Abstract
Long non-coding RNAs (lncRNAs) can be specifically expressed in different tissues and cancers. By controlling the gene expression at the transcriptional and translational levels, lncRNAs have been reported to be involved in tumor growth and metastasis. Recent data demonstrated that multiple lncRNAs have a crucial role in renal cell carcinoma (RCC) progression—the most common malignant urogenital tumor. In the present study, we found a trend towards increased PROX1 antisense RNA 1 (PROX1-AS1) expression in RCC specimens compared to non-tumoral margins. Next, we found a positive correlation between PROX1-AS1 expression and the occurrence of distant and lymph node metastasis, higher tumor stage (pT1 vs. pT2 vs. pT3–T4) and high-grade (G1/G2 vs. G3/G4) clear RCC. Furthermore, global demethylation in RCC-derived cell lines (769-P and A498) and human embryonic kidney 293 (HEK293) cells induced a significant increase of PROX1-AS1 expression level, with the most remarkable change in HEK293 cells. In line with this evidence, bisulfite sequencing analysis confirmed the specific demethylation of bioinformatically selected CpG islands on the PROX1-AS1 promoter sequence in the HEK293 cell line but not in the tumor cells. Additionally, the human specimen analysis showed the hemimethylated state of CG dinucleotides in non-tumor kidney tissues, whereas the tumor samples presented the complete, partial, or no demethylation of CpG-islands. In conclusion, our study indicated that PROX1-AS1 could be associated with RCC progression, and further investigations may define its role as a new diagnostic marker and therapeutic target.
1. Introduction
Renal cell carcinoma (RCC) is a deadly genitourinary malignancy characterized by metastases and chemotherapy resistance [1]. The major RCC histological variants are clear cell renal cell carcinoma (KIRC), accounting for 70% of cases, papillary renal cell carcinoma (KIRP) ~10–15%, and chromophobe cell carcinoma (KICH) ~5% and others ~15% [2,3]. Tyrosine kinase inhibitors, such as sunitinib, axitinib, and pazopanib, inhibiting the vascular endothelial growth factor receptor cascade, represent the first-line treatment for this disease. Unfortunately, RCC eventually develops resistance to these medications, so novel therapeutic strategies and approaches are urgently required [4]. Understanding RCC molecular basis is paramount for discovering new therapeutic interventions against this disease [1,5].
Long non-coding RNAs (lncRNAs) represent a group of molecules with a length of over 200 nucleotides that do not code for proteins, despite having a similar structure to functional mRNA [6]. LncRNAs display tissue-specific expression patterns and have been shown to affect a broad range of biological functions [7]. LncRNAs have initially been considered to regulate gene expression at the post-transcriptional level, but a growing body of evidence indicates they also play an important role in epigenetic control [8,9]. On the other hand, lncRNA expression can be regulated by epigenetic mechanisms, including methylation [10].
It was already shown that lncRNAs play a vital role in the development of renal diseases, including fibrosis [11], autosomal dominant polycystic kidney disease [12], diabetic kidney disease [13], and RCC [14,15]. Recent data indicated that a series of lncRNA covers a regulatory function and may have a prognostic value in RCC (Table 1).

Table 1.
Long non-coding RNAs and their implications in renal cell carcinoma.
In this scenario, understanding lncRNA biology in kidney cancer disease may provide new diagnostic and therapeutic opportunities.
The most common renal cancer metastatic sites are the lungs (45%), followed by the bones (30%) and lymph nodes (22%) [22]. It was estimated that RCC tumor lymph node infiltration is associated with distant metastases occurrence, significantly reducing patient 5-year survival [23]. In this context, one of the primary lymphatic markers—Prospero homeobox 1 (PROX1)—was detected in RCC-derived cells as a factor correlated with elevated tumor aggressiveness and lymph node invasion [24].
Both PROX1 and its antisense strand—PROX1-AS1 (3399 bp) located on human chromosome 1q32.3 (Figure 1), can be actively involved in tumor progression [25,26,27,28].
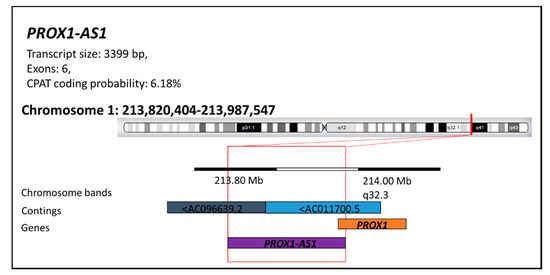
Figure 1.
The PROX1 antisense RNA 1 structure (PROX1-AS1). The PROX1-AS1 presents a transcript of 3399 bp, including six exons with a faint (6.18%) probability of protein translation (according to the Coding-Potential Assessment Tool (CPAT)). LncRNA-PROX1-AS1 is located on the long arm of chromosome 1 at position 1q32.3 (start position: 213820404; end position: 213987547) in the regions AC096639.2-AC011700.5 of the DNA segments. This information is available at LNCipedia [29], RNA central [30], and Ensemble [31] databases.
Previous studies demonstrated a significantly increased PROX1-AS1 expression in papillary thyroid cancer, prostate, and ovarian tumor specimens compared to adjacent non-tumoral tissues. Moreover, an enhanced PROX1-AS1 expression increased the invasion of different cancer cells in vitro [28,32,33].
Herein, we investigated and correlated RCC specimen PROX1-AS1 expression to control tissues, distant and lymph node metastatic events, higher tumor stages, and high-grade tumors. Furthermore, the bioinformatic simulation and data derived from in vitro and ex-vivo experiments indicate that an epigenetic mechanism could be involved in PROX1-AS1 expression.
