Abstract
The taxonomy of the genus Fusarium has been in a flux because of ambiguous circumscription of species-level identification based on morphotaxonomic criteria. In this study, multigene phylogeny was conducted to resolve the evolutionary relationships of 88 Indian Fusarium isolates based on the internal transcribed spacer region, 28S large subunit, translation elongation factor 1-alpha, RNA polymerase second largest subunit, beta-tubulin and calmodulin gene regions. Fusarium species are well known to produce metabolites such as beauvericin (BEA) and enniatins. These identified isolates were subjected to fermentation in Fusarium-defined media for BEA production and tested using TLC, HPLC and HRMS. Among 88 isolates studied, 50 were capable of producing BEA, which varied from 0.01 to 15.82 mg/g of biomass. Fusarium tardicrescens NFCCI 5201 showed maximum BEA production (15.82 mg/g of biomass). The extract of F. tardicrescens NFCCI 5201 showed promising antibacterial activity against Staphylococcus aureus MLS16 MTCC 2940 and Micrococcus luteus MTCC 2470 with MIC of 62.5 and 15.63 µg/mL, respectively. Similarly, the F. tardicrescens NFCCI 5201 extract in potato dextrose agar (40 µg/mL) exhibited antifungal activity in the food poison technique against plant pathogenic and other fungi, Rhizoctonia solani NFCCI 4327, Sclerotium rolfsii NFCCI 4263, Geotrichum candidum NFCCI 3744 and Pythium sp. NFCCI 3482, showing % inhibition of 84.31, 49.76, 38.22 and 35.13, respectively. The antibiotic effect was found to synergize when Fusarium extract and amphotericin B (20 µg/mL each in potato dextrose agar) were used in combination against Rhizopus sp. NFCCI 2108, Sclerotium rolfsii NFCCI 4263, Bipolaris sorokiniana NFCCI 4690 and Absidia sp. NFCCI 2716, showing % inhibition of 50.35, 79.37, 48.07 and 76.72, respectively. The extract also showed satisfactory dose-dependent DPPH radical scavenging activity with an IC50 value of 0.675 mg/mL. This study reveals the correct identity of the Indian Fusarium isolates based on multigene phylogeny and also throws light on BEA production potential, suggesting their possible applicability in the medicine, agriculture and industry.
1. Introduction
Members of the genus Fusarium are ubiquitous and found in soil, air and on plants [1]. Fusarium is an important group of plant-pathogenic fungi which affect a vast diversity of crops in all climatic zones across the globe [2]. Being a genus of great concern, accurate and correct identification of its species becomes a necessity. However, its taxonomy has been in flux due to ambiguous circumscription based on morphotaxonomic criteria. Over the past two decades, many phylogenetic studies have established that morphological species recognition frequently fails to distinguish many Fusarium species [3]. Phylogenetic studies have revealed that nuclear ribosomal DNA (nrDNA) is nearly useless for species-level recognition in the case of Fusarium and related genera; these markers are useful only in the discrimination of Fusarium species complexes [4]. Discordance on using the nuclear rDNA internal transcribed spacer 2 (ITS2) gene region is because of the highly divergent ITS2 rDNA xenologs or paralogs discovered in many species’ complexes [5,6]. Many protein-coding genes have been explored for identification and taxonomic purposes in Fusarium and related genera. The two primary genes commonly used nowadays for identification that offer high discriminatory power and are well represented in public databases are translation elongation factor 1-alpha (tef-1α) and RNA polymerase second largest subunit (rpb2). Tef1-α is the most common marker, which has very good resolution power for most species [7], and Rpb2 provides enhanced discrimination between closely related species [8,9,10]; however, PCR amplification and sequencing success rate is better for Tef1-α than for Rpb2. In multigene phylogenetic analyses, additional genetic markers are often used with the previously mentioned genes, including β-tub, CaM and Rpb1; these markers vary in resolution depending on species complexes [11].
