Abstract
Lactic acid bacteria (LAB) resist sodium selenite of concentrations greater than 100 mg/L in fermentation media. Selenium affects the growth rate, but once the microorganism absorbs selenium, this element is converted through a complex mechanism into selenocysteine and then into a selenoprotein structure. This study verified the presence of selenocysteine in Enterococcus faecium ABMC-05. The microorganism was cultivated in a medium enriched with a minimum inhibitory concentration of sodium selenite (184 mg/L). The concentration of selenium absorbed and the bioconversion into selenocysteine were determined by inductively coupled plasma optical emission spectrometry (ICP-OES) and reverse-phase high-performance chromatography (RP-HPLC), respectively. The presence of the selD, selA, and cysK genes was determined by amplifying the 16S rDNA through polymerase chain reaction (PCR). The microorganism accumulated inorganic selenium, and part was transformed into selenocysteine. The growth curves were atypical for a lactic acid bacterium with a stationary phase greater than 70 h. Determining the genetic expression showed only the presence of the cysK gene and the absence of the selD and the selA genes. The results demonstrate that this microorganism produces selenocysteine through a mechanism independent of the SelA and SelD pathways in contrast to other LAB.
1. Introduction
Selenium (Se) is an essential trace element for humans and living forms. This element occurs naturally as selenate and selenite; however, these inorganic chemical forms of Se are often toxic [1,2]. Some microorganisms (bacteria and yeasts) transform inorganic Se into seleno-amino acids, such as selenocysteine (Sec) and selenomethionine (SeM) [3,4]. Within these microorganisms, LAB has been shown to incorporate inorganic Se, transforming it into Sec [3,5,6].
The route by which LAB achieves this transformation needs to be studied further. However, advances in research have shown that biotransformation is due to Sec being incorporated into the polypeptide chains of specific proteins [5]. The mechanism begins with the aminoacylation of tRNASec (SelC) with an adenylated serine residue by the action of the enzyme seryl-tRNA synthetase (SerS) [7,8]. That is why the presence of serine in the medium to initiate the transformation process has been studied [9].
After aminoacylation, a selenophosphate is incorporated as a Se donor via selenocysteine synthase (SelA) to load serine-tRNASec with Se, generating selenocystyl-tRNASec [8]. The selD gene encodes selenophosphate synthetase (SelD), which produces an “activated form of Se”, which is a selenophosphate formed from selenide and ATP [10]. This results in selenocystyl-tRNASec inserting Sec into polypeptide chains on the ribosome. Therefore, Sec is inserted in response to the UGA codon, usually a stop signal. During translation, the Sec-specific elongation factor (SelB in bacteria) carries Sec-tRNASec to the ribosome at the UGA codon, which encodes Sec after recognition of a secondary structure (SECIS) in the mRNA [11,12].
Accordingly, the selA and selD genes are part of the main pathway for Sec formation and its insertion into selenoprotein structures. Studies have revealed the presence of both genes in LAB during Sec formation [13]. However, some LAB do not survive the presence of small concentrations of Se, or their growth is inhibited or slowed down, but they form Sec [9,14,15,16,17,18]. Therefore, other alternative pathways could be proposed or studied. One of the mechanisms is the specific pathway involving the cysK gene, which codes for O-acetyl serine (thiol)lyase. Only CysK of the cys gene pool is known to be used as a key enzyme as the entry point of selenium in the bioconversion to selenocysteine [7]. Recently, CysK was reported in Bacillus subtillis, which follows an alternate route for producing selenocysteine. This route catalyzes selenide with O-acetylserine (OAS) to synthesize SeCys [19].
Due to the importance of different routes to biotransformation of inorganic Se into Sec, this work aimed to verify whether the formation of Sec by E. faecium ABMC-05 isolated from a traditional Mexican fermented beverage is not dependent on the presence of the selA and selD genes but on the gene cysK during cell selenization. With this gene identification, this study explains the biogenic formation of Sec in a microorganism resistant at high concentrations of inorganic selenium.
2. Materials and Methods
2.1. Chemicals and Reagents
Man Rogosa Sharpe broth was supplied by Becto Dickinson Difco (Maryland, USA), bacteriological agar was purchased of Bioxon, BD Lab. (Edo. México, México), and glycerol was purchased by Fermont (Monterrey, México). Sodium selenite, dithiothreitol, Se standard solution, seleno-L-cystine, nitric acid, potassium borohydride, β-mercaptoethanol, iodoacetic acid, borate, formic acid, standard solvent (acetonitrile 50%, water 47.5%, and trifluoroacetic acid 2.5%), sodium acetate, ethanol, ethidium bromide, and agarose were supplied by Sigma-Aldrich Co. (St. Louis, MO, USA). O-phthaldehyde was provided by Pickering Lab (Mountain View, CA, USA), and α-cyano-4-hydroxycinnamic acid (HCCA) was purchased from Bruker Daltonics (Bremen, Germany). HPLC grade acetonitrile and methanol, NaOH, and HCl were supplied by JT Baker, Thermo Fisher Scientific (CDMX Mexico). DNA: A DNeasy UltraClean Microbial Kit was provided by Quiagen (Hilden, Germany). PCR: GO Taq Flexi DNA polymerase, dNTPs, MgCl2, buffer (5x), nuclease-free water, and a ladder of 100 pb were provided by Promega (Madison, WI, USA). TAE buffer (Tris-acetate-EDTA) and BigDye were supplied by Thermo Fisher Scientific (Vilnius, Lithuania). Gene amplification: Taq DNA polymerase was provided by Invitrogen (Carlsbad, CA, USA). MgCl2, 10 X PCR buffer, and a dNTP mix were purchased from Thermo Scientific (Vilnius, Lithuania). GelRed (Biotium, Fisher Biotec, Wembley, Australia).
2.2. Growing Conditions
The lactic acid bacterium in this study was previously isolated from a typical Mexican fermented beverage called “tepache” and others [20]. The strain was activated three times from a frozen glycerol stock (−20 °C) in Man Rogosa Sharpe broth (MRS, BD Difco Laboratories, Thermo Fisher Scientific Inc.) and incubated at 37 °C for 12 h under anaerobic conditions. Cells were distributed on the surface of solid MRS agar (BD Difco) in a Petri dish, and after 24 h of incubation at 37 °C, a colony was placed in 10 mL of MRS broth to incubate at 37 °C for 12 h. Subsequently, the culture was divided into vials, and 15% (v/v) sterile glycerol was added and stored at −20 °C to be used as a working strain. A Gram stain test was performed to verify its morphology and the viable count to determine the concentration of the initial culture.
2.3. Bacterium Identification
The bacterium was reported previously by our research group [21], but the identification methodology has yet to be reported. Thus, the microorganism was identified by combining MALDI mass spectrometry (matrix-assisted laser desorption/ionization), TOF (time of flight), and MALDI BioTyper software. 16S rDNA and phylogenetic analyses were performed to validate the information.
