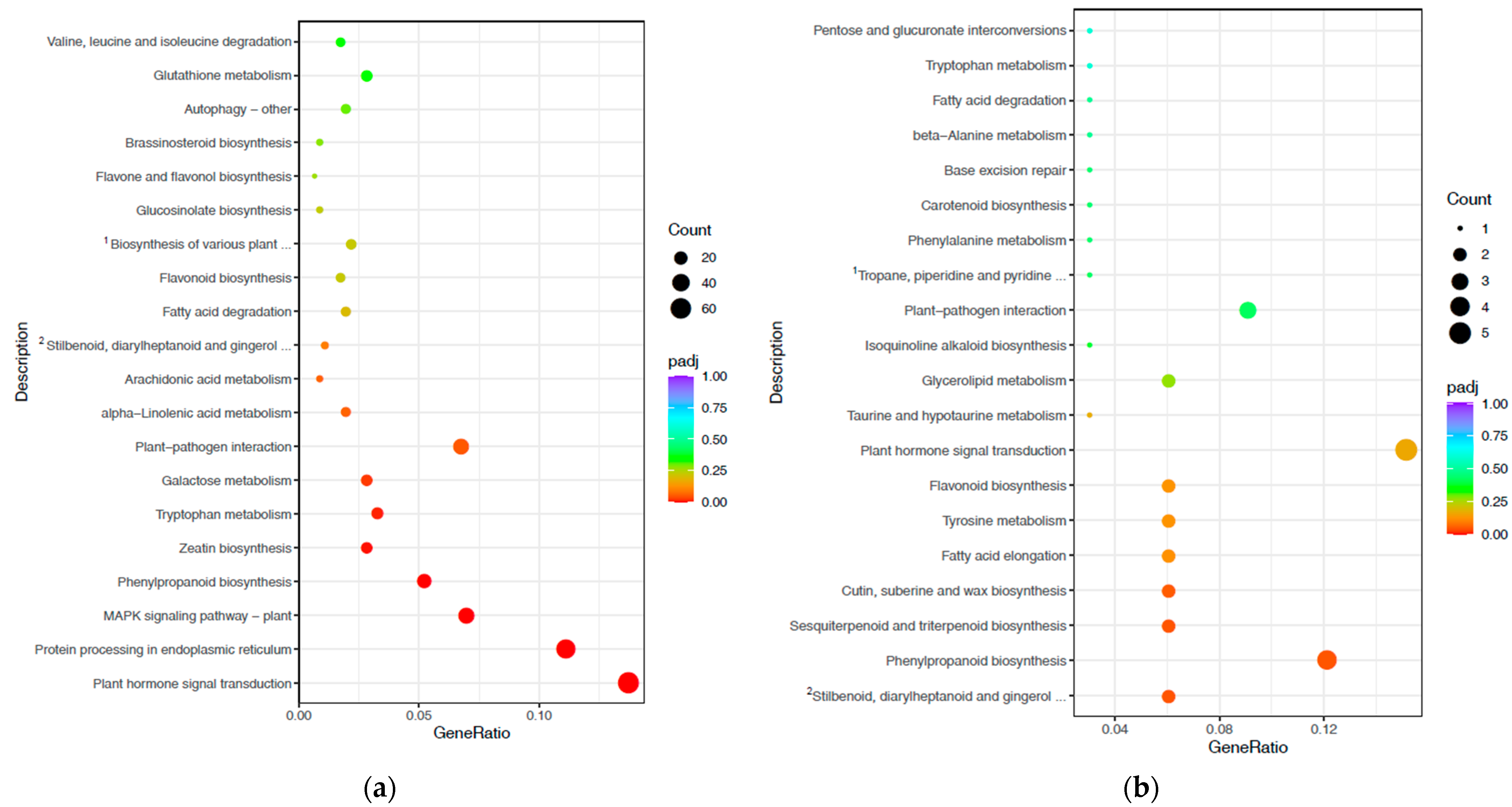
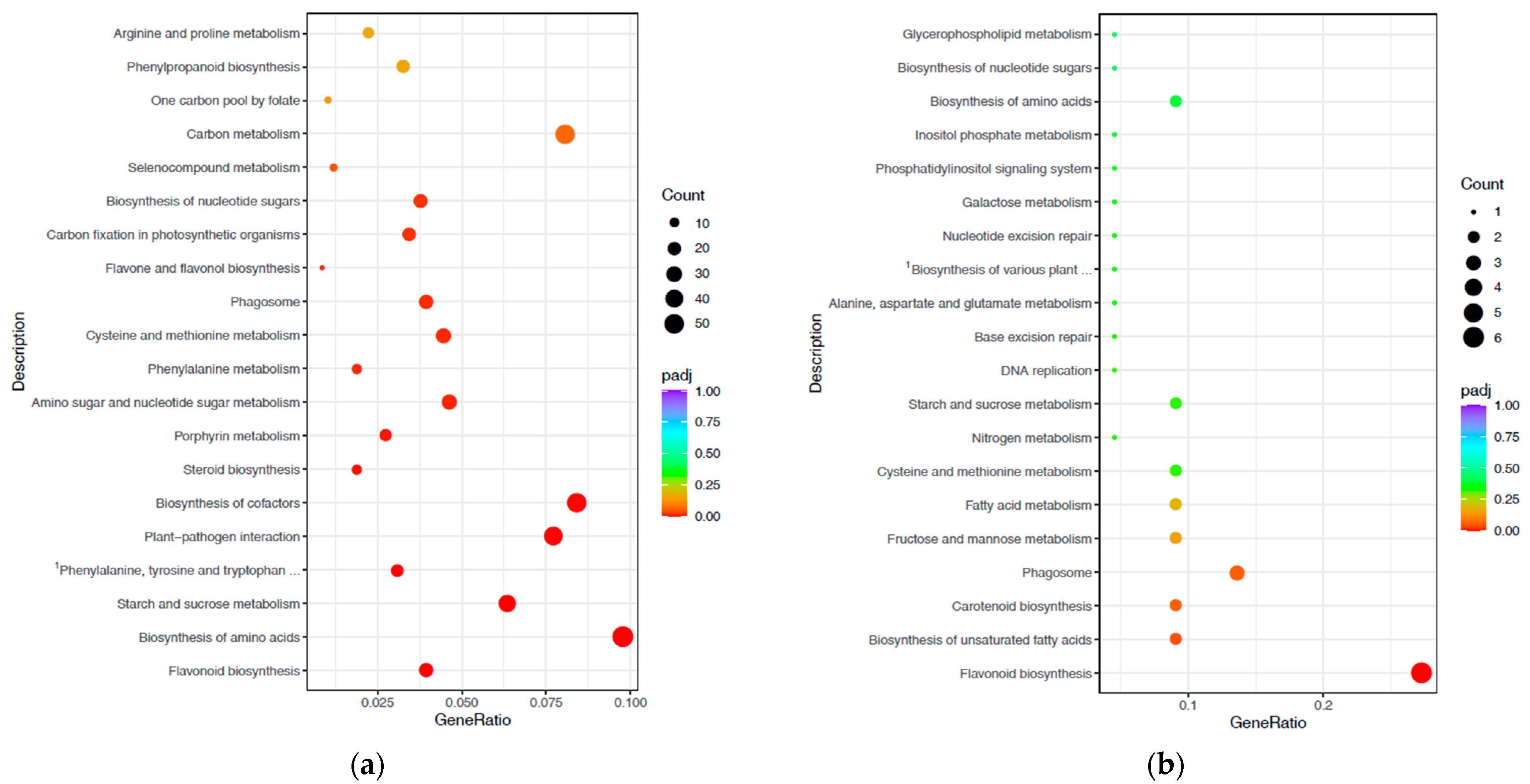
3.3. Gene Ontology and Kyoto Encyclopaedia of Genes and Genome-Enrichment Analyses
3.4. Pathways Involved in Drought and Waterlogging Stress Response
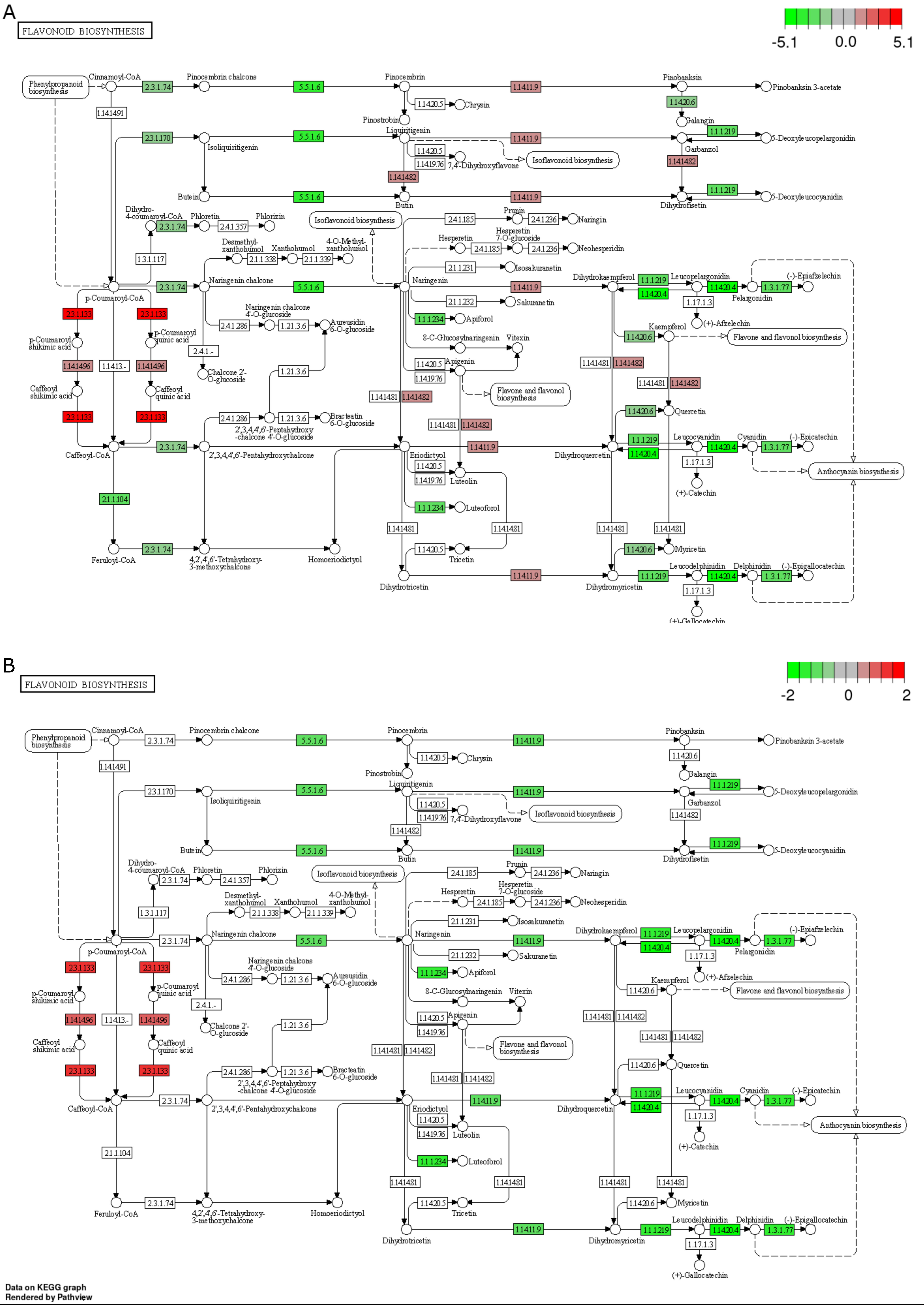
3.4.1. Effect of Drought and Waterlogging on Genes Involved in Secondary Metabolites Pathways
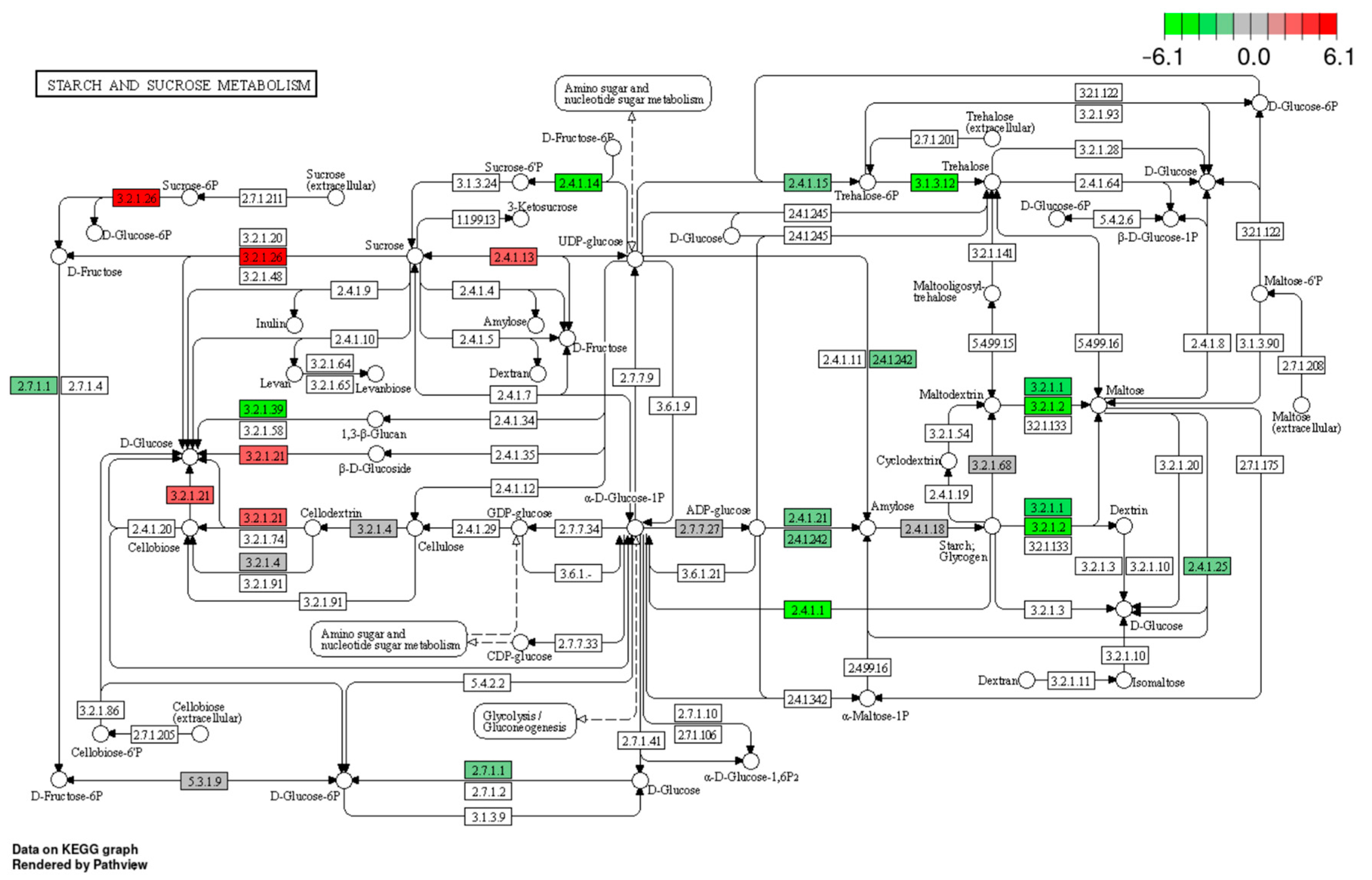
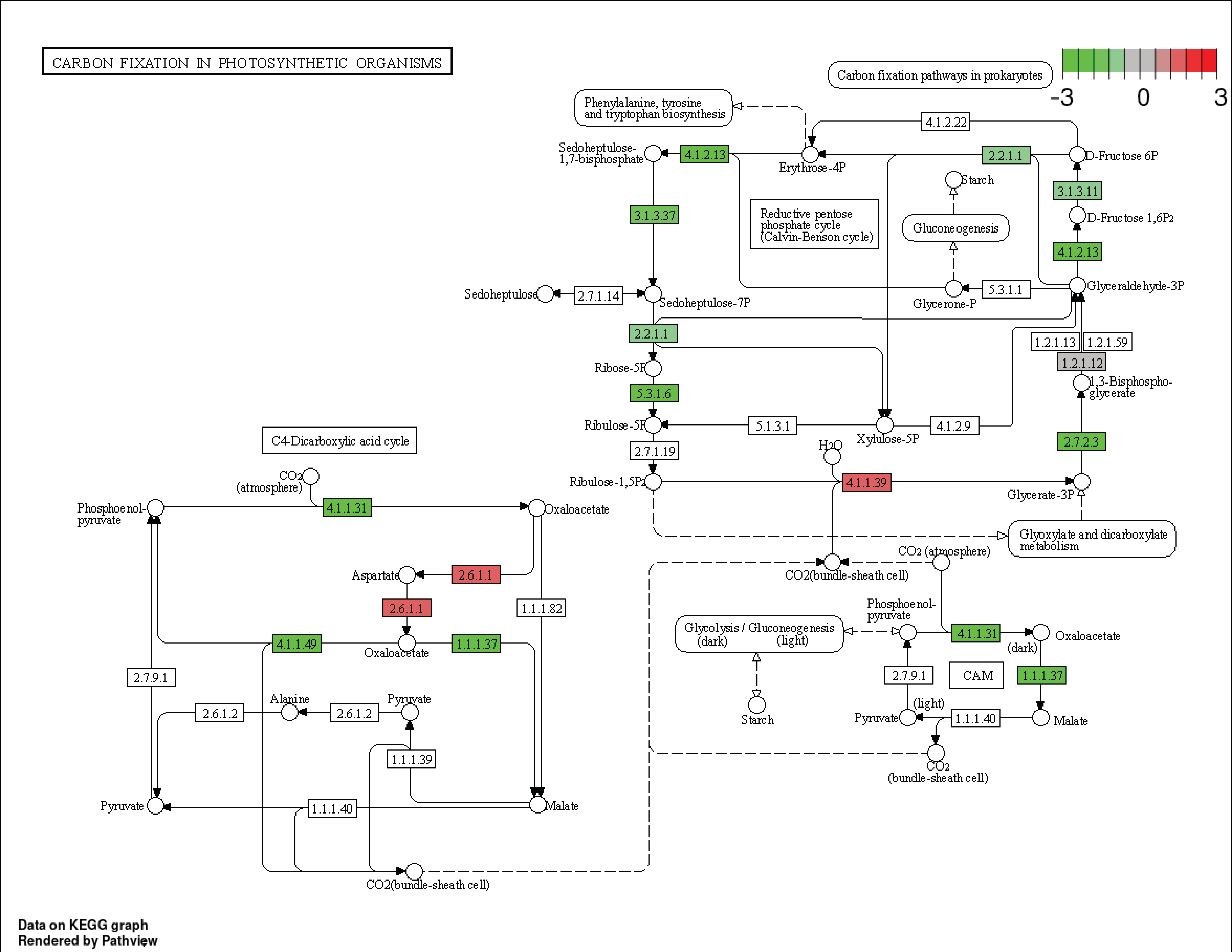
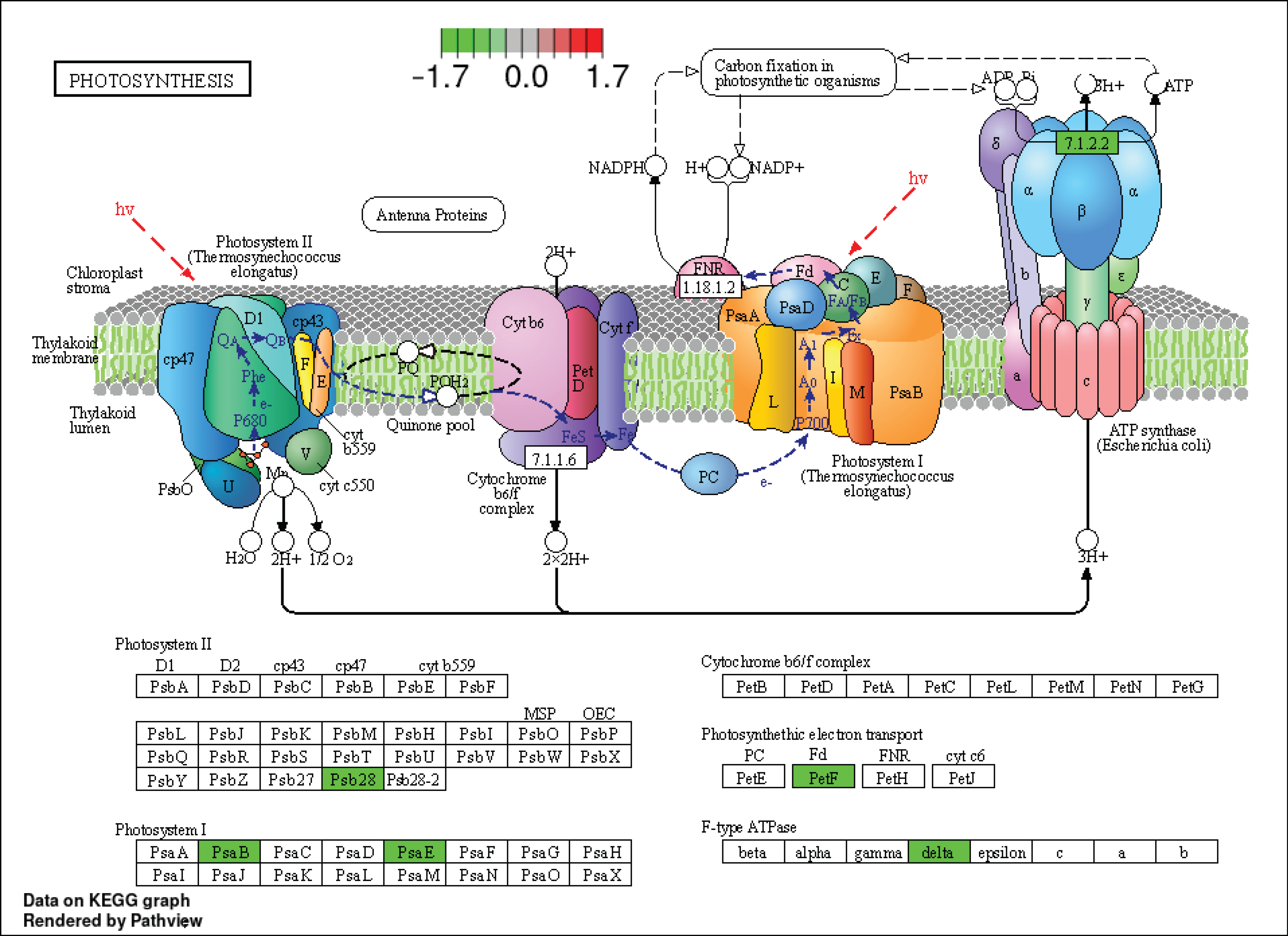
3.4.2. Effect of Drought and Waterlogging on Genes Involved in Carbon and Sugar Metabolism, Carbon Fixation, and Photosynthesis
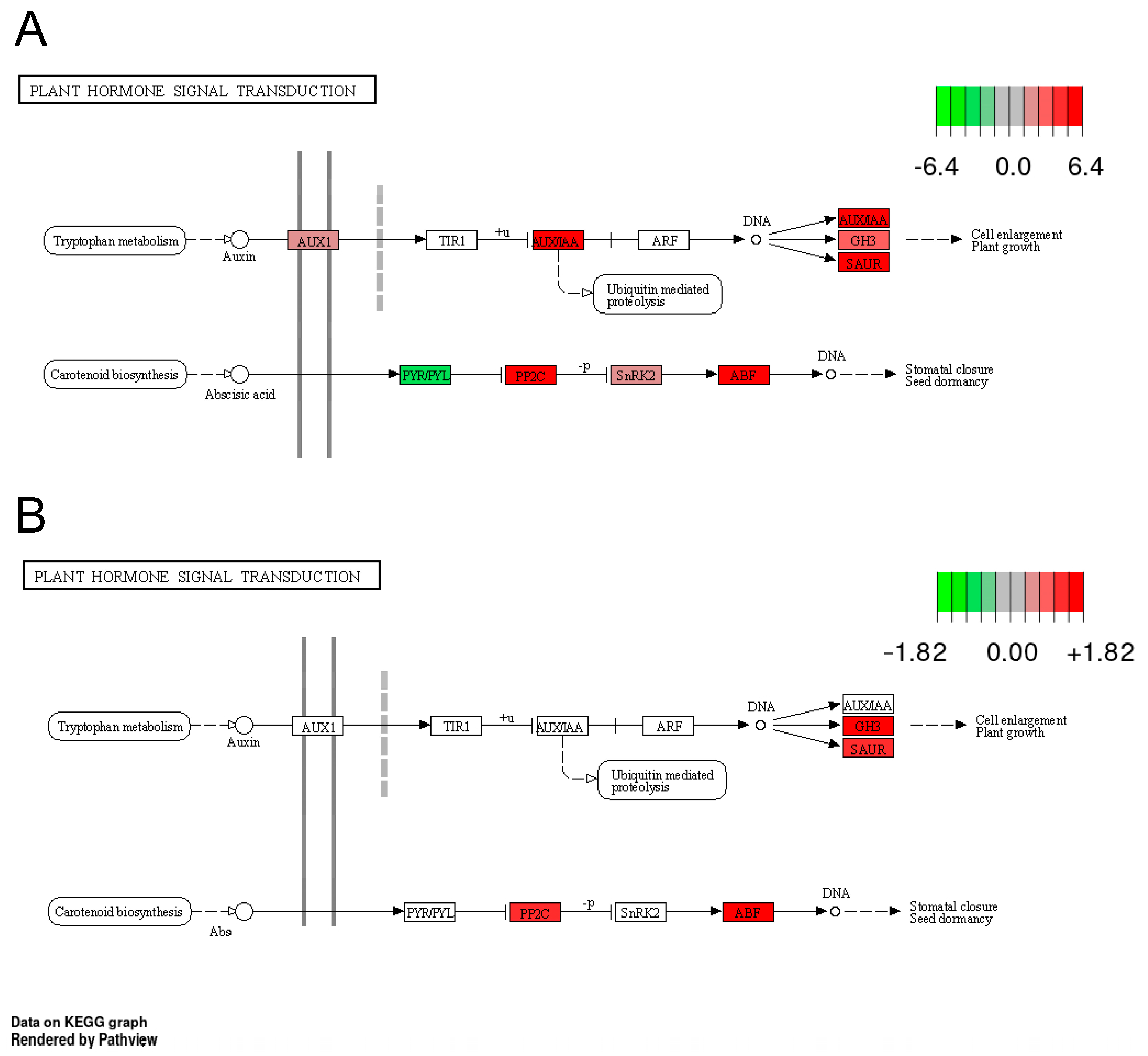
3.4.3. Effect of Drought and Waterlogging on Genes Involved in Hormone Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zandalinas, S.I.; Sengupta, S.; Fritschi, F.B.; Azad, R.K.; Nechushtai, R.; Mittler, R. The impact of multifactorial stress combination on plant growth and survival. New Phytol. 2021, 230, 1034–1048. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.C.; Griffiths, M.; Golebiowska, A.; Mairhofer, S.; Burr-Hersey, J.; Goh, T.; von Wangenheim, D.; Atkinson, B.; Sturrock, C.J.; Lynch, J.P.; et al. Shaping 3D root system architecture. Curr. Biol. 2017, 27, R919–R930. [Google Scholar] [CrossRef] [PubMed]
- Jarzyniak, K.M.; Jasiński, M. Membrane transporters and drought resistance—A complex issue. Front. Plant Sci. 2014, 5, 687. [Google Scholar] [CrossRef] [PubMed]
- Daszkowska-Golec, A.; Szarejko, I. Open or close the gate–stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 2013, 4, 138. [Google Scholar] [CrossRef] [PubMed]
- Good, A.G.; Zaplachinski, S.T. The effects of drought stress on free amino acid accumulation and protein synthesis in Brassica napus. Physiol. Plant. 1994, 90, 9–14. [Google Scholar] [CrossRef]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Bacelar, E.L.; Moutinho-Pereira, J.M.; Gonçalves, B.M.; Brito, C.V.; Gomes-Laranjo, J.; Ferreira, H.M.; Correia, C.M. Water use strategies of plants under drought conditions. In Plant Responses to Drought Stress: From Morphological to Molecular Features; Aroca, R., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 145–170. [Google Scholar] [CrossRef]
- Liu, X.; Gao, T.; Liu, C.; Mao, K.; Gong, X.; Li, C.; Ma, F. Fruit crops combating drought: Physiological responses and regulatory pathways. Plant Physiol. 2023, 192, 1768–1784. [Google Scholar] [CrossRef] [PubMed]