Fusarium spp. are known to produce several secondary metabolites including beauvericin (BEA) and enniatins. BEA is a cyclic hexadepsipeptide that consists of alternating D-2-hydroxyisovaleric acid and N-methylphenylalanine [12]. BEA is a potent bioactive compound that exhibits very effective anticancer, cytotoxic, antiplatelet aggregation, antimicrobial, leishmanicidal and insecticidal activities [13]. These activities are primarily due to its ionophoric properties that generally disrupt the normal physiological concentration of cations across the plasma membrane [14,15,16]. It releases Ca2+ from internal Ca2+ stores of the endoplasmic reticulum [17].
Studies prove that BEA can exhibit promising antibacterial activity against gram-positive or gram-negative bacteria, for example, Agrobacterium tumefaciens, Bacillus cereus, B. pumilus, B. sphaericus, B. subtilis, B. mycoides, Bifidobacterium adolescentis, Clostridium perfringens, Escherichia coli, Eubacterium biforme, Mycobacterium tuberculosis, Paenibacillus azotofixans, P. alvei, P. pulvifaciens, P. validus P. macquariensis, Peptostreptococcus productus, P. anaerobius, Pseudomonas lachrymans, Salmonella typhimurium, Staphylococcus haemolyticus, Vibrio fisheri and Xanthomonas vesicatoria are inhibited by BEA [18,19,20,21,22,23].
There has been public concern about certain opportunistic fungi that can cause infections; particularly, cases of Candida albicans are greater than before. Patients with compromised immune systems, such as those receiving organ transplants and cancer chemotherapy or those infected by human immunodeficiency virus (HIV), are more prone to such opportunistic infections [24]. BEA alone does not exhibit any antifungal activity; however, it has been found to enhance miconazole activity against C. albicans (wild and fluconazole-resistant) [25]. The activity of azole antifungals becomes enhanced against azole-resistant Candida isolates by BEA via inhibition of multidrug efflux [26]. The BEA with ketoconazole is known to synergize the antifungal effect against Candida albicans and C. tropicalis, signifying BEA’s possible use as a co-drug for antifungal infections [23,27,28]. A promising strategy using BEA, which involves simultaneous targeting drug resistance and morphogenesis, can boost antifungal efficacy against human fungal pathogens to combat life-threatening fungal infections.
In this study, 88 isolates of Fusarium and related genera from various substrates were identified by multigene phylogeny based on the internal transcribed spacer region, translation elongation factor 1-alpha, 28S large subunit, RNA polymerase second largest subunit, beta-tubulin and calmodulin gene regions. Further, these isolates were screened for beauvericin production. The isolate which showed maximum BEA production was subjected to large-scale fermentation, and its extract was used to study antibacterial, antifungal and antioxidant potential.
2. Materials and Methods
2.1. Isolates and Fungarium Specimens
First, 64 fungal strains identified as Fusarium based on morphology were procured from the National Fungal Culture Collection of India (NFCCI-WDCM 932) Table 1. The bacterial isolates used in this study, Escherichia coli MTCC 739, Bacillus subtilis MTCC 121, Micrococcus luteus MTCC 2470, Raoultella planticola MTCC 530, Pseudomonas aeruginosa MTCC 2453, Staphylococcus aureus MLS16 MTCC 2940 and Staphylococcus aureus MTCC 96, were procured from Microbial Type Culture Collection (MTCC). Additionally, fungal pathogens Pythium sp. NFCCI 3482, Geotrichum candidum NFCCI 3744, Rhizoctonia solani NFCCI 4327, Rhizopus sp. NFCCI 2108, Sclerotium rolfsii NFCCI 4263, Bipolaris sorokiniana NFCCI 4690 and Absidia sp. NFCCI 2716 were also procured from the National Fungal Culture Collection of India (NFCCI) to study the antifungal activity.

Table 1.