2.4. MALDI-TOF MS
Sample preparation. The bacterium was cultivated on MRS agar plates at 37 °C for 36 h under anaerobic conditions. The MALDI Biotyper protocol (MALDI Biotyper 3.1 manual) was used to identify the selected colonies. One colony (approximately 10 ng) was transferred to a microtube containing 300 μL of ultrapure water and 900 μL of pure ethanol. The suspension was centrifuged (Sorvall fresco, Thermo Fisher Scientific, Waltham, MA, USA) twice at 7700× g for 2 min, and then the supernatant was removed. The pellet was allowed to dry at room temperature to remove the residual ethanol. Next, 10 μL of 70% aqueous formic acid was added, and acetonitrile (1:1) was added after vortex mixing. The suspension was centrifuged (Sorvall fresco, Thermo Fisher Scientific, Waltham, MA, USA) at 7700× g for 15 min, and 1 μL of the supernatant was transferred to a clean space in the equipment’s plate and allowed to dry at room temperature. The preparation was covered with 1 μL of the saturated matrix solution of an α-cyano-4-hydroxycinnamic acid (HCCA) matrix solution in 50% acetonitrile and 2.5% trifluoroacetic acid (Bruker Daltonik). It was left to dry at room temperature until the crystallization of the experimental sample.
Sample analysis. Mass spectra and analysis were performed with an auto flex speed mass spectrometer (Bruker Daltonik, Billerica, MA, USA) using MALDI BioTyper 3.1 software (Bruker Daltonik, Billerica, MA, USA). The mass spectra obtained from each isolate were imported into BioTyper software and analyzed by standard pattern matching, expressed according to the manufacturer, with scores between 0 and 3. Scores below 1.7 were considered unreliable identification; a score ≥ 1.7 was considered the identification of the genus, while a score of ≥2.0 indicated identification of the species.
2.5. 16S rDNA Identification and Phylogenetic Analysis
DNA extraction. A total of 20 µL of the medium with the bacteria previously activated was inoculated in 10 mL of MRS broth for 24 h (three times). The culture medium was centrifuged at 7700× g for 5 min in sterile Falcon tubes. The pellet was transferred to a 2 mL microtube for DNA extraction performed using a DNeasy UltraClean Microbial Kit following the manufacturer’s instructions. DNA was quantified by micro-drop spectrophotometry (NanoDrop One) and 1% agarose gel electrophoresis for 30 min at 120 V.
Amplification of the 16S rDNA gene. The extracted DNA was used to amplify the variable region V1–V3 of the 16S rRNA gene (approximately 510 bp) using the primers 8F (5′-AGAGTTTGATCCTGCCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) according to Turner [7]. The PCR reactions were conducted in a thermocycler (Thermo Fisher, Arktik model). The reaction mixture (total volume of 50 µL) consisted of 3 µL (10–20 ng/ µL) of genomic DNA, 0.25 µL (1.25 U) of GO Taq Flexi DNA polymerase (Promega), 1 µL (10 µM) of 8F primer, 1 µL (10 µM) of 1492R primer, 0.5 µL (10 mM) of dNTPs, 2.4 µL (25 mM) of MgCl2, 10 µL of buffer (5x), and 31.85 µL of nuclease-free water. The temperature cycle for the PCR consisted of an initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 0.5 min, 55 °C for 0.5 min, and 72 °C for 0.5 min with a final extension of 10 min at 72 °C. To detect DNA, 3 μL of the PCR product were loaded on 1% agarose gels stained with Texas red (0.2 μL) and electrophoresed in 1X TAE buffer (Tris-Acetate-EDTA) for 30 min at 100 V. The gel was visualized on a transilluminator. A 100 bp DNA molecular weight marker (Invitrogen) was used on each gel.
16S rRNA gene sequencing and analysis. The 16S rRNA gene amplification was purified with BigDye terminator v3.1 and sequenced on a 3730xl DNA Analyzer (Applied Biosystems, Waltham, MA, USA). The nucleotide sequence was visualized and analyzed using Python version 3.6.7 (Wilmington, DE, USA). Three databases (NCBI, RDP, and SILVA) trained with the RDP Classifier algorithm were used to investigate the similarity of the 16S sequence obtained against other sequences deposited in GenBank.
Molecular Phylogenetic Analysis. CLUSTAL 1.2.4 (Dublin, Ireland) software was used to subject the sequences to multiple sequence alignment. A phylogenetic tree was constructed using the maximum likelihood method based on the Kimura two-parameter model [22], using the 16S rRNA sequence and those obtained in the database (five sequences from Enterococcus, five sequences from Lactobacillus, two of Lactococcus, and one of Streptococcus). Molecular Evolutionary Genetics Analysis Version 7.0 (MEGA7) (Canterbury, Kent, UK) was used to infer the maximum likelihood and generate a phylogenetic tree [23].
2.6. Minimum Inhibitory Concentration of Na2SeO3
With some modifications, the minimum inhibitory concentration (MIC) of Se in lactic acid bacteria was determined according to Castañeda-Ovando et al. [9]. The bacteria were inoculated (0.1 mL, 1 × 10 9 CFU/mL) in 5 mL of MRS broth supplemented with sodium selenite (Na2SeO3) at different concentrations: 0, 100, 200, 300, 400, and 500 mg/L. A stock solution of 1000 mg/L of Na2SeO3 in deionized water was prepared to obtain selenite concentrations. This solution was sterilized through a 0.22 µm sterile syringe filter (Millipore) and supplemented with MRS medium in the tubes to incubate them at 37 °C for 36 and 48 h under anaerobic conditions. The viable count was carried out by a plate count on MRS agar (BD Difco, MD, USA) using the micro-drop technique [24]. The control consisted of MRS supplemented with Na2SeO3 without bacteria. The experiment was carried out in separate tubes. With the viability data obtained, the MIC was determined according to the graphic method of Talmadge and Fitch reported by Escobar-Ramírez et al. [21].
2.7. Selenization of E. faecium ABMC-05
The bacterium was inoculated (100 μL; 1 × 109 cells/mL) in tubes with 5 mL of MRS broth and Na2SeO3 at the calculated minimum inhibitory concentration. The fermentation was developed at 37 °C for 120 h under anaerobic conditions. The culture inoculated in MRS broth without Na2SeO3 was used as a control, sampling from independent tubes every 24 h. Total Se was determined and viability was analyzed from each sample using a plate count on MRS agar (BD Difco, MD, USA).
2.8. Quantification of Total Selenium Content by Inductively Coupled Optical Emission Spectrometry (ICP-OES)
Biomass separation. A total of 1 mL of the fermented material was centrifuged (Sorvall fresco, Thermo Fisher Scientific, Waltham, MA, USA) at 7700× g for 15 min at 4 °C to separate the cells from the culture medium. To remove the Se adhered to the membrane, the cells were washed in 100 µL of dithiothreitol (DTT; Sigma-Aldrich) at 0.3% (w/v) and centrifuged at 7700× g for 15 min at 4 °C. The first and second centrifugation supernatants were mixed to determine residual Se. The cells were dried at 60 °C for 24 h in a convection oven, and the dry biomass was stored in a desiccator until Sec analysis.
Total Se content. A total of 1 mL of the supernatant mixture and 10 mL of concentrated nitric acid were digested in a microwave accelerated reaction system (MARS 5 microwave, CEM Corporation, Matthews, NC, USA) under the following conditions: 175 °C for 5.5 min and 175–180 °C for 4.5 min with a pressure limit of 110 psi (7.0307 kg/cm2). After digestion, it was brought to a final volume of 25 mL with deionized water. The calibration curve was performed in a range of 0.2–4 mg/L from a 50 mg/L Se standard solution (Se standard, Sigma-Aldrich) in 5% HNO3. The samples were analyzed by inductively coupled plasma (ICP) with optical emission spectrometry (OES) on an Optima 8300 ICP-OES Spectrometer (PerkinElmer, Waltham, MA, USA) at a wavelength of 166 nm. The concentration of Se accumulated was calculated by the difference between the concentrations of Se at time 0 (T0) and the sample at the different fermentation times T24, T48, T72, T96, and T102.