- Kalladan, R.; Lasky, J.R.; Chang, T.Z.; Sharma, S.; Juenger, T.E.; Verslues, P.E. Natural variation identifies genes affecting drought-induced abscisic acid accumulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 11536–11541. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Bailey-Serres, J.; Ashikari, M.; Atwell, B.J.; Colmer, T.D.; Fagerstedt, K.; Fukao, T.; Geigenberger, P.; Hebelstrup, K.H.; Hill, R.D.; et al. Community recommendations on terminology and procedures used in flooding and low oxygen stress research. New Phytol. 2017, 214, 1403–1407. [Google Scholar] [CrossRef]
- Salvatierra, A.; Toro, G.; Mateluna, P.; Opazo, I.; Ortiz, M.; Pimentel, P. Keep calm and survive: Adaptation strategies to energy crisis in fruit trees under root hypoxia. Plants 2020, 9, 1108. [Google Scholar] [CrossRef]
- Habibi, F.; Liu, T.; Shahid, M.A.; Schaffer, B.; Sarkhosh, A. Physiological, biochemical, and molecular responses of fruit trees to root zone hypoxia. Environ. Exp. Bot. 2023, 206, 105179. [Google Scholar] [CrossRef]
- Xie, R.; Zheng, L.; Jiao, Y.; Huang, X. Understanding physiological and molecular mechanisms of citrus rootstock seedlings in response to root zone hypoxia by RNA-Seq. Environ. Exp. Bot. 2021, 192, 104647. [Google Scholar] [CrossRef]
- Domingo, R.; Pérez-Pastor, A.; Ruiz-Sánchez, M.C. Physiological responses of apricot plants grafted on two different rootstocks to flooding conditions. J. Plant Physiol. 2002, 159, 725–732. [Google Scholar] [CrossRef]
- McGee, T.; Shahid, M.A.; Beckman, T.G.; Chaparro, J.X.; Schaffer, B.; Sarkhosh, A. Physiological and biochemical characterization of six Prunus rootstocks in response to flooding. Environ. Exp. Bot. 2021, 183, 104368. [Google Scholar] [CrossRef]
- Baldi, E.; Quartieri, M.; Chiarelli, G.; Larocca, G.N.; Masroleo, M.; Xylogiannis, E.; Falsone, G.; Toselli, M. Water retention potentials of Italian soils and physiological responses of potted golden kiwifruit. Sci. Hort. 2024, 329, 113013. [Google Scholar] [CrossRef]