2. Materials and Methods
2.1. Tissues Samples
The samples included in this study comprised RCC (T) and the surrounding non-cancerous kidney (NT) tissues. Fresh samples were snap-frozen in liquid nitrogen and stored at −80 °C. A partial nephrectomy was performed for localized disease, including 3–5 mm of peritumoral margin collected outside the cancer tissue and tumor capsule. All removed structures were morphologically examined. The samples derived from patients with pT1-2N0M0 renal masses treated with laparoscopic and robotic partial nephrectomy (n = 28 (67%)) and patients with pT3-4 N0-1 M0-1 renal masses treated with laparoscopic radical nephrectomy (n = 14 (33%)) at the Institute for Urology and Reproductive Health, Sechenov University, Moscow, Russia between 2019 and 2020.
The research was carried out following the ethical proceedings approved by the Ethical Committees of Sechenov University, Moscow, Russia. The samples were collected from 42 patients, including 22 males and 20 females, with a median age of 58.8 years (range 26–80 years). Seven patients developed metastasis, and three had lymph node tumor invasion. Lymph node metastasis were detected before surgery via CT scan examination. If the lymph nodes were more than 1.0–2.0 cm, lymphadenectomy followed by histopathological examination was performed. RCC sample postoperative histopathological verifications were performed according to the International Agency for Research on Cancer (2016) WHO classification of tumors of the urinary system and male genital organs (IARC WHO classification of tumors), 4th ed. Written informed consents were received from all participant patients before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee at Sechenov University, Moscow, Russia of N04–12.
2.2. Cell Lines
Human 786-P, A498 RCC cell lines, and embryonic kidney cells HEK293 were kindly provided by Dr. Vadim Pokrovsky (purchased from American Type Culture Collection). The cells were grown in RPMI 1640 or DMEM/F12 medium supplemented with 10% fetal bovine serum and 1% mixture of antibiotics penicillin-streptomycin (all from Gibco, Waltham, MA, USA) at 5% CO2 and 37 °C in a humidified atmosphere containing 5% CO2. Cells were cultivated as it was previously described [34]. All tested cell lines were authenticated by STR DNA Profiling Analysis (GORDIZ, Moscow, Russia).
2.3. RNA Isolation and Real-Time (RT)-qPCR
Tissues were disintegrated and fragmentized using TissueLyser LT or TissueLyser II and steel beads (Qiagen, Hilden, Germany). Total RNA was extracted from kidney specimens and cultured cells using the Total RNA isolation kit (Evrogen, Moscow, Russia) according to the manufacturer’s protocol. RNA quality and concentration were evaluated using NanoDrop spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). Samples with the A260/A280 ratio ~2.0 were used for further analyses and stored at −80 °C.
Complementary DNA (cDNA) was transcribed from mRNA using cDNA synthesis kit (Evrogen, Moscow, Russia), according to the manufacturer’s protocols. For RT reaction 1 µg of total RNA was used with optical density OD260/OD280 1.7–2.0 measured with NanoDrop One (Thermo Fisher Scientific, Waltham, MA, USA). Expression of the human genes was quantified by RT-qPCR using the cDNAs as a template in reactions containing the double-stranded DNA-specific dye BioMaster HS-qPCR SYBR Blue (2x) (BiolabMix, Novosibirsk, Russia) and specific oligonucleotide primers: (1) PROX1-AS1: F-5′-CTAGTTAGCAGGGGCAGCAC-3′, R-5′-AACAGAGAGGCGTGGAAGAA-3′; (2) GAPDH F-5′-AGAAGGCTGGGGCTCATTTG-3′, AGGGGCCATCCACAGTCTTC-3′. PCR reactions were performed in triplicates with the following conditions: 95 °C/30 s, 40 cycles of 95 °C/5 s, 60 °C/15 s and 72 °C/10 s in iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The cycle threshold (Ct) values estimated for analyzed genes were normalized against corresponding Ct values of GAPDH gene. The relative quantification value (RQ) was calculated as the relative change in PROX1-AS1 transcript expression level compared to the PROX1-AS1 level in the internal control represented by a mixture of RNA extracted from control samples.
2.4. Methylation Analysis
The methylation simulation was performed in the 1q32.3 region (start position chr1: chr1: 213,985,153–end position: 214,000,000 on the Homo sapiens chromosome 1, GRCh38.p13 Primary Assembly) harboring the lncRNA-PROX1-AS1 (ncRNA: NR_037850.2) exon 1 and the gene’s promoter localized 3kB upstream from the PROX1-AS1 transcription start point. CpG sites prediction was performed using the online UCSC Genome Browser tool (http://genome.ucsc.edu; accessed on 9 April 2021).
2.5. In Vitro Treatment of Cultured Cells with 5-Aza-2′-Deoxycytidine
Cells were seeded at a density of 1.0 × 106 cells in T25 flasks and placed at 37 °C overnight in a 5% CO2 incubator. The next day, the cell culture medium was replaced with a fresh medium containing 5 μM 5-aza-2′-deoxycytidine 5-AZA and cultured for the next 48 h.
2.6. DNA Isolation, Bisulfite Modification and Sequencing PCR
Genomic DNA from kidney cancer patients and cell cultures HEK293, 786-P, and A498 were isolated using a standard phenol-chloroform method. Bisulfite conversion was performed using the EpiTect Bisulfite kit (Qiagen, Hilden, Germany) to distinguish the methylated from non-methylated cytosines in the DNA sequence.