2.9. Selenocysteine Determination
The determination was made according to the methodology proposed by Castañeda-Ovando et al. [9] from carboxymethyl-selenocysteine synthesis as the Sec standard, with some modifications.
Carboxymethylation. In a male Schlenk flask (Anorsur), 50 mg of seleno-l-cystine (Sigma-Aldrich), 15 mg of KBH4 (Sigma-Aldrich), and 375 μL of deionized water were added. The reaction was conducted under a nitrogen atmosphere at 50 °C for 1 h. Then, 1 mL of iodoacetic acid (0.4 mol/L (Sigma-Aldrich) at pH 8.5 adjusted with 1 M NaOH (JT Baker, Thermo Fisher Scientific)) was added to the flask. The reaction was carried out under a nitrogen atmosphere at 37 °C for 1 h in the dark. Subsequently, 50 μL of β-mercaptoethanol (Sigma-Aldrich) was added and left to stand in the refrigerator and the dark overnight. To end the reaction, 50 μL of 6 mol/L HCl (JT Baker, Thermo Fisher Scientific) was added under a nitrogen atmosphere at 110 °C for 24 h. The dried cells were treated the same way as the standard, with prior sonication in an ultrasonic bath for 30 min [25].
Derivatization. A total of 1 mL of borate buffer (0.4 mol/L, pH 9.5), 30 μL of the sample or carboxymethyl selenocysteine standard, and 30 μL of OPA solution [26]. The OPA solution was prepared with 50 mg of O-phthaldehyde (OPA, Pickering Laboratories) dissolved in 4 mL of methanol, followed by the addition of 50 μL of β-mercaptoethanol and 500 μL of 0.4 mol/L borate buffer. The solution was prepared before each analysis due to the instability [27,28].
RP-HPLC. Sec determination was performed by reverse-phase HPLC (PerkinElmer Series 200, Madrid, Spain). The mobile phases used were sodium acetate buffer (0.1 mol/L, pH 7.2) with 0.1% acetonitrile (phase A) and methanol (phase B). All separations were performed on a C18 Zorbax Eclipse column (Agilent Technologies Inc., Santa Clara, CA, USA; 250 × 4.6 mm, 5 μm) by elution gradient 25% A for 10 min and 100% B for 5 min. Detection was performed through a diode array detector at 340 nm. The injection volume was 20 μL at a 1.0 mL/min flow rate.
2.10. Gene Amplification: selA, selD, and cysK
The primers selA and cysK were designed from consensus sequences using an algorithm in Python language developed in the Biotechnology laboratory (N104) of the Universidad Autónoma Metropolitana, Unidad Xochimilco. This algorithm can predict and design primers from reference sequences to identify genes in non-sequenced organisms. The parameters for the prediction and design of the primers were the following: Optimum melting temperature of 55 (with a tolerance of 0.5 and a standard deviation of less than 0.05), GC content between 40% and 60%, amplicon length between 490 and 510 bp, and primer size of 21 or 22 bases. Phylogenetic relationships were inferred for the cysK and selA genes using the maximum likelihood method based on the Kimura two-parameter model [22]. The bootstrap consensus tree was inferred from 1000 replicates [29]. The phylogenetic analysis was conducted in MEGA7 [23].
The PCR reaction was conducted on a final volume of 25 µL, containing 0.1 µL of Taq DNA polymerase (5 U/µL), 2.5 µL of 10X PCR buffer without MgCl2, 1.25 µL of each primer, 0.75 µL of 50 mM MgCl2, 0.5 µL of 10 mM dNTP mix, and 10 ng of E. faecium DNA and amplifications were carried out in an Axygen MaxyGene II Thermal Cycler (Axygen a Corning Brand, USA). The PCR cycles included an initial denaturation of 5 min at 94 °C, followed by 35 cycles of denaturation for 45 s at 94 °C, annealing for 40 s at 55.2 °C and an extension of 45 s at 72 °C, with a final extension of 10 min at 72 °C. The PCR products were analyzed by gel electrophoresis run in TAE buffer in a GelRed (Biotium) stained 1.5% agarose gel for 40 min at 75 volts and visualized using the AlphaImager Digital Imaging System (Alpha Innotech Corp., San Leandro, CA, USA).
selD gene sequences were not found in E. faecium, but other Enterococcus species were used for the primer design using the Primer-BLAST program. The PCR reactions were conducted in a thermocycler (Thermo Fisher, Arktik model). The reaction mixture of a total volume of 50 µL consisted of 5 µL (10 ng/ µL) of genomic DNA, 1 µL (1.25 U) of Taq DNA polymerase (ThermoFisher Scientific), 2 µL (100 ng/µL) of each primer, 1 µL (10 mM) of dNTPs, 1.5 µL (50 mM) of MgCl2, and 5 µL of 10X Taq buffer and nuclease-free water. The PCR conditions were as follows: Initial denaturation step at 94 °C for 5 min, 30 cycles of 94 °C for 45 s, 50 °C for 45 s, and 72 °C for 45 s, with a final extension of 72 °C for 10 min. The products amplified by PCR were analyzed by electrophoresis in agarose gel at 1% at 120 V for 30 min.
2.11. Statistical Analysis
Microbiological data were analyzed using the one-factor ANOVA test. Se accumulation data were analyzed with repeated measurements over time. Tukey’s test (p < 0.05) determined significant differences between mean values using the Minitab 18 program.
3. Results and Discussion
3.1. Identification
The ABMC-05 strain isolated from a traditional fermented beverage, “tepache”, was characterized as a Gram-positive bacterium in the form of a coccus; the colonies measured approximately 2–3 mm in diameter, circular in shape with a defined convex edge and white color (Figure 1).
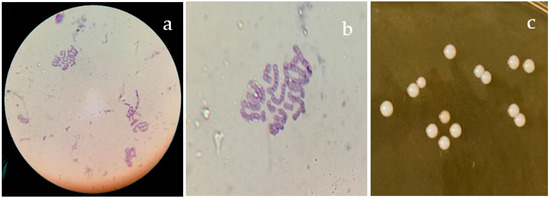
Figure 1.
Gram stain of E. faecium ABMC-05. (a) Image at 40×; (b) image at 100×; (c) colonies on MRS agar plates.
In this study, the ABMC-05 strain was identified as E. faecium by MALDI BioTyper software and by the conventional 16S rDNA method. The identification score of the ABMC-05 strain with the MALDI BioTyper software was more significant than 2.0 in all of the samples analyzed. The 16S rDNA sequence of strain ABMC-05 showed 99% similarity to several species of E. faecium available from NCBI (Figure 2). This is consistent with the established criteria for evaluating individual samples, in which the difference in nucleotide sequence between the reference strain in the database and the considered model should not differ by more than 2% (98% probability) [30]. Therefore, the isolated ABMC-05 strain was designated as E. faecium ABMC-05. The sequence was deposited in GenBank under accession number OL413240.