Sixty-four Fusarium isolates procured from the National Fungal Culture Collection of India (NFCCI), along with other details such as host, place of collection and date of isolation, are listed.
2.2. Collection, Isolation and Morphological Identification
During the course of this study, Fusarium and other related fungi were isolated from various substrates collected from different regions of India. Table 2 enlists the 24 isolates which were isolated afresh in this study along with details of their host, place of collection, date of collection and date of isolation. Pure cultures of all 24 isolates were raised using a single spore isolation technique.

Table 2.
Twenty-four isolates isolated in this study along with their accession nos., host, place of collection, date of collection and date of isolation details.
Colony characteristics of all Fusarium isolates were studied on multiple media, such as potato dextrose agar (PDA), malt extract agar (MEA), potato carrot agar (PCA), oatmeal agar (OMA), cornmeal agar (CMA) and synthetic nutrient agar (SNA). Microscopic structures of these isolates were observed using lactophenol-cotton blue under a Carl Zeiss Image Analyzer 2 (Germany) microscope. Measurements and photomicrographs of the fungal structures were recorded using Axiovision Rel 4.8 software and Digi-Cam with an attached Carl Zeiss Image Analyzer 2 microscope (Figure 1 and Figure 2). The pure cultures are deposited and accessioned in the National Fungal Culture Collection of India (NFCCI 5189-5212) (Table 2).
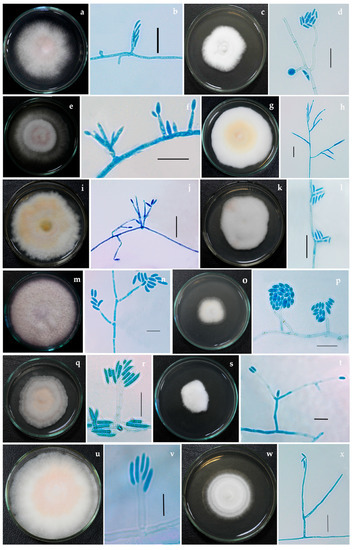
Figure 1.
Fusarium isolates with phialides bearing conidia (a,b) Fusarium nirenbergiae NFCCI 5189 (c,d) Neocosmospora solani NFCCI 2315 (e,f) F. mangiferae NFCCI 2885 (g,h) F. annulatum NFCCI 2962 (i,j) F. microconidium NFCCI 3020 (k,l) F. sacchari NFCCI 3147 (m,n) N. oblonga NFCCI 2150 (o,p) Albonectria rigidiuscula NFCCI 5202 (q,r) F. irregulare NFCCI 2460 (s,t) Neocosmospora suttoniana NFCCI 2961 (u,v) F. annulatum NFCCI 2964 (w,x) Neocosmospora vasinfecta NFCCI 2972. Scale bars: b, d, f, h, j, l, p, r, t, v, x = 20 μm; n = 10 μm.
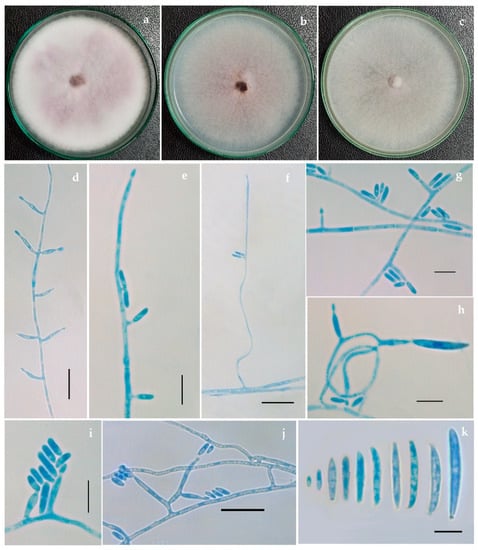
Figure 2.