Bisulfite Sequencing PCR
The fragments of CpG- islands (1, 2, 3) were amplified from the PROX1-AS1 promoter region, using specific primers for bisulfite sequencing PCR (Table 2). Bisulfite sequencing PCR was performed using BigDye™ Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, USA). DNA methylation status of the selected CpG islands were validated with bisulfite Sanger sequencing—on the automatic genetic analyzer 3500 (Thermo Fisher Scientific) according to the manufacturer’s protocols.

Table 2.
The sequence of primers for bisulfite sequencing PCR.
2.7. Statistical Analysis
Statistical analysis was performed using Statistica13.1 (StatSoft, Tulsa, OK, USA). When comparing independent groups, ANOVA Kruskal–Wallis (for comparison of two groups), U Mann–Whitney tests (for comparison of three and more groups) with additional multiple comparison (two-sided) test were used to evaluate the relationship between lncRNA expression level (RQ) and patients’ characteristics (age and sex of the patient) and clinical features of the tumor (staging according to Tumour Node Metastasis (TNM), American Joint Committee on Cancer Staging (AJCC) and histopathological RCC subtypes. Correlation of lncRNA expression in RCCs and non-cancerous kidney tissues within groups were performed using Spearman’s rank correlation. The results of relative expression analysis (RQ values) are presented as mean ± SD for normal distribution. p < 0.05 was considered statistically significant.
3. Results
3.1. LncRNA PROX1-AS1 Expression Is Higher in RCC Specimens and Positively Correlates with Metastasis, Lymph Node Invasion, Tumor Stage, and Grade
To evaluate the prognostic role of PROX1-AS1 in RCC progression, we investigated its expression in 42 RCC (T) samples and the corresponding surrounding non-cancerous kidney tissues (NT) (Table 3).

Table 3.
Patient clinical and pathological features and PROX1-AS1 lncRNA expression. The relative quantification value (RQ) was calculated as the relative change in PROX1-AS1 transcript expression level compared to the PROX1-AS1 level in the internal control represented by a mixture of RNA extracted from control samples.
PROX1-AS1 expression analysis showed an increase in T compared to paired NT tissues in the case of KIRC and KICH groups and a decrease in KIRP and renal angiomyolipoma (AML). However, the revealed differences were found to be not statistically significant. A positive correlation between PROX1-AS1 expression in T and NT was notified for all studied groups, containing KIRC, KICH, KIRP, and AML patients (Spearman’s rank correlation; R = 0.307, p < 0.05). Additionally, a positive correlation of PROX1-AS1 relative expression was observed in KIRC patients with metastasis (M1) between T and NT (Spearman’s rank correlation; R = 0.48, p < 0.05). Interestingly in women, the PROX1-AS1 level was higher in T than in NT, while in men, it was the opposite; however, changes were determined to be not statistically significant.
The correlation with the clinicopathological data showed a remarkably higher relative expression of PROX1-AS1 in patients with distant metastasis (U Mann-Whitney test, p = 0.0435; Figure 2A) and lymph node invasion (U Mann-Whitney test, p = 0.04; Figure 2B). We also observed that PROX-AS1 expression increased in pT2-pT4 compared to pT1. PROX-AS1 expression was significantly elevated in pT2 (compared to pT1, p = 0.03), but not in pT3-4 (Figure 2C), showing the typical bell-shaped distribution of PROX1-AS1 expression for tumor stage. Furthermore, among KIRC patients, PROX1-AS1 expression in T positively correlated with the higher grades G3/G4 compared to G1/G2 (Wilcoxon pair order test, p = 0.04, Figure 2D).
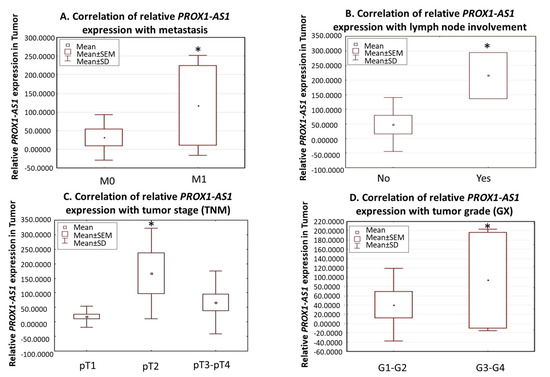
Figure 2.
Expression of PROX1-AS1 in human renal cell carcinomas. (A) PROX1-AS1 expression is higher in patients with metastases (M1) compared to patients without metastasis (M0). (B) Increased PROX1-AS1 expression corresponded to lymph node tumor infiltration. (C) PROX1-AS1 increased in the higher tumor stages (pT1 vs. pT2 and pT3–4). (D) PROX1-AS1 significantly increased in G3–G4 vs. G1–G2 grades in clear renal cell carcinomas. * p < 0.05.
3.2. Methylation Could Regulate PROX1-AS1 Expression in Healthy Kidney and Renal Carcinoma Cells
To investigate the possible relationship between the methylation and lncRNA PROX1-AS1 expression we performed a bioinformatic analysis of the lncRNA PROX1-AS1 promotor region, searching for CpG islands (Figure 3).
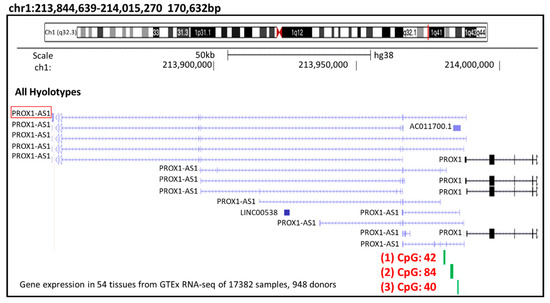
Figure 3.