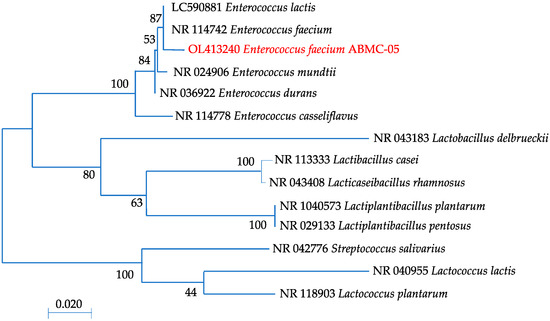
Figure 2.
Phylogenetic maximum likelihood analysis of the partial 16S gene for bacterial strains based on the nucleotide sequences available in GenBank. The numbers on the nodes are supported by starting values from 1000 replications.
Several studies have shown the importance of tepache as a matrix for isolating LAB [31]. Moreno-Terraza [32] isolated two probiotic LAB from tepache. Similarly, Cervantes and Pedroza [33] separated an exopolysaccharide-producing Leuconostoc species with probiotic characteristics. Likewise, de la Fuente-Salcido et al. [34] showed that LAB isolated from tepache can produce metabolites that inhibit the growth of pathogenic bacteria such as E. coli. Escobar-Ramírez et al. [20] reported the isolation of five different LAB from tepache, one of which showed probiotic characteristics. However, few studies have demonstrated the selenization of LAB isolated from traditional Mexican fermented beverages [21].
3.2. Minimum Concentration of the Inhibition and Selenization of E. faecium ABMC-05
Table 1 shows the results of the effect of sodium selenite concentration on the viability during the growth of E. faecium ABMC-05 expressed as the concentration in colony-forming units (CFU/mL) (Table S1).

Table 1.
Viable count of E. faecium ABMC-05 in MRS culture medium at different concentrations of sodium selenite during 36 and 48 h of incubation.
From 100 mg/L of Na2SeO3, a significant decrease (p < 0.05) in the cell concentration was observed. Three log units reduced the number of viable cells at both experimental times (36 and 48 h). Since in previous studies, concentrations lower than 100 mg/L of Na2SeO3 were tested and it was observed that from 20 mg/L, a decrease in cell concentration was generated without reaching total inhibition (data not shown), the experiment was carried out at concentrations higher than 100 mg/L of Na2SeO3. Thus, in the experiments with 500 mg/L of Na2SeO3, the number of living cells decreased by only 3.5 log units. This indicates that the bacteria could tolerate concentrations higher than 500 mg/L without reaching total inhibition. This behavior has been observed in some LAB where the selenization process is carried out first by adaptation of the microorganism to high concentrations of selenite, as an adaptation process. Therefore, it presents resistance to the presence of salt [35]. Some LABs are known to tolerate different selenium concentrations in the medium. The most studied case is Bifidobacterium animalis ssp., which resists up to 1000 mg/L of sodium selenite in a culture medium, reaching an accumulation of 60 mg/g of dry weight. Before reaching the maximum tolerated concentration, Se enrichment (Se enrichment rate) gradually increases with increasing inorganic Se concentration, while the LAB biomass decreases [36].
In general, LAB has shown a low tolerance to selenite stress and a low ability to reduce high concentrations of selenite [9,15,21,36,37]. However, Estrada et al. [13] showed that Enterococcus sp. isolated from a Mexican cheese can tolerate up to 600 mg/L of Na2SeO3 and maintain a high cell count (109 CFU/mL) after 20 h of incubation. Recently E. faecium CCDM 922A showed an increase in bacterial cell count (8 to 9 log CFU/mL) after 24 h of incubation in a medium containing different concentrations of sodium selenite (1, 5, 10, 30, and 50 mg/L) [38]. Several reports have indicated that stress tolerance is related to glutathione (GSH) synthesis by Enterococcus spp. [35,39]. GSH is a low molecular weight antioxidant thiol that plays a role in maintaining intracellular redox homeostasis to protect cells against oxidative stress damage [36,40]. The detoxification mechanism is the main process by which GSH directly reduces selenium species to hydrogen selenide and Se0 [19].
The minimum concentration of Na2SeO3 to inhibit the growth of E. faecium ABMC-05 was calculated using data obtained from cell growth in the presence of Se. The results of Talmage and Fitch demonstrated that the curve obtained at 48 h of fermentation presented the best correlation (R2 = 0.9339), so the equation of this curve was used for the calculation of the MIC, which was 184 mg/L of Na2SeO3, using it for the selenization of E. faecium ABMC-05. This salt concentration was higher than the one used by Yang et al. [6]. They determined that at 80 mg/L of Na2SeO3, the cell concentration of different LAB growing in a similar medium decreased. However, this concentration was the same as that calculated using the OD method to measure the microbial growth of E. faecium ABMC-05 in previous work [21]. In addition, in this case, the first inflection in the curve of the inhibitory experiment was calculated at 36 and 48 h because the microorganism concentration decreased from 8 to 5 log CFU/mL at the two first concentrations of Na2SeO3 assayed (100 and 200 mg/L, respectively) (Table 1). The critical point calculated from the data obtained at 36 h of incubation was 102 mg/L using the same graphical method (Talmage and Fitch). To observe differences in the growth of the LAB tested, the selenization was carried out at 80, 102, and 184 mg/L of selenite and the growth curves of E. faecium ABMC-05 at the different concentrations of Na2SeO3 are shown in Figure 3.
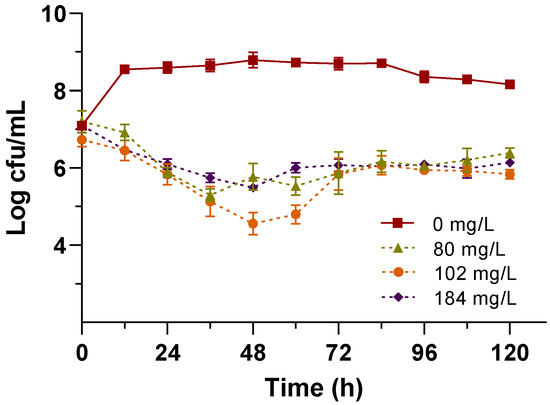
Figure 3.
Growth of E. faecium ABMC-05 at 80, 102, and 184 mg/L of Na2SeO3 and the control (0 mg/L) after 120 h of incubation.
Significant differences (p < 0.05) were found in the growth profile of the curves between the bacteria enriched with Se and the control (0 mg/L of Na2SeO3). The concentration of viable cells decreased in the first 48 h of fermentation in the three concentrations tested. Still, the most significant difference was found at 102 mg/L of Na2SeO3 at 48 h, with an increase in viable cells maintained up to 72 h. After this time, stabilization was observed until the end of the fermentation. After 120 h of incubation, the cultures with selenite had a cell concentration ranging from 5.39 to 6.84 log CFU/mL. This behavior is not consistent with that reported for other LAB. Studies have demonstrated that the growth of lactobacilli is favored at low concentrations of selenite, but once the maximum tolerance level is reached, the viability decreases significantly [6,9,15,36,41].