Fusarium tardicrescens (NFCCI 5201) culture grown on (a) Potato dextrose agar (b) Malt extract agar (c) Cornmeal agar (d–j) Phialides arising from mycelial hyphae producing micro and macroconidia (k) Micro and macroconidia. Scale bars: d, f, j = 20 μm; e, g, h, i, k = 10 μm.
2.3. DNA Extraction, PCR Amplification and DNA Sequencing
Genomic DNA was isolated from pure colonies of all 88 isolates growing on Potato Dextrose Agar (PDA) petri-plates, incubated for a week’s time using a simple and rapid DNA extraction protocol using FastPrep®24 tissue homogenizer (MP Biomedicals GmbH, Eschwege, Germany) [29]. Partial gene sequences of isolates were determined for six gene markers, i.e., ITS, tef1-α, LSU, rpb2, β-tub and CaM. The primer sets used to amplify particular gene region are summarized in Table 3.

Table 3.
PCR primer sets used in this study for amplification as well as sequencing.
PCR was carried out in a 25 μL reaction using 12.5 μL 2X Invitrogen Platinum SuperFi PCR Mastermix, 2 μL template DNA (10–20 ng), 1.5 μL 10 pmol primer, 5 μL 5X GC enhancer and H2O (Sterile Ultra-Pure Water, Sigma, St. Louis, MO, USA), with the volume made to 25 μL. The conditions of the thermo-cycling involved:
An initial denaturation at 94 °C for 5 min, 35 cycles of 1 min at 94 °C, 30 s at 52 °C, 1 min at 72 °C and a final extension at 72 °C for 8 min for ITS gene region;
An initial denaturation of 5 min at 94 °C, 30 cycles of 45 s at 94 °C, 30 s at 57 °C and 1 min at 72 °C followed by a final 7 min extension at 72 °C for Tef1-α;
5 min denaturation at 94 °C, 35 cycles of 1 min at 94 °C, 50 s at 52 °C and 1.2 min at 72 °C with a final 8 min extension at 72 °C for LSU;
5 min denaturation at 95 °C, 35 cycles of 45 s at 95 °C, 1 min at 52 °C and 1.5 min at 72 °C with a final 10 min extension at 72 °C for Rpb2;
2 min denaturation at 94 °C, 40 cycles of 35 s at 94 °C, 55 s at 52 °C and 2 min at 72 °C with a final 10 min extension at 72 °C for β-tub;
An initial denaturation of 5 min at 94 °C, 30 cycles of 1 min at 94 °C, 30 s at 55 °C and 2 min at 72 °C followed by a final 7 min extension at 72 °C for CaM.
The PCR amplicons were purified with a FavorPrep™ PCR purification kit as per the manufacturer’s instructions. Purified PCR products of all marker genes were checked on 1.2% agarose electrophoresis gels stained with ethidium bromide and were further subjected to a sequencing PCR reaction using a BigDye®Terminator v3.1 Cycle Sequencing Kit, as per manufacturer’s instructions.
The sequencing PCR reaction of 20 µL included 4 µL of 5× sequencing buffer, 2 µL of BigDye™ Terminator premix, 4 µL of primer (5 pmol), 4 µL of the purified amplicon and H2O (Sterile Ultra-Pure Water, Sigma), with the volume made to 20 μL. Thermal cycling conditions consisted of an initial denaturing at 96 °C for 3 min, followed by 30 cycles of 94 °C for 10 s, 50 °C for 40 s and 60 °C for 4 min. The BigDye® terminators and salts were removed from using The BigDye Xterminator® Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) as per the manufacturer’s instructions. The purified sequencing products were transferred into a 96-well microplate. The sequence was elucidated on Applied Biosystems SeqStudio Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Sequences obtained were submitted in the NCBI GenBank (accession numbers in Table S1).