- Smith, G.S.; Judd, M.J.; Miller, S.A.; Buwalda, J.G. Recovery of kiwifruit vines from transient waterlogging of the root system. New Phytol. 1990, 115, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bai, D.; Zhong, Y.; Abid, M.; Qi, X.; Hu, C.; Fang, J. Physiological responses of two contrasting kiwifruit (Actinidia spp.) rootstocks against waterlogging stress. Plants 2021, 10, 2586. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Huang, S.N.; Mo, Z.H.; Xuan, J.P.; Jia, X.D.; Wang, G.; Guo, Z.R. De novo transcriptome sequencing and comparative analysis of differentially expressed genes in kiwifruit under waterlogging stress. Mol. Breed. 2015, 35, 208. [Google Scholar] [CrossRef]
- Li, Z.; Bai, D.; Zhong, Y.; Lin, M.; Sun, L.; Qi, X.; Hu, C.; Fang, J. Full-length transcriptome and RNA-seq analyses reveal the mechanisms underlying waterlogging tolerance in kiwifruit (Actinidia valvata). Int. J. Mol. Sci. 2022, 23, 3237. [Google Scholar] [CrossRef]
- Arismendi, M.J.; Almada, R.; Pimentel, P.; Bastias, A.; Salvatierra, A.; Rojas, P.; Salvatierra, A.; Rojas, P.; Hinrichsen, P.; Pinto, M.; et al. Transcriptome sequencing of Prunus sp. rootstocks roots to identify candidate genes involved in the response to root hypoxia. Tree Genet. Genomes 2015, 11, 11. [Google Scholar] [CrossRef]
- Naor, A.; Klein, I.; Doron, I. Stem water potential and apple fruit size. J. Am. Soc. Hort. Sci. 1995, 120, 577–582. [Google Scholar] [CrossRef]
- Turner, N.C. Measurement of plant water status by the pressure chamber technique. Irrig. Sci. 1988, 9, 289–308. [Google Scholar] [CrossRef]
- Pilkington, S.M.; Crowhurst, R.; Hilario, E.; Nardozza, S.; Fraser, L.; Peng, Y.; Gunaseelan, K.; Simpson, R.; Tahir, J.; Deroles, S.C.; et al. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genom. 2018, 19, 257. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Prot. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yao, Q.; Patil, G.B.; Agarwal, G.; Deshmukh, R.K.; Lin, L.; Wang, B.; Wang, Y.; Prince, S.J.; Song, L.; et al. Identification and comparative analysis of differential gene expression in soybean leaf tissue under drought and flooding stress revealed by RNA-Seq. Front. Plant Sci. 2016, 7, 1044. [Google Scholar] [CrossRef] [PubMed]
- Nanjo, Y.; Maruyama, K.; Yasue, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Komatsu, S. Transcriptional responses to flooding stress in roots including hypocotyl of soybean seedlings. Plant Mol. Biol. 2011, 77, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef]
- Hamel, L.P.; Nicole, M.C.; Duplessis, S.; Ellis, B.E. Mitogen-activated protein kinase signaling in plant-interacting fungi: Distinct messages from conserved messengers. Plant Cell 2012, 24, 1327–1351. [Google Scholar] [CrossRef]