Unlike other species of the genus Enterococcus reported by other authors [13,35], the behavior of E. faecium ABMC-05 is very similar to that reported by Lampis et al. [42] for Stenotrophomonas maltophilia SeITE02. These authors subjected this microorganism to different concentrations of sodium selenite 0.5, 2.0, 3.0, and 5.0 mM (86, 346, 519, and 865 mg/L). The maximum cell yield was observed after 192 h of fermentation. As in our study, in that report, no total inhibition of the growth of the microorganism was found in any concentration of sodium selenite during the 120 h of fermentation. Additionally, after incubation of E. faecium ABMC-05 in the presence of Na2SeO, the culture medium and biomass turned red. The color became more intense as the fermentation time elapsed, suggesting that this bacterium, like other LAB, reduces Na2SeO3 to Se0, the less toxic form of inorganic selenium [6,43,44,45].
3.3. Determination of Selenium Accumulation in Bacteria by ICP
The concentration of Se bioaccumulated by the bacteria determined by ICP in each of the samples collected during the 120 h of fermentation is shown in Table 2.

Table 2.
Accumulation of Se by E. faecium ABMC-05 at different concentrations of Na2SeO3 for 120 h.
The analyses showed that the accumulation of Se by E. faecium ABMC-05 significantly increased (p < 0.05) with the increasing Na2SeO3 concentration in the medium and the fermentation time. The highest concentration of accumulated Se (4.82 µg Se/log CFU) was reached at 120 h with a concentration of 184 mg/L of Na2SeO3 in the medium. LAB were reported to accumulate Se and then transform into organic selenite compounds such as selenocysteine through detoxification [7,36,46]. Thus, the detoxification process is directly related to the degree of toxicity that Se exerts on the bacteria, and it is known that the detoxification speed increases at higher toxicity levels [6,43,44,45,46,47].
The maximum concentration of accumulated Se found in this study is similar to that reported by other researchers who demonstrated a high capacity for bioaccumulation by Enterococcus spp. Recently Krausova et al. [38] found a high accumulation of Se in E. faecium AADM 922A (6491 g/g of the dry cell) when there was 50 mg/L of Na2SeO3 in the medium. Similarly, Estrada et al. [13] reported a wide range of bioaccumulation in Enterococcus spp. from 0.01796 to 0.11264 g/g dry weight when 600 mg/L of Na2SeO3 was tested during 20 h of fermentation. Pieniz et al. [44] found that as the concentration of Na2SeO3 increased, the content of Se in the medium also increased, reaching 0.017243 g/L when the highest concentration was tested (120 mg/L of Na2SeO3).
Previous studies have reported that Se accumulation occurs in the logarithmic and/or exponential phases of growth [42,47,48]. However, this study found that although the initial concentration of selenite did not favor the growth of E. faecium ABMC-05, there was an increase in Se accumulation concerning fermentation time. In the case of other LAB, it has been observed that there is an increase in the growth rate at low concentrations of selenite. In contrast, at high concentrations, the growth of the microorganism is inhibited, activating the detoxification process [36,41,49]. Similarly, it has also been proposed that bacteria that reduce selenite to Se0 can insert Se into proteins, exopolysaccharides, and nucleic acids, and excess selenite is released by the detoxification mechanism as Se0 [50,51,52,53].
Although there is no detailed research regarding the reduction of selenite in Gram-positive bacteria, it has been proposed that glutathione (GSH) is involved in Se metabolism through the formation of selenodiglutathione (GS-Se-GS), mainly in the reduction of selenite to hydrogen selenide. It has been shown that only some LAB possess the glutathione biosynthetic pathway, and most of them import GSH from the growth medium [43,54]. Such as some LAB, E. faecium synthesizes GSH, and it has shown glutathione reductase (RG) enzyme activity in parental and mutant strains with a high tolerance to sodium selenite [35]. Although in this study, the activity of the enzymes involved in the reduction of selenite to hydrogen selenide was not determined, in research carried out by other authors, the genes (gshF or gshAB) that synthesize GSH have already been identified in E. faecium when it is grown on MRS or other complex media [39].
3.4. Selenocysteine Determination
Once Se accumulation was confirmed, Sec was determined in E. faecium ABMC-05 through HPLC. Sec was identified through the synthesized standard of carboxymethyl selenocysteine. The Sec identification chromatograms obtained from bacterial growth at the lowest (80 mg/L) and highest (184 mg/L) selenite concentrations assayed after 120 h of fermentation are shown in Figure 4.
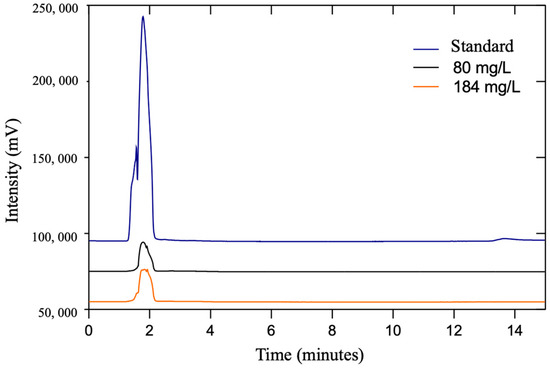
Figure 4.
The carboxymethyl-selenocysteine chromatogram in the standard and E. faecium ABMC-05 cells were exposed to 80 and 184 mg/L Na2SeO3 after 120 h of incubation.
The standard chromatogram shows that carboxymethyl-selenocysteine had a retention time of 1.75 min, which was the reference for the signals of the samples evaluated at 1.79 min (80 mg/L) and 1.75 min (184 mg/L). In this way, the presence of Sec in the biomass of the bacteria was verified after 120 h of fermentation for the two concentrations evaluated, even though the concentration of Se accumulated by the cell was significantly different in both studies (3.40 and 4.82 µg Se/log CFU). This indicates that E. faecium ABMC-05 is capable of biotransforming inorganic Se into Sec regardless of the initial concentration of Se in the medium. The presence of Sec was determined previously and reported by our research group [21]; however, in this case, the presence of the biogenic compound at two different concentrations related to the accumulation was determined.
The results obtained coincide with those reported by Palomo-Siguero et al. [47] and Martinez et al. [43] for other LAB (L. brevis, L. plantarum, Lactococcus lactis, Enterococcus casseliflavus, Frutobacillus trapeoli, Weissella cibaria, and L. bulgaricus). These authors detected Se species in selenocystine (SeCys2) by LC-ICPMS effectively that correspond to Sec by carbamidomethylation of this compound. Recently, Krausova et al. [38] published that the only species found in the biomass of E. faecium CCDM 922A were SeMet and MeSeCys, although in a low proportion (5–10% and 2–10% of the total Se in the extract, respectively). In this study, no other peak was observed in the chromatogram that could be related to any other Se species.
3.5. Determination of the Presence of the Genes selD, selA, and cysK in E. faecium ABMC-05
Due to the behavior of the microorganism, the presence of the selD gene that codes for an ATP-dependent selenophosphate synthetase (SelD, EC 2.7.9.3) and the selA gene that codes for a selenocysteine synthase (SelA, EC. 2.9. 1.1), which are involved in the incorporation of Se into proteins, were determined. The study revealed no amplification of the selD and selA genes (Figure 5). SelD is responsible for the conversion of selenide into selenophosphate [55], while SelA is responsible for loading Se in the form of selenophosphate to serine-tRNASec to generate selenocystyl-tRNASec [12,55,56]. This is a primordial step for the incorporation of Sec into bacterial selenoproteins.