PROX1-AS1 promoter region CpG island analysis. In the methylation simulation performed in the lncRNA-PROX1-AS1 exon 1 and the gene’s promoter (start position chr1: 213,985,153–end position: 214,000,000, accession data: >NC_000001.11:213985153-214000000 Homo sapiens chromosome 1, GRCh38.p13 Primary Assembly), three CpG islands (1: CpG 42; 2: CpG 84 and 3: CpG 40) were selected with UCSC Genome Browser (http://genome-euro.ucsc.edu/; accessed on 9 April 2021).
In the methylation simulation conducted in the lncRNA-PROX1-AS1 exon 1 and the gene’s promoter (localized 3 kB upstream from the PROX1-AS1 transcription start point) using UCSC Genome Browser (http://genome-euro.ucsc.edu/; accessed on 9 April 2021), three CpG islands located in the PROX1-AS1 promoter sequence were selected for further analysis (Table 4).

Table 4.
The location and characteristics of the islands.
The treatment of human RCC-derived (769-P and A498) and embryonic kidney cells (HEK293) with the demethylating agent 5-aza-2′-deoxycytidine resulted in a notable increase of PROX1-AS1 expression level (Figure 4) with the most significant change in HEK293 cells. These results indicated that methylation could contribute to PROX1-AS1 expression regulation in renal carcinomas and non-tumor kidney cells.
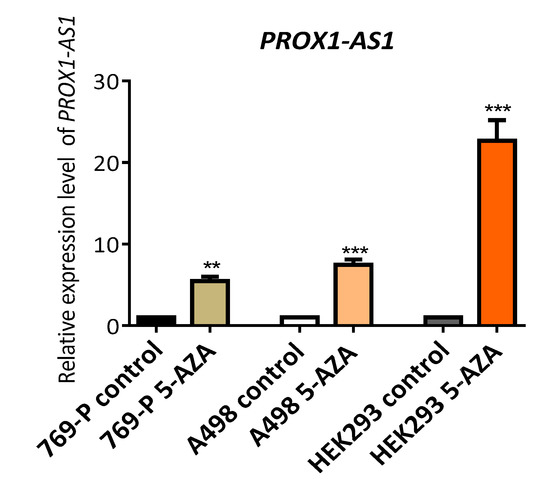
Figure 4.
The demethylation induced by 5-aza-2′-deoxycytidine increased the expression of PROX1-AS1 in human renal carcinoma cells (769-P and A498) and embryonic kidney 293 cells (HEK293). PROX1-AS1 expression was determined by qRT-PCR following exposure of cells with 5 uM 5-aza-2′-deoxycytidine for 48 h. ** p ≤ 0.01, *** p ≤ 0.001.
After bisulfite sequencing of the selected CpG-islands, 1 (CpG 42), 2 (CpG 84), and 3 (CpG 40), the presence of cytosine hypermethylation was observed (Table 5.).

Table 5.
CpG-islands methylation (methC) and demethylation (demethC) before and after treatment with the demethylating agent 5-aza-2′-deoxycytidine.
In particular, cytosine demethylation in CG dinucleotides was detected in CpG-islands 1 and 3 in the HEK293 line, confirming the hypothesis of a relationship between cytosine demethylation and overexpression of PROX-AS1 (Figure 5). The increase of PROX1-AS1 expression in RCC-derived cells (796-P and A498) after the same treatment could indicate that this effect was due to other phenomena (i.e., transcription factors, other non-coding RNA, etc.).
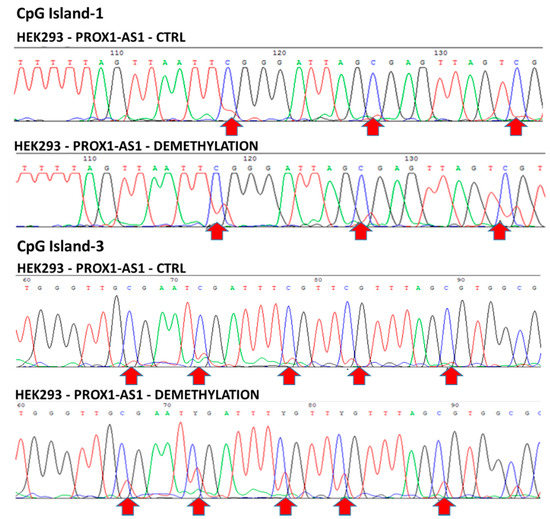
Figure 5.
CpG-islands 1 and 3 methylation in HEK293 before and after 5-aza-2′-deoxycytidine treatment. CpG island 1 and CpG island 3 before (top line) and after (bottom line) HEK293 treatment with 5-aza-2′-deoxycytidine. Arrows indicate the location of the demethylated cytosines after the treatment with the demethylating agent (the appearance of red peaks in the bottom line corresponds to cytosine demethylation).
To investigate the methylation status of all the three islets of the PROX1-AS1 gene in patient tumor tissues, we performed bisulfite sequencing of nine paired T and NT tissues. We detected CG dinucleotide hemimethylated in normal non-tumor tissues. On the other hand, in some tumor samples, complete CpG-island 3 demethylation and partial CpG-island 1 demethylation were observed (Figure 6). In the other tumor samples, no demethylation was detected for CpG-islands 1 and 3. These results support our observation of decreased expression of PROX1-AS1 in control samples and variable higher expression in tumor specimens.