2.4. Phylogenetic Analysis
To determine the phylogenetic status of all 88 isolates, the Internal transcribed spacer region of the nrDNA (ITS), translation elongation factor 1-alpha (tef1-α), 28S large subunit of the nrDNA (LSU), RNA polymerase second largest subunit (rpb2), beta-tubulin (β-tub) and calmodulin (CaM) loci were used to compare the isolates used in this study with already known authentic strains in the genus Fusarium. The sequences of the type/reference strains were retrieved from NCBI. A total of 186 isolates of Fusarium and related genera were used in the phylogenetic analysis (Table S1). Atractium stilbaster CBS 410.67 was selected as the outgroup taxon. The chosen strains used in the construction of the phylogenetic tree, along with their accession numbers and other related details, are listed in Table S1. Each gene region was individually aligned using MAFFT v. 6.864b [37]. The alignments were checked and adjusted manually using Aliview [38]. Further, alignments were concatenated and subjected to phylogenetic analyses. The best substitution model out of 286 DNA models was figured using ModelFinder [39]. Additionally, windows version IQ-tree tool v.1.6.11 [40] was used to construct the phylogenetic tree. The reliability of the tree branches was assessed and tested based on 1000 ultrafast bootstrap (UFBoot) support replicates and a SH-like approximate likelihood ratio test (SH-like aLRT) with 1000 replicates. The constructed phylogenetic tree was visualized in FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 20 February 2022).
2.5. Fermentation for Beauvericin Production
Fusarium isolates were subcultured onto PDA and incubated at 25 °C for a week. Once the cultures were mature enough, spore suspensions were prepared separately for all isolates. Then, 10 µL of Tween 20 was added to 10 mL of normal saline (0.89%) and mixed adequately, saline solution was added to the culture and the spores were harvested carefully with the help of a sterile cotton swab and vortexed. The spore suspension’s optical density (OD) was adjusted between 0.08 and 0.1 at 530 nm [41]. Next, 1 mL of this spore suspension was added to 100 mL of Fusarium defined media (FDM) (Sucrose 25 g, Sodium nitrate (NaNO3) 4.25 g, Sodium chloride (NaCl) 5 g, Magnesium sulfate heptahydrate (MgSO4·7H2O) 2.5 g, Potassium dihydrogen phosphate (KH2PO4) 1.36 g, Ferrous sulphate heptahydrate (FeSO4·7H2O) 0.01 g, Zinc sulfate heptahydrate (ZnSO4·7H2O) 0.0029 g/L; pH 5.5) [42] in a 250 mL Erlenmeyer flask and was incubated in a shaking incubator at 25 °C at 150 rpm for 7 days (Figure S1).
2.6. Extraction of Beauvericin
Beauvericin was extracted from the fermented broth as per the protocol described earlier, with slight modification [43]. The fermented broth was filtered using Whatman filter paper no. 4. Biomass was collected, washed twice with distilled water and was allowed to dry at 50 °C. Dried biomass was extracted overnight in 25 mL of acetonitrile:water 90:10 (v/v). Further, the mixture was sonicated twice for 15 min. The mixture was filtered; filtrate was extracted with 25 mL of heptane and separated using a separating funnel. The bottom layer was evaporated to dryness in a rotary evaporator (Heidolph, Schwabach, Germany). The dry residue was dissolved in 25 mL of methanol:water, 60:40 (v/v) and extracted twice with 25 mL of dichloromethane. The dichloromethane phase containing beauvericin (BEA) was collected and evaporated to dryness in a rotary evaporator. The evaporated extract containing beauvericin was dissolved in 1 mL of Acetonitrile.
2.7. Detection of Beauvericin
The presence of beauvericin was initially qualitatively analyzed by thin-layer chromatography (TLC) and quantitatively by high-performance liquid chromatography (HPLC). In addition, high-resolution mass spectrometry (HRMS) was carried out for a few samples for further confirmation.