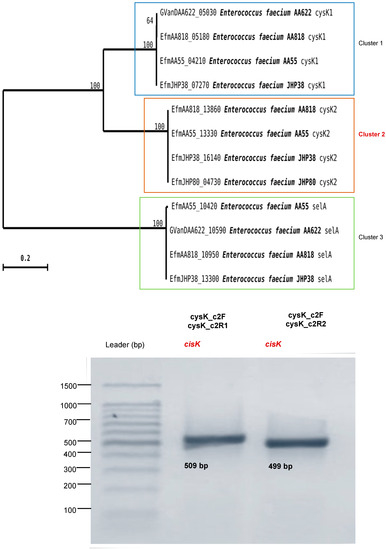
Figure 5.
Maximum likelihood (ML) phylogenetic analysis of the cysk and selA genes for E. faecium strains based on nucleotide sequences available in GenBank. Numbers at the nodes are bootstrap values supported by 1000 replications. On the right side are fragments of the cysk gene of 509 (lane 2) and 499 (lane 3) bp, respectively, obtained with the primers cysK_c2F (5′-TAATCATCGTGATGCCTGAGAC-3′), cysK_c2R1 (5′-TTCTTTTCGCCCAACTTCTCT-3′), and cysK_c2R2 (5′-CCAACTTCTCTAGCTGTTTCCA-3′).
Due to the absence of the selA and selD genes, the results demonstrated that Se was incorporated into Sec through an alternative pathway to selenoprotein synthesis. An in silico study was carried out to determine an alternative route of Se incorporation through which E. faecium ABMC-05 synthesizes Sec. One of the routes studied is based on the production of Sec as a free amino acid, carried out by the presence of O-acetyl serine sulfhydrylase (CysK, EC: 2.5.1.47) [7,57]. In E. coli, CysK can generate Cys and Sec from sulfide (HS) and selenide (HSe), respectively, while cysteinyl-tRNA synthetase (CysRS) can mistakenly load tRNACys with Sec instead of Cys [56,57,58]. Therefore, these non-specific incorporation pathways replace Cys residues by Sec [19,58]. In this study, the presence of CysK was identified by PCR amplification, indicating the possibility of selenocysteine production as a free amino acid by the enzyme CysK, which is incorporated in the metabolism of E. faecium ABMC-05.
Other investigations have reported the presence of SelD and SelA in enterococcus [12,13]. However, in the in silico analysis with the NCBI database accessed in 2021, selD was not found in the complete genomes of E. faecium. Despite not finding the presence of the two main proteins of the selenium to selenocysteine bioconversion pathway, more studies are needed to completely determine their absence. Some isoenzymes with low homology of SelD and SelA could be present. However, there are no reported studies in this regard, which opens the possibility of deeper exploration for the metabolic pathways involved in producing organic selenium by E. faecium ssp. Likewise, these results demonstrate the importance of microorganisms isolated from traditional fermented foods and those that are associated, in many cases, with health benefits. Additionally, the selenization of these microorganisms and the knowledge of the metabolic routes involved are essential in their biotechnological implications.
4. Conclusions
The E. faecium ABMC-05 strain isolated from a fermented beverage could tolerate up to 500 mg/L of Na2SeO3, accumulating Se under unidentified growth conditions in other LAB and transforming it into Sec. The presence of the cysK gene in its DNA indicated another possible route to produce Sec as a free amino acid, which has not yet been studied in species of Enterococcus. The results obtained in this research highlight the importance of E. faecium ABMC-05 in transforming inorganic Se to selenocysteine with the possibility of selenium nanoparticle production through an selD-independent route. It is also very important to determine the presence of enzymes such as glutathione, thioredoxin, thioredoxin reductase, glutathione reductase, selenodiglutathione, and glutathioselenol to gain certainty about the detoxifying capacity of E. faecium ABMC-05. According to the results, it is important to highlight that organic selenium in the cell could be advantageous, especially in preparing functional foods or in pharmacy and medicine.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9070684/s1, Table S1. Viable count of E. faecium ABMC-05 in MRS culture medium at different concentrations of sodium selenite (from 0 to 100 mg/L) during 36 h of incubation
Author Contributions
Conceptualization, L.G.G.-O. and G.M.R.-S.; methodology, M.C.E.-R. and E.Z.-L.; software, E.Z.-L.; validation, M.A.G.-M. and G.M.R.-S.; formal analysis, M.C.E.-R. and E.Z.-L.; investigation, M.C.E.-R.; resources, L.G.G.-O. and G.M.R.-S.; data curation, M.C.E.-R. and E.P.-E.; writing—original draft preparation, M.C.E.-R. and E.P.-E.; writing—review and editing, L.G.G.-O. and G.M.R.-S.; visualization, L.G.G.-O. and E.Z.-L.; supervision, M.A.G.-M. and G.M.R.-S.; project administration, M.C.E.-R. and L.G.G.-O.; funding acquisition, M.C.E.-R. and L.G.G.-O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CONACYT-SAGARPA, grant number 2010-01-144591. SIGI 1318392121, and the same governmental agency, supported the APC.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within this article.
Acknowledgments
The authors thank the National Institute for Agricultural and Livestock Forest Research for the facilities granted to the student Meyli Escobar to carry out doctoral studies and the support of CONACYT for the scholarship awarded. Special thanks to Dra. Laura Márquez and Dra. Nelly López from LaNaBio at UNAM for their support of the genetic analysis. This paper is part of the Basic Science 2014 project (CB-2014-241333) funded by Consejo Nacional de Ciencia y Tecnología (CONACYT). The authors thank the Sistema Nacional de Investigadores (CDMX, Mexico) and CONACYT for the stipend received.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Misra, S.; Boylan, M.; Selvam, A.; Spallholz, J.E.; Björnstedt, M. Redox-active selenium compounds from toxicity and cell death to cancer treatment. Nutrients 2015, 7, 3536–3556. [Google Scholar] [CrossRef] [PubMed]
- Vinceti, M.; Filippini, T.; Wise, L.A. Environmental selenium and human health: An update. Curr. Environ. Health Rep. 2018, 5, 464–485. [Google Scholar] [CrossRef] [PubMed]
- Pescuma, M.; Gomez-Gomez, B.; Perez-Corona, T.; Font, G.; Madrid, Y.; Mozzi, F. Food prospects of selenium enriched-Lactobacillus acidophilus CRL 636 and Lactobacillus reuteri CRL 1101. J. Funct. Foods 2017, 35, 466–473. [Google Scholar] [CrossRef]
- Maseko, T.; Callahan, D.L.; Dunshea, F.R.; Doronila, A.; Kolev, S.D.; Ng, K. Chemical characterisation and speciation of organic selenium in cultivated selenium-enriched Agaricus bisporus. Food Chem. 2013, 141, 3681–3687. [Google Scholar] [CrossRef] [PubMed]