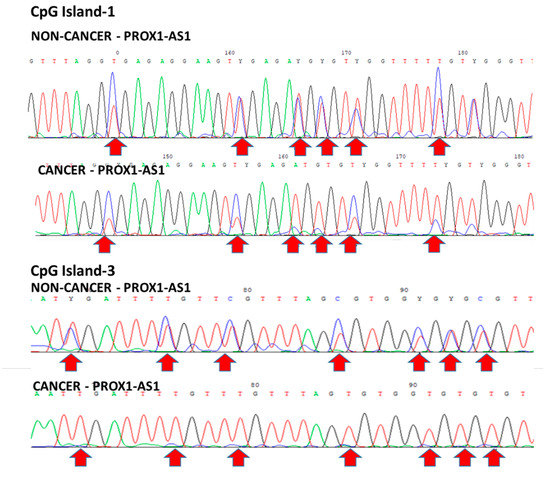
Figure 6.
Bisulfite sequencing of paired non-tumor and tumor tissues. CpG island 1: Top-line—hemimethylation in NT (blue and red peaks). Bottom line—differential methylation (blue or red peaks) in some CG in T. CpG island 3—hemimethylation in NT (blue and red peaks) and demethylation (only red peaks) in T.
4. Discussion
RCC is one of the most common malignant urogenital tumors with a high metastatic behavior. Tyrosine kinase inhibitors represent the first-line treatment for this disease even though drug resistance phenomena often occur against these therapeutic strategies [35]. Therefore, investigations regarding RCC biology may clarify the exact mechanisms underlying these processes, identify novel molecular targets and valuable diagnostic markers [36].
Increasing evidence suggests that lncRNAs play crucial roles in carcinogenesis, and their dysregulation is closely related to the tumor invasion process [37,38]. Recent studies have shown that lncRNAs may be targeted for tackling RCC growth and metastasis [39]. The dysregulation of different lncRNA expression was correlated with tumor proliferation, apoptosis, metastasis, and RCC patient outcome [14,15,17,18].
A few papers have reported the potential roles of PROX1-AS1 in cancer progression and its higher expression was noticed in ovarian, prostate, and papillary thyroid carcinoma specimens compared to control non-tumoral samples [28,32,33]. In vitro, PROX1-AS1 overexpression or silencing regulated the proliferation, migration, and invasion of different cancer cells. To provide more insights into the role of PROX1-AS1 in RCC, we correlated its expression in patient tumor samples with clinical data. This approach was justified by recent studies based on the coding gene—PROX1—and its regulatory role in RCC development and lymph node spreading [28].
In our analysis, the PROX1-AS1 expression in tumor tissues compared to healthy control samples derived from the paired cancer lesion burden varied between kidney cancer subtypes. However, the differences were insignificant. Additionally, the observed PROX1-AS1 increase in KIRP and AML controls can indicate a tissue-dependent expression manner.
The more important observation was the positive correlation (evaluated by Spearman’s rank-order correlation) between PROX1-AS1 expression in cancer and non-cancer tissues in the entire patient cohort analyzed and in the KIRC subtype. Its expression increase, observed in both tissues, may result from the growing cancer lesion influence on the surrounding healthy tissue. It is worth mentioning that the tumor margin is morphologically unchanged, but at the molecular level, it is affected by the growing lesion [40,41].
We observed differences in PROX1-AS1 expression level among gender and age groups; however, this was determined to not be statistically significant. Interestingly, PROX-AS1 expression in tumor and non-tumoral samples among gender showed very different trends between women and men, and this phenomenon could be caused by the differential hormonal regulation on long non-coding RNA expression [42,43].
PROX1-AS1 expression was significantly increased in patients with lymph node tumor infiltration and distant metastasis. In addition, compared to pT1 tumor stage, it increased in pT2 samples, while it decreased in pT3-pT4 samples. This bell-shaped distribution may be attributed to necrosis that occurs in higher tumor stages, affecting global measures of gene expression [44].
Furthermore, in KIRC subtype, PROX1-AS1 expression was elevated in higher tumor grade (G3–G4 vs. G1–G2), highlighting the role of PROX1-AS1 in tumor dissemination. This phenomenon was already observed for the coding PROX1 gene, regulating cancer cell dissemination via lymph and angiogenesis regulation. This evidence supports the hypothesis that PROX1-AS1 acts simultaneously on the lesion and the surrounding tissue, favoring further tumor growth.
All these observations suggest that lncRNA-PROX1-AS1 could be considered as RCC diagnostic marker, and more analysis is needed to determine its precise involvement in different RCC phenotypes and its potential role as a therapeutic target.
The genome-wide DNA methylation study in RCC identified increased global methylation in more aggressive cancers [45,46,47]. DNA methylation was shown in RCC as a potential risk factor associated with malignant transformation [48]. Furthermore, the aberrant hypermethylation correlated with the more advanced tumor stage and grade in KIRC [49]. On the other hand, cancer-associated DNA hypomethylation was as well registered, and related with the transcriptional activation of oncogenes and consequent tumor progression [50].
It was shown that lncRNAs could regulate gene expression at the transcriptional, post-transcriptional, and epigenetic levels [8,9], while epigenetic modifications, including hyper-/hypomethylation, can modulate lncRNA expression [10]. Functional analyses established that specific lncRNAs are epigenetically activated in tumors by the loss of methylation in CpG sites of their promoter region, and a few special lncRNAs are deactivated because of increased methylation [51,52]. In the context of RCC, genomic analysis of normal kidney tissues and RCC samples revealed a series of lncRNA which expression was regulated by hyper- or hypomethylation and they were associated with poor patient outcome [53,54].