- Czékus, Z.; Szalai, G.; Tari, I.; Khan, M.I.R.; Poór, P. Role of ethylene in ER stress and the unfolded protein response in tomato (Solanum lycopersicum L.) plants. Plant Physiol. Biochem. 2022, 181, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Manghwar, H.; Li, J. Endoplasmic reticulum stress and unfolded protein response signaling in plants. Int. J. Mol. Sci. 2022, 23, 828. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.L.; Rudaz, S.; Hae Choi, Y.; Kyong Kim, H. Plant metabolomics: From holistic data to relevant biomarkers. Curr. Med. Chem. 2013, 20, 1056–1090. [Google Scholar] [CrossRef] [PubMed]
- Parida, A.K.; Panda, A.; Rangani, J. Metabolomics-guided elucidation of abiotic stress tolerance mechanisms in plants. In Plant Metabolites and Regulation under Environmental Stress; Academic Press: Cambridge, MA, USA, 2018; pp. 89–131. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant flavonoids—Biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.C.; Zhang, X.D.; Gao, Z.Q.; Hu, T.; Liu, Y. The research progress of chalcone isomerase (CHI) in plants. Mol. Biotechnol. 2019, 61, 32–52. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cui, W.; Qi, X.; Lin, M.; Qiao, C.; Zhong, Y.; Hu, C.; Fang, J. MicroRNA858 negatively regulates anthocyanin biosynthesis by repressing AaMYBC1 expression in kiwifruit (Actinidia arguta). Plant Sci. 2020, 296, 110476. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Wang, Y.; Xie, H.; Qiu, C.; Zhang, S.; Xiao, J.; Li, H.; Chen, L.; Li, X.; Ding, Z. Drought stress triggers proteomic changes involving lignin, flavonoids and fatty acids in tea plants. Sci. Rep. 2020, 10, 15504. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Cano, L.; Gomez-Cano, F.; Dillon, F.M.; Alers-Velazquez, R.; Doseff, A.I.; Grotewold, E.; Gray, J. Discovery of modules involved in the biosynthesis and regulation of maize phenolic compounds. Plant Sci. 2020, 291, 110364. [Google Scholar] [CrossRef]
- Cesarino, I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef]
- Bhardwaj, R.; Handa, N.; Sharma, R.; Kaur, H.; Kohli, S.; Kumar, V.; Kaur, P. Lignins and abiotic stress: An overview. In Physiological Mechanisms and Adaptation Strategies in Plants under Changing Environment; Ahmad, P., Wani, M., Eds.; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2014; Volume 1, pp. 267–296. [Google Scholar] [CrossRef]