- Galano, E.; Mangiapane, E.; Bianga, J.; Palmese, A.; Pessione, E.; Szpunar, J.; Lobinski, R.; Amoresano, A. Privileged incorporation of selenium as selenocysteine in Lactobacillus reuteri proteins demonstrated by selenium-specific imaging and proteomics. Mol. Cell Proteom. 2013, 12, 2196–2204. [Google Scholar] [CrossRef]
- Yang, J.; Wang, J.; Yang, K.; Liu, M.; Qi, Y.; Zhang, T.; Fan, M.; Wei, X. Antibacterial activity of selenium-enriched lactic acid bacteria against common food-borne pathogens in vitro. J. Dairy Sci. 2017, 101, 1930–1942. [Google Scholar] [CrossRef]
- Turner, R.T.; Weiner, J.H.; Taylor, D.E. Selenium metabolism in Escherichia coli. Biometals 1998, 11, 223–227. [Google Scholar] [CrossRef]
- Rigger, L.; Schmidt, R.L.; Holman, K.M.; Simonović, M.; Micura, R. The synthesis of methylated, phosphorylated, and phosphonated 3′-aminoacyl-tRNASec mimics. Chem. Eur. J. 2013, 19, 15872–15878. [Google Scholar] [CrossRef]
- Castañeda-Ovando, A.; Segovia-Cruz, J.A.; Flores-Aguilar, J.F.; Rodríguez-Serrano, G.M.; Salazar-Pereda, V.; Ramírez-Godínez, J.; Contreras-López, E.; Jaimez-Ordaz, J.; González-Olivares, L.G. Serine-enriched minimal medium enhances conversion of selenium into selenocysteine by Streptococcus thermophilus. J. Dairy Sci. 2019, 102, 6781–6789. [Google Scholar] [CrossRef]
- Mariotti, M.; Ridge, P.G.; Zhang, Y.; Lobanov, A.V.; Pringle, T.H.; Guigo, R.; Hatfield, D.L.; Gladyshev, V.N. Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS ONE 2012, 7, e33066. [Google Scholar] [CrossRef]
- Santesmasses, D.; Mariotti, M.; Guigó, R. Computational identification of the selenocysteine tRNA (tRNASec) in genomes. PLoS Comput. Biol. 2017, 13, e1005383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Turanov, A.A.; Hatfield, D.L.; Gladyshev, V.N. In silico identification of genes involved in selenium metabolism: Evidence for a third selenium utilization trait. BMC Genom. 2008, 9, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Estrada, A.M.; Olivares, L.G.G.; López, E.C.; Serrano, G.R. SelA and SelD genes involved in selenium absorption metabolism in lactic acid bacteria isolated from Mexican cheeses. Int. Dairy J. 2020, 103, 104629. [Google Scholar] [CrossRef]
- Palomo, M.; Gutiérrez, A.M.; Pérez-Conde, M.C.; Cámara, C.; Madrid, Y. Se metallomics during lactic fermentation of Se-enriched yogurt. Food Chem. 2014, 164, 371–379. [Google Scholar] [CrossRef]
- Deng, Y.; Man, C.; Fan, Y.; Wang, Z.; Li, L.; Ren, H.; Cheng, W.; Jiang, Y. Preparation of elemental selenium-enriched fermented milk by newly isolated Lactobacillus brevis from kefir grains. Int. Dairy J. 2015, 44, 31–36. [Google Scholar] [CrossRef]
- Calomme, M.R.; Van den Branden, K.; Vanden Berghe, D.A. Selenium and Lactobacillus species. J. Appl. Microbiol. 1995, 79, 331–340. [Google Scholar] [CrossRef]
- Alzate, A.; Cañas, B.; Pérez-Munguía, S.; Hernández-Mendoza, H.; Pérez-Conde, C.; Gutiérrez, A.M.; Cámara, C. Evaluation of the inorganic selenium biotransformation in selenium-enriched yogurt by HPLC-ICP-MS. J. Agric. Food Chem. 2007, 55, 9776–9783. [Google Scholar] [CrossRef]
- Krittaphol, W.; Wescombe, P.A.; Thomson, C.D.; McDowell, A.; Tagg, J.R.; Fawcett, J.P. Metabolism of L-selenomethionine and selenite by probiotic bacteria: In vitro and in vivo studies. Biol. Trace Elem. Res. 2011, 144, 358–1369. [Google Scholar] [CrossRef]
- Yin, X.; Zhou, Y.; Yang, H.; Liao, Y.; Ma, T.; Wang, F. Enhanced selenocysteine biosynthesis for seleno-methylselenocysteine production in Bacillus subtilis. Appl. Microbiol. Biotechnol. 2023, 107, 2843–2854. [Google Scholar] [CrossRef]
- Escobar-Ramírez, M.C.; Jaimez-Ordaz, J.; Escorza-Iglesias, V.A.; Rodríguez-Serrano, G.M.; Contreras-López, E.; Ramírez-Godínez, J.; Castañeda-Ovando, A.; Morales-Estrada, A.I.; Felix-Reyes, N.; González-Olivares, L.G. Lactobacillus pentosus ABHEAU-05: An in vitro digestion resistant lactic acid bacterium isolated from a traditional fermented Mexican beverage. Rev. Argent Microbiol. 2020, 52, 305–314. [Google Scholar] [CrossRef]
- Escobar-Ramírez, M.C.; Rodríguez-Serrano, G.M.; Salazar-Pereda, V.; Castañeda-Ovando, A.; Pérez-Escalante, E.; Jaimez-Ordaz, J.; González-Olivares, L.G. Biogenic production of selenocysteine by Enterococcus faecium ABMC-05: An indigenous lactic acid bacterium from fermented Mexican beverage. Food Sci. Technol. 2022, 43, e63622. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Strahsburger, E.; Retamales, P.; Estrada, J.; Seeger, M. Microdot method: Used with chromogenic agar is a useful procedure for sanitary monitoring in aquaculture. Lat. Am. J. Aquat. Res. 2016, 44, 742–749. [Google Scholar] [CrossRef]
- Turło, J.; Gutkowska, B.; Malinowska, E. Relationship between the selenium, selenomethionine, and selenocysteine content of submerged cultivated mycelium of Lentinula edodes (Berk.). Acta Chromatogr. 2007, 18, 36–48. [Google Scholar]
- Vázquez-Ortíz, F.A.; Caire, G.; Higuera-Ciapara, I.; Hernández, G. High-performance liquid chromatographic determination of free amino acids in shrimp. J. Liq. Chromatogr. 1995, 18, 2059–2068. [Google Scholar] [CrossRef]
- Jones, B.N. Amino acid analysis by o-phthaldialdehyde precolumn derivatization and reverse-phase HPLC. In Methods Protein Microcharacterization: Biological Methods, 1st ed.; Shively, J.E., Ed.; Humana Press: Totowa, NJ, USA, 1986; pp. 121–151. [Google Scholar] [CrossRef]
- Castillo-Portela, G.; Villar-Delgado, J.; Montano-Martínez, R.; Martínez, C.; Pérez-Alfocea, F.; Albacete, A.; Sánchez-Bravo, J.; Acosta-Echeverria, M. Cuantificación por HPLC del contenido de aminoácidos presentes en el FOTOMAS-E. ICIDCA Sobre Los Deriv. Caña Azúcar 2011, 45, 64–67. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic Note: A Place for DNA-DNA Reassociation and 16s rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Gutiérrez-Sarmiento, W.; Peña-Ocaña, B.A.; Lam-Gutiérrez, A.; Guzmán-Albores, J.M.; Jasso-Chávez, R.; Ruíz-Valdiviezo, V.M. Microbial community structure, physicochemical characteristics and predictive functionalities of the Mexican tepache fermented beverage. Microbiol. Res. 2022, 260, 127045. [Google Scholar] [CrossRef]
- Moreno-Terrazas, R.D. Determinación de las características microbiológicas, bioquímicas y sensoriales para la estandarización del proceso de elaboración de tepache. Doctoral Dissertation, Universidad Autónoma Metropolitana (Unidad Xochimilco), México City, Mexico, 27 January 2005. [Google Scholar]