For this reason, we analyzed the CpG islands in the promoter sequence detecting three CpG islands, which can mute the PROX1-AS1 expression. To confirm the epigenetic PROX1-AS1 regulation, we performed in vitro global demethylation. The treatment with 5-aza-2′-deoxycytidine significantly enhanced PROX1-AS1 expression in all the tested cell lines (769-P, A498, and HEK293) with the greatest change in human kidney HEK293 cells. We confirmed the hypermethylation of PROX1-AS1 in the selected CpG-islands in HEK293, 769-P, and A498 cell lines by bisulfite sequencing. However, after treatment with 5-aza-2′-deoxycytidine, cytosine demethylation in CG dinucleotides was detected only in HEK293 cells (CpG-islands 1 and 3), proving specific demethylation effect in kidney cells. We assume that the enhanced PROX1-AS1 expression after demethylation in tumor cell lines (769-P and A498) can be connected with other regulators, including transcription factors, non-coding RNAs, or genes activated upon global demethylation [55,56].
Next, we analyzed selected human specimens and detected (i) hemimethylated state of CG dinucleotides in all analyzed non-tumoral tissues, and (ii) complete, partial methylation or no methylation of CpG islands in RCC patients’ samples. These data can explain the lower PROX1-AS1 expression in controls and variable higher expression in RCC tissues. Those findings are in concordance with the previous observations of non-coding RNA regulation via hyper- and hypomethylation in both renal cell carcinoma and healthy kidney tissues [53].
All these observations pointed out the occurrence of a possible lnc-PROX1-AS1 epigenetic regulation and its active role in RCC progression.
5. Conclusions
In summary, the above results suggest the potential value of PROX1-AS1 in RCC development and its possible epigenetic modifications, which can affect its expression pattern. The presented data may contribute to a better understanding of PROX1-AS1 role in renal carcinoma development. Furthermore, our observations indicated that PROX1-AS1 could be considered as a new diagnostic/prognostic marker and a potential molecular target, which should be investigated in further studies.
To better understand the role of PROX1-AS1 in RCC progression, it is necessary to investigate the molecular mechanisms by which PROX1-AS1 regulates RCC malignant behavior in vitro and in vivo.
Author Contributions
Conceptualization, M.R.; methodology, M.R., S.V.M., Y.S., E.B.K.; data analysis, M.R., K.H.C.-C., E.B.K., M.V.N.; sample collections; D.O.K., A.Z.V., N.P.; draft preparation, M.R., K.H.C.-C., M.V.N.; proofreading, A.P.; supervision, A.A.Z.J.; funding acquisition, A.A.Z.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation (grant # 16-15-10410).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee at Sechenov University, Moscow, Russia of N04–12 on 11 April 2012.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data is not publicly available due to privacy restrictions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Buti, S.; Bersanelli, M.; Sikokis, A.; Maines, F.; Facchinetti, F.; Bria, E.; Ardizzoni, A.; Tortora, G.; Massari, F. Chemotherapy in metastatic renal cell carcinoma today? A systematic review. Anti-Cancer Drugs 2013, 24, 535–554. [Google Scholar] [CrossRef]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs—Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef]
- Low, G.; Huang, G.; Fu, W.; Moloo, Z.; Girgis, S. Review of renal cell carcinoma and its common subtypes in radiology. World J. Radiol. 2016, 8, 484–500. [Google Scholar] [CrossRef]
- Golovastova, M.O.; Korolev, D.O.; Tsoy, L.V.; Varshavsky, V.A.; Xu, W.-H.; Vinarov, A.Z.; Zernii, E.Y.; Philippov, P.P.; Zamyatnin, A.A. Biomarkers of Renal Tumors: The Current State and Clinical Perspectives. Curr. Urol. Rep. 2017, 18, 3. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Stamatakis, L.; Singer, E.A.; Srinivasan, R. Renal cell carcinoma. Curr. Opin. Oncol. 2014, 26, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.-W.; Huang, K.; Yang, C.; Kang, C.-S. Non-coding RNAs as regulators in epigenetics. Oncol. Rep. 2016, 37, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Tang, B. Inference of Crosstalk Effects between DNA Methylation and lncRNA Regulation in NSCLC. BioMed. Res. Int. 2018, 2018, 1–6. [Google Scholar] [CrossRef]
- Li, Z.; Tan, H.; Yu, H.; Deng, Z.; Zhou, X.; Wang, M. DNA methylation and gene expression profiles characterize epigenetic regulation of lncRNAs in colon adenocarcinoma. J. Cell. Biochem. 2019, 121, 2406–2415. [Google Scholar] [CrossRef]
- Zhou, Q.; Chung, A.C.; Huang, X.R.; Dong, Y.; Yu, X.; Lan, H.Y. Identification of Novel Long Noncoding RNAs Associated with TGF-β/Smad3-Mediated Renal Inflammation and Fibrosis by RNA Sequencing. Am. J. Pathol. 2014, 184, 409–417. [Google Scholar] [CrossRef]