- Boncan, D.A.T.; Tsang, S.S.; Li, C.; Lee, I.H.; Lam, H.M.; Chan, T.F.; Hui, J.H. Terpenes and terpenoids in plants: Interactions with environment and insects. Int. J. Mol. Sci. 2020, 21, 7382. [Google Scholar] [CrossRef] [PubMed]
- Tabaeizadeh, Z. Drought-induced responses in plant cells. Int. Rev. Cytol. 1998, 182, 193–247. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; He, S.; Liu, D.; Patil, G.B.; Zhai, H.; Wang, F.; Stephenson, T.J.; Wang, Y.; Wang, B.; Nguyen, H.T.; et al. A sweet potato geranylgeranyl pyrophosphate synthase gene, IbGGPS, increases carotenoid content and enhances osmotic stress tolerance in Arabidopsis thaliana. PLoS ONE 2015, 10, e0137623. [Google Scholar] [CrossRef]
- Lemoine, R.; Camera, S.L.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Thvenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [PubMed]
- Livingston, D.P.; Hincha, D.K.; Heyer, A.G. Fructan and its relationship to abiotic stress tolerance in plants. Cell. Mol. Life Sci. 2009, 66, 2007–2023. [Google Scholar] [CrossRef]
- Santarius, K.A. The protective effect of sugars on chloroplast membranes during temperature and water stress and its relationship to frost, desiccation and heat resistance. Planta 1973, 113, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yu, B. Changes in content of free, conjugated and bound polyamines and osmotic adjustment in adaptation of vetiver grass to water deficit. Plant Physiol. Biochem. 2010, 48, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, M.; Pazmino, D.; Seung, D.; Horrer, D.; Nigro, A.; Meier, T.; Kölling, K.; Pfeifhofer, H.W.; Zeeman, S.C.; Santelia, D. Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 2016, 28, 1860–1878. [Google Scholar] [CrossRef]
- Zadražnik, T.; Moen, A.; Egge-Jacobsen, W.; Meglič, V.; Šuštar-Vozlič, J. Towards a better understanding of protein changes in common bean under drought: A case study of N-glycoproteins. Plant Physiol. Biochem. 2017, 118, 400–412. [Google Scholar] [CrossRef]
- Gregorova, Z.; Kovacik, J.; Klejdus, B.; Maglovski, M.; Kuna, R.; Hauptvogel, P.; Matusikova, I. Drought-induced responses of physiology, metabolites, and PR proteins in Triticum aestivum. J. Agric. Food Chem. 2015, 63, 8125–8133. [Google Scholar] [CrossRef] [PubMed]