- Cervantes-Contreras, M.; Pedroza, A.M. Caracterización microbiológica del pulque y cuantificación de su contenido de etanol mediante espectroscopia Raman. Superf. Y Vacio. 2008, 21, 1–5. [Google Scholar]
- de la Fuente-Salcido, N.M.; Castañeda-Ramírez, J.C.; García-Almendárez, B.E.; Bideshi, D.K.; Salcedo-Hernández, R.; Barboza-Corona, J.E. Isolation and characterization of bacteriocinogenic lactic bacteria from M-Tuba and Tepache, two traditional fermented beverages in México. Food Sci. Nutr. 2015, 3, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Pusztahelyi, T.; Kovács, S.; Pócsi, I.; Prokisch, J. Selenite-stress selected mutant strains of probiotic bacteria for Se source production. J. Trace Elem. Med. Biol. 2015, 30, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Wang, C. Factors affecting selenium-enrichment efficiency, metabolic mechanisms and physiological functions of selenium-enriched lactic acid bacteria. J. Future Foods 2022, 2, 285–293. [Google Scholar] [CrossRef]
- Martínez, F.G.; Cuencas-Barrientos, M.E.; Mozzi, F.; Pescuma, M. Survival of selenium-enriched lactic acid bacteria in a fermented drink under storage and simulated gastro-intestinal digestion. Food Res. Int. 2019, 123, 115–124. [Google Scholar] [CrossRef]
- Krausova, G.; Kana, A.; Hyrslova, I.; Mrvikova, I.; Kavkova, M. Development of selenized lactic acid bacteria and their selenium bioaccummulation capacity. Fermentation. 2020, 6, 91. [Google Scholar] [CrossRef]
- Kim, E.K.; Cha, C.J.; Cho, Y.J.; Cho, Y.B.; Roe, J.H. Synthesis of gama-glutamylcysteine as a major low-molecular-weight thiol in lactic acid bacteria Leuconostoc spp. Biochem. Biophys. Res. Commun. 2008, 369, 1047–1051. [Google Scholar] [CrossRef]
- Masip, L.; Veeravalli, K.; Georgiou, G. The many faces of glutathione in bacteria. Antioxid. Redox Signal. 2006, 8, 753–762. [Google Scholar] [CrossRef]
- Xia, S.K.; Chen, L.; Liang, J.Q. Enriched selenium and its effects on growth and biochemical composition in Lactobacillus bulgaricus. J. Agric. Food Chem. 2007, 55, 2413–2417. [Google Scholar] [CrossRef]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Cecconi, D.; Monti, F.; Micaroni, M.; Turner, R.J.; Butler, C.S.; Vallini, G. Selenite biotransformation and detoxification by Stenotrophomonas maltophilia SeITE02: Novel clues on the route to bacterial biogenesis of selenium nanoparticles. J. Hazar. Mater. 2017, 324, 3–14. [Google Scholar] [CrossRef]
- Martínez, F.G.; Moreno-Martin, G.; Pescuma, M.; Madrid-Albarrán, Y.; Mozzi, F. Biotransformation of selenium by lactic acid bacteria: Formation of seleno-nanoparticles and seleno-amino acids. Front. Bioeng. Biotechnol. 2020, 8, 506. [Google Scholar] [CrossRef] [PubMed]
- Pieniz, S.; Andreazza, R.; Mann, M.B.; Camargo, F.; Brandelli, A. Bioaccumulation and distribution of selenium in Enterococcus durans. J. Trace Elem. Med. Biol. 2017, 40, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Stabnikova, O.; Khonkiv, M.; Kovshar, I.; Stabnikov, V. Biosynthesis of selenium nanoparticles by lactic acid bacteria and areas of their possible applications. World J. Microbiol. Biotechnol. 2023, 39, 230. [Google Scholar] [CrossRef]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biogenic selenium nanoparticles: Current status and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 2555–2566. [Google Scholar] [CrossRef] [PubMed]
- Palomo-Siguero, M.; Gutiérrez, A.M.; Pérez-Conde, C.; Madrid, Y. Effect of selenite and selenium nanoparticles on lactic bacteria: A multi-analytical study. Microchem. J. 2016, 126, 488–495. [Google Scholar] [CrossRef]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Bernardi, P.; Butler, C.S.; Vallini, G. Delayed formation of zero-valent selenium nanoparticles by Bacillus mycoides SeiTE01 as a consequence of selenite reduction under aerobic conditions. Microb. Cell Factories 2014, 13, 35–49. [Google Scholar] [CrossRef]
- Fernández-Llamosas, H.; Castro, L.; Blázquez, M.L.; Díaz, E.; Carmona, M. Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci. Rep. 2017, 7, 16046. [Google Scholar] [CrossRef]
- Kousha, M.; Yeganeh, S.; Amirkolaie, A.K. Effect of sodium selenite on the bacteria growth, selenium accumulation, and selenium biotransformation in Pediococcus acidilactici. Food Sci. Biotechnol. 2017, 26, 1013–1018. [Google Scholar] [CrossRef]
- Sarret, G.; Avoscan, L.; Carrière, M.; Collins, R.; Geoffroy, N.; Carrot, F.; Covès, J.; Gouget, B. Chemical forms of selenium in the metal-resistant bacterium Ralstonia metallidurans CH34 exposed to selenite and selenate. Appl. Environ. Microbiol. 2005, 71, 2331–2337. [Google Scholar] [CrossRef]
- Kessi, J.; Hanselmann, K.W. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J. Biol. Chem. 2004, 279, 50662–50669. [Google Scholar] [CrossRef]
- Kessi, J.; Ramuz, M.; Wehrli, E.; Spycher, M.; Bachofen, R. Reduction of selenite and detoxification of elemental selenium by the phototrophic bacterium Rhodospirillum rubrum. Appl. Environ. Microbiol. 1999, 65, 4734–4740. [Google Scholar] [CrossRef] [PubMed]
- Pophaly, S.D.; Singh, R.; Pophaly, S.D.; Kaushik, J.K.; Tomar, S.K. Current status and emerging role of glutathione in food grade lactic acid bacteria. Microb. Cell Factories 2012, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Scortecci, J.F.; Serrão, V.H.B.; Fernandes, A.F.; Basso, L.G.M.; Gutierrez, R.F.; Araujo, A.P.U.; Neto, M.O.; Thiemann, O.H. Initial steps in selenocysteine biosynthesis: The interaction between selenocysteine lyase and selenophosphate synthetase. Int. J. Biol. Macromol. 2020, 156, 18–26. [Google Scholar] [CrossRef]
- Manzine, L.R.; Cassago, A.; da Silva, M.T.A.; Thiemann, O.H. An efficient protocol for the production of tRNA-free recombinant Selenocysteine Synthase (SELA) from Escherichia coli and its biophysical characterization. Protein Expr. Purif. 2013, 88, 80–84. [Google Scholar] [CrossRef]
- Müller, S.; Heider, J.; Böck, A. The path of unspecific incorporation of selenium in Escherichia coli. Arch. Microbiol. 1997, 168, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Young, P.A.; Kaiser, I.I. Aminoacylation of Escherichia coli cysteine tRNA by selenocysteine. Arch. Biochem. Biophys. 1975, 171, 483–489. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).