- Aboudehen, K.; Farahani, S.; Kanchwala, M.; Chan, S.C.; Avdulov, S.; Mickelson, A.; Lee, D.; Gearhart, M.D.; Patel, V.; Xing, C.; et al. Long noncoding RNA Hoxb3os is dysregulated in autosomal dominant polycystic kidney disease and regulates mTOR signaling. J. Biol. Chem. 2018, 293, 9388–9398. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-L.; Hu, F.; Xue, M.; Jia, Y.-J.; Zheng, Z.-J.; Li, Y.; Xue, Y.-M. Early growth response protein-1 upregulates long noncoding RNA Arid2-IR to promote extracellular matrix production in diabetic kidney disease. Am. J. Physiol. Physiol. 2019, 316, C340–C352. [Google Scholar] [CrossRef]
- Xiao, Z.-D.; Han, L.; Lee, H.; Zhuang, L.; Zhang, Y.; Baddour, J.; Nagrath, D.; Wood, C.G.; Gu, J.; Wu, X.; et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lin-Hui, W.; Chen, C.; Liu, J.-Y.; Shi, J.-Z.; Liu, S.-P.; Liu, B.; Wu, D.-S.; Fang, Z.-Y.; Bao, Y.; Jiang, M.-M.; et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol. Cancer 2017, 16, 1–14. [Google Scholar] [CrossRef]
- Chen, Z.; Zhuang, Q.; Cheng, K.; Ming, Y.; Zhao, Y.; Ye, Q.; Zhang, S. Long non-coding RNA TCL6 enhances preferential toxicity of paclitaxel to renal cell carcinoma cells. J. Cancer 2020, 11, 1383–1392. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, P.; Lv, W.; Han, X.; Yang, J.; Qin, S. Long noncoding RNA lnc-DILC stabilizes PTEN and suppresses clear cell renal cell carcinoma progression. Cell Biosci. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Zhai, W.; Zhu, R.; Ma, J.; Gong, D.; Zhang, H.; Zhang, J.; Chen, Y.; Huang, Y.; Zheng, J.; Xue, W. A positive feed-forward loop between LncRNA-URRCC and EGFL7/P-AKT/FOXO3 signaling promotes proliferation and metastasis of clear cell renal cell carcinoma. Mol. Cancer 2019, 18, 1–15. [Google Scholar] [CrossRef]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Ding, J.; Chen, C.; Wu, Z.; Liu, B.; Gao, Y.; Chen, W.; Liu, F.; Sun, W.; Li, X.-F.; et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 2016, 29, 653–668. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, W.; Ma, T.; Lin, Z.; Liu, C.; Liu, Y.; Hou, F.F. lncRNA lnc-TSI Inhibits Metastasis of Clear Cell Renal Cell Carcinoma by Suppressing TGF-β-Induced Epithelial-Mesenchymal Transition. Mol. Ther. Nucleic Acids 2020, 22, 1–16. [Google Scholar] [CrossRef]
- Shields, L.B.; Kalebasty, A.R. Metastatic clear cell renal cell carcinoma in isolated retroperitoneal lymph node without evidence of primary tumor in kidneys: A case report. World J. Clin. Oncol. 2020, 11, 103–109. [Google Scholar] [CrossRef]
- Tadayoni, A.; Paschall, A.K.; Malayeri, A.A. Assessing lymph node status in patients with kidney cancer. Transl. Androl. Urol. 2018, 7, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Lv, T.; Liu, Y.; Zhang, J.; Xu, L.; Zhu, Y.; Yin, H.; An, H.; Lin, Z.; Xie, Y.; Chen, L. Impact of an Altered PROX1 Expression on Clinicopathology, Prognosis and Progression in Renal Cell Carcinoma. PLoS ONE 2014, 9, e95996. [Google Scholar] [CrossRef]
- Rudzińska, M.; Grzanka, M.; Stachurska, A.; Mikula, M.; Paczkowska, K.; Stępień, T.; Paziewska, A.; Ostrowski, J.; Czarnocka, B. Molecular Signature of Prospero Homeobox 1 (PROX1) in Follicular Thyroid Carcinoma Cells. Int. J. Mol. Sci. 2019, 20, 2212. [Google Scholar] [CrossRef] [PubMed]
- Rudzinska, M.; Ledwon, J.K.; Gawel, D.; Sikorska, J.; Czarnocka, B. The role of prospero homeobox 1 (PROX1) expression in follicular thyroid carcinoma cells. Oncotarget 2017, 8, 114136–114155. [Google Scholar] [CrossRef][Green Version]
- Rudzińska, M.; Mikula, M.; Arczewska, K.D.; Gajda, E.; Sabalińska, S.; Stępień, T.; Ostrowski, J.; Czarnocka, B. Transcription Factor Prospero Homeobox 1 (PROX1) as a Potential Angiogenic Regulator of Follicular Thyroid Cancer Dissemination. Int. J. Mol. Sci. 2019, 20, 5619. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Liao, C.H.; Tan, B.F.; Chen, Y.F.; Dang, B.W.; Chen, J.L.; Liu, C.B. LncRNA PROX1-AS1 promotes proliferation, invasion, and migration in papillary thyroid carcinoma. Biosci. Rep. 2018, 24, 2938–2944. [Google Scholar] [CrossRef]
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- RNAcentral Consortium; Sweeney, A.B.; Petrov, I.A.; Ribas, E.C.; Finn, R.D.; Bateman, A.; Szymanski, M.; Karlowski, W.M.; Seemann, E.S.; Gorodkin, J.; et al. RNAcentral 2021: Secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021, 49, D212–D220. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2020. Nucleic Acids Res. 2019, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, J.-F.; Tong, X.-J. Long noncoding RNA PROX1-AS1 promoted ovarian cancer cell proliferation and metastasis by suppressing KLF6. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6561–6568. [Google Scholar] [PubMed]
- Qian, C.; Liao, C.-H.; Tan, B.-F.; Chen, Y.-F.; Dang, B.-W.; Chen, J.-L.; Liu, C.-B. LncRNA PROX1-AS1 promotes proliferation, invasion, and migration in prostate cancer via targeting miR-647. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2938–2944. [Google Scholar]