- Cardenas-Manríquez, G.; Vega-Muñoz, I.; Villagómez-Aranda, A.L.; León-Galvan, M.F.; Cruz-Hernandez, A.; Torres-Pacheco, I.; Rangel-Cano, R.M.; Rivera-Bustamante, R.F.; Guevara-Gonzalez, R.G. Proteomic and metabolomic profiles in transgenic tobacco (N. tabacum Xanthi nc) to CchGLP from Capsicum chinense BG-3821 resistant to biotic and abiotic stresses. Environ. Exp. Bot. 2016, 130, 33–41. [Google Scholar] [CrossRef]
- Chen, S.; Ding, G.; Wang, Z.; Cai, H.; Xu, F. Proteomic and comparative genomic analysis reveals adaptability of Brassica napus to phosphorus-deficient stress. J. Proteom. 2015, 117, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, D.; Pandey, A.; Choudhary, M.K.; Datta, A.; Chakraborty, S.; Chakraborty, N. Comparative proteomics analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol. Cell. Proteom. 2007, 6, 1868–1884. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.B.; Rai, Y.; Gayali, S.; Chakraborty, S.; Chakraborty, N. Plant organellar proteomics in response to dehydration: Turning protein repertoire into insights. Front. Plant Sci. 2016, 7, 460. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.J.; Sauter, A.; Wichert, K.; Messdaghi, D.; Hartung, W. Extracellular β-glucosidase activity in barley involved in the hydrolysis of ABA glucose conjugate in leaves. J. Exp. Bot. 2000, 51, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Hayat, K.; Iqbal, A.; Xie, L. Implications of abscisic acid in the drought stress tolerance of plants. Agronomy 2020, 10, 1323. [Google Scholar] [CrossRef]
- Wu, J.; Wang, J.; Hui, W.; Zhao, F.; Wang, P.; Su, C.; Gong, W. Physiology of plant responses to water stress and related genes: A review. Forests 2022, 13, 324. [Google Scholar] [CrossRef]
- Todaka, D.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. ABA-responsive gene expression in response to drought stress: Cellular regulation and long-distance signalling. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2019; Volume 92, pp. 83–113. [Google Scholar] [CrossRef]
- Arbona, V.; Zandalinas, S.I.; Manzi, M.; González-Guzmán, M.; Rodriguez, P.L.; Gómez-Cadenas, A. Depletion of abscisic acid levels in roots of flooded Carrizo citrange (Poncirus trifoliata L. Raf.× Citrus sinensis L. Osb.) plants is a stress-specific response associated to the differential expression of PYR/PYL/RCAR receptors. Plant Mol. Biol. 2017, 93, 623–640. [Google Scholar] [CrossRef]
- Hsu, F.C.; Chou, M.Y.; Peng, H.P.; Chou, S.J.; Shih, M.C. Insights into hypoxic systemic responses based on analyses of transcriptional regulation in Arabidopsis. PLoS ONE 2011, 6, e28888. [Google Scholar] [CrossRef]
- Jackson, M.B.; Young, S.F.; Hall, K.C. Are roots a source of abscisic acid for the shoots of flooded pea plants? J. Exp. Bot. 1988, 39, 1631–1637. [Google Scholar] [CrossRef]
- Rodríguez-Gamir, J.; Ancillo, G.; González-Mas, M.C.; Primo-Millo, E.; Iglesias, D.J.; Forner-Giner, M.A. Root signaling and modulation of stomatal closure in flooded citrus seedlings. Plant Physiol. Biochem. 2011, 49, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, X.; Jiu, S.; Zhang, K.; Wang, C.; Fang, J. Analysis of the regulation networks in grapevine reveals response to waterlogging stress and candidate gene-marker selection for damage severity. R. Soc. Open Sci. 2018, 5, 172253. [Google Scholar] [CrossRef]
- Smith, G.S.; Miller, S.A. Effects of root anoxia on the physiology of kiwifruit vines. Acta Hort. 1991, 297, 401–408. [Google Scholar] [CrossRef]
- Creelman, R.A.; Gage, D.A.; Stults, J.T.; Zeevaart, J.A. Abscisic acid biosynthesis in leaves and roots of Xanthium strumarium. Plant Physiol. 1987, 85, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Lan, W.; Buchanan, B.B.; Luan, S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21419–21424. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Lim, C.W.; Lan, W.; He, K.; Luan, S. ABA signaling in guard cells entails a dynamic protein–protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol. Plant 2013, 6, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.H.; Lee, S.C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef]
- Jung, H.; Lee, D.K.; Do Choi, Y.; Kim, J.K. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 2015, 236, 304–312. [Google Scholar] [CrossRef]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 2019, 10, 4021. [Google Scholar] [CrossRef]
- Liu, Q.; Feng, Z.; Xu, W.; Vetukuri, R.R.; Xu, X. Exogenous melatonin-stimulated transcriptomic alterations of Davidia involucrata seedlings under drought stress. Trees 2021, 35, 1025–1038. [Google Scholar] [CrossRef]