- Rudzińska, M.; Parodi, A.; Maslova, V.D.; Efremov, Y.M.; Gorokhovets, N.V.; Makarov, V.A.; Popkov, V.A.; Golovin, A.V.; Zernii, E.Y.; Zamyatnin, J.A.A. Cysteine Cathepsins Inhibition Affects Their Expression and Human Renal Cancer Cell Phenotype. Cancers 2020, 12, 1310. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.J.; Purdue, M.P.; Signoretti, S.; Swanton, C.; Albiges, L.; Schmidinger, M.; Heng, D.Y.; Larkin, J.; Ficarra, V. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017, 3, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Eichelberg, C.; Junker, K.; Ljungberg, B.; Moch, H. Diagnostic and Prognostic Molecular Markers for Renal Cell Carcinoma: A Critical Appraisal of the Current State of Research and Clinical Applicability. Eur. Urol. 2009, 55, 851–863. [Google Scholar] [CrossRef]
- Zhao, J.; Li, L.; Han, Z.-Y.; Wang, Z.-X.; Qin, L.-X. Long noncoding RNAs, emerging and versatile regulators of tumor-induced angiogenesis. Am. J. Cancer Res. 2019, 9, 1367–1381. [Google Scholar]
- Balas, M.M.; Johnson, A.M. Exploring the mechanisms behind long noncoding RNAs and cancer. Non-Coding RNA Res. 2018, 3, 108–117. [Google Scholar] [CrossRef]
- Martens-Uzunova, E.S.; Böttcher, R.; Croce, C.M.; Jenster, G.; Visakorpi, T.; Calin, G.A. Long Noncoding RNA in Prostate, Bladder, and Kidney Cancer. Eur. Urol. 2014, 65, 1140–1151. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, A.J. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Miano, V.; Ferrero, G.; Reineri, S.; Caizzi, L.; Annaratone, L.; Ricci, L.; Cutrupi, S.; Castellano, I.; Cordero, F.; De Bortoli, M. Luminal long non-coding RNAs regulated by estrogen receptor alpha in a ligand-independent manner show functional roles in breast cancer. Oncotarget 2016, 7, 3201–3216. [Google Scholar] [CrossRef]
- Huang, P.-S.; Chang, C.-C.; Wang, C.-S.; Lin, K.-H. Functional roles of non-coding RNAs regulated by thyroid hormones in liver cancer. Biomed. J. 2020, 27, 2319–4170. [Google Scholar] [CrossRef]
- Bredholt, G.; Mannelqvist, M.; Stefansson, I.M.; Birkeland, E.; Bø, T.H.; Oyan, A.M.; Trovik, J.; Kalland, K.-H.; Jonassen, I.; Salvesen, H.B.; et al. Tumor necrosis is an important hallmark of aggressive endometrial cancer and associates with hypoxia, angiogenesis and inflammation responses. Oncotarget 2015, 6, 39676–39691. [Google Scholar] [CrossRef]
- Arai, E.; Kanai, Y. Genetic and epigenetic alterations during renal carcinogenesis. Int. J. Clin. Exp. Pathol. 2010, 4, 58–73. [Google Scholar] [PubMed]
- Arai, E.; Ushijima, S.; Tsuda, H.; Fujimoto, H.; Hosoda, F.; Shibata, T.; Kondo, T.; Imoto, I.; Inazawa, J.; Hirohashi, S.; et al. Genetic Clustering of Clear Cell Renal Cell Carcinoma Based on Array-Comparative Genomic Hybridization: Its Association with DNA Methylation Alteration and Patient Outcome. Clin. Cancer Res. 2008, 14, 5531–5539. [Google Scholar] [CrossRef] [PubMed]
- Arai, E.; Chiku, S.; Mori, T.; Gotoh, M.; Nakagawa, T.; Fujimoto, H.; Kanai, Y. Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis 2012, 33, 1487–1493. [Google Scholar] [CrossRef]
- Pérez, J.M.-; Gu, J.; Herrera, L.A.; Tannir, N.M.; Matin, S.F.; Karam, J.A.; Huang, M.; Chang, D.W.; Wood, C.G.; Wu, X. Genomic DNA Hypomethylation and Risk of Renal Cell Carcinoma: A Case–Control Study. Clin. Cancer Res. 2016, 22, 2074–2082. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, N.; Vallumsetla, N.; Zou, Y.; Galeas, J.N.; Shrivastava, M.; Hu, C.; Susztak, K.; Verma, A. Role of DNA methylation in renal cell carcinoma. J. Hematol. Oncol. 2015, 8, 1–13. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA methylation in cancer: Too much, but also too little. Oncogene 2002, 21, 5400–5413. [Google Scholar] [CrossRef] [PubMed]
- Morlando, M.; Fatica, A. Alteration of Epigenetic Regulation by Long Noncoding RNAs in Cancer. Int. J. Mol. Sci. 2018, 19, 570. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Zhao, L.; Zhao, X.; Li, Q.; An, Y.; Li, L.; Li, D. Genome-wide DNA methylation regulation analysis of long non-coding RNAs in glioblastoma. Int. J. Mol. Med. 2020, 46. [Google Scholar] [CrossRef]
- Malouf, G.; Zhang, J.; Tannir, N.M.; Thompson, E.; Spano, J.-P.; Khayat, D.; Su, X. Charting DNA methylation of long non-coding RNA in clear-cell renal cell carcinoma. J. Clin. Oncol. 2015, 33, 432. [Google Scholar] [CrossRef]
- Posa, I.; Carvalho, S.; Tavares, J.; Grosso, A.R. A pan-cancer analysis of MYC-PVT1 reveals CNV-unmediated deregulation and poor prognosis in renal carcinoma. Oncotarget 2016, 7, 47033–47041. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, X.; Liu, L.; Deng, H.; Zhang, J.; Xu, Q.; Cen, B.; Ji, A. Regulation of lncRNA expression. Cell. Mol. Biol. Lett. 2014, 19, 561–575. [Google Scholar] [CrossRef] [PubMed]
- López-Urrutia, E.; Montes, L.P.B.; Cervantes, D.L.D.G.; Pérez-Plasencia, C.; Campos-Parra, A.D. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front. Oncol. 2019, 9, 669. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).