- Du, H.; Wu, N.; Chang, Y.; Li, X.; Xiao, J.; Xiong, L. Carotenoid deficiency impairs ABA and IAA biosynthesis and differentially affects drought and cold tolerance in rice. Plant Mol. Biol. 2013, 83, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Baek, D.; Park, H.C.; Chun, H.J.; Oh, D.H.; Lee, M.K.; Cha, Y.J.; Kim, W.Y.; Kim, M.C.; Chung, W.S.; et al. Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol. Plant 2013, 6, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. Linking development to defense: Auxin in plant–pathogen interactions. Trends Plant Sci. 2009, 14, 373–382. [Google Scholar] [CrossRef]
- Shi, H.; Chen, L.; Ye, T.; Liu, X.; Ding, K.; Chan, Z. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol. Biochem. 2014, 82, 209–217. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Hassan, M.J.; Li, Z.; Peng, Y. Indole-3-acetic acid improves drought tolerance of white clover via activating auxin, abscisic acid and jasmonic acid related genes and inhibiting senescence genes. BMC Plant Biol. 2020, 20, 150. [Google Scholar] [CrossRef]
| TREATMENT | Pn | gs | Ci | E | Stem Ψw |
|---|---|---|---|---|---|
| Control | 13.2 a | 0.174 a | 244 b | 4.09 a | −0.438 a |
| Waterlogging | 0.155 c | 0.063 b | 480 a | 0.787 b | −0.563 a |
| Drought | 3.64 b | 0.036 b | 266 b | 1.27 b | −1.18 b |
| Significance | *** | ** | * | *** | *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baldi, E.; Pastore, C.; Chiarelli, G.; Quartieri, M.; Spinelli, F.; Toselli, M. Molecular Responses to Drought and Waterlogging Stresses of Kiwifruit (Actinidia chinensis var. deliciosa) Potted Vines. Horticulturae 2024, 10, 834. https://doi.org/10.3390/horticulturae10080834
Baldi E, Pastore C, Chiarelli G, Quartieri M, Spinelli F, Toselli M. Molecular Responses to Drought and Waterlogging Stresses of Kiwifruit (Actinidia chinensis var. deliciosa) Potted Vines. Horticulturae. 2024; 10(8):834. https://doi.org/10.3390/horticulturae10080834
Chicago/Turabian StyleBaldi, Elena, Chiara Pastore, Giacomo Chiarelli, Maurizio Quartieri, Francesco Spinelli, and Moreno Toselli. 2024. "Molecular Responses to Drought and Waterlogging Stresses of Kiwifruit (Actinidia chinensis var. deliciosa) Potted Vines" Horticulturae 10, no. 8: 834. https://doi.org/10.3390/horticulturae10080834
APA StyleBaldi, E., Pastore, C., Chiarelli, G., Quartieri, M., Spinelli, F., & Toselli, M. (2024). Molecular Responses to Drought and Waterlogging Stresses of Kiwifruit (Actinidia chinensis var. deliciosa) Potted Vines. Horticulturae, 10(8), 834. https://doi.org/10.3390/horticulturae10080834