2.7.1. Thin-Layer Chromatography (TLC)
About 5 to 10 µL of the extracts were spotted onto TLC Silica gel plates (TLC Silica gel 60 F254; Merck life science Pvt. Ltd., Bengaluru, Karnataka, India) along with standard Beauvericin (BEA) (500 mg/mL) (Sigma) with the help of glass capillary tube. TLC plates were developed in petroleum ether:ethyl acetate (1:1) as a solvent system. The plates were air-dried, and consequently, the spots formed on the TLC plates were detected using iodine vapours. Retention factor (Rf) values for the standard and extracts were measured as described earlier [44,45]. The extracts were further subjected to high-performance liquid chromatography (HPLC).
2.7.2. High-Performance Liquid Chromatography (HPLC)
The amount of BEA in the extracts was determined by HPLC (Waters Corporation, Milford, MA, USA) using a C18 column and acetonitrile: H2O (85:15 v/v) as the mobile phase at a flow rate of 1 mL/min under isocratic conditions and U.V. detection at 210 nm [46]. A stock solution of standard beauvericin with a concentration of 1 mg/mL was prepared in acetonitrile and was further diluted to 15.6 µg/mL, 31.25 µg/mL, 62.5 µg/mL, 125 µg/mL, 250 µg/mL and 500 µg/mL. The retention time for BEA was found to be 9.1 min. The injection volume was 20 µL. The peak area was calibrated to the amount of BEA standard (Sigma). The amount of BEA produced was calculated with respect to the area under the peak.
2.7.3. High-Resolution Mass Spectrometry (HRMS)
The mass of beauvericin was determined in selected extracts; high-resolution product ion spectra were acquired using ESI-UHR-Q-TOF (Bruker Impact II Ultra-High Resolution-TOF) to confirm the presence of Beauvericin. The system ionization source type was ESI with positive ion polarity. The instrument was run in the full-scan mode (m/z 150–1200). Nitrogen was used for drying (200 °C; 7 L/min), and the nebulizer gas pressure was 1.7 Bar. Other typical operating parameters were set as capillary voltage—4500 V, end plate offset—500 V, and charging voltage—2000 V.
2.8. Large Scale Fermentation and Extraction of Beauvericin from Fusarium tardicrescens NFCCI 5201
Among the 88 tested isolates, the highest beauvericin producer, F. tardicrescens NFCCI 5201, was subjected to flask scale fermentation in a total of 3.5 L of Fusarium defined medium (FDM). Each flask was inoculated with 1% spore suspension of O.D. 0.08–0.1, as described earlier in Section 2.5, and incubated at 25 °C with 150 rpm for one week. Extraction of beauvericin was carried out as described in Section 2.6. The final dry residue was dissolved in 36 mL of acetonitrile. This extract was used for further study.
2.9. Determination of the Antibacterial Activity
The Fusarium tardicrescens NFCCI 5201 extract containing BEA dissolved in acetonitrile (500 µg/mL) was used to check antimicrobial activity. The antibacterial activity was tested against a panel of test organisms, including Escherichia coli MTCC 739, Bacillus subtilis MTCC 121, Micrococcus luteus MTCC 2470, Raoultella planticola MTCC 530, Pseudomonas aeruginosa MTCC 2453, Staphylococcus aureus MLS16 MTCC 2940 and Staphylococcus aureus MTCC 96. Cultures were grown on nutrient agar plates and incubated at 37 °C for 48 h. After incubation, one to two pure colonies of each culture were transferred to 10 mL Muller hinton broth (MHB) and incubated for 4–5 h at 37 °C. Further, optical density was adjusted to 1 using broth. The cultures were diluted to a cell density of 1 × 106 cells/mL. The test was carried out using a resazurin-based turbidometric assay in a 96-well microtiter plate. All wells of rows (A–G) were filled with 100 µL Muller hinton broth. The first well of each row was filled with 100 µL of Fusarium tardicrescens NFCCI 5201 extract containing BEA (500 µg/mL), which was mixed well, and then 100 µL of the mixture from the first well was transferred to the second well of the vertical row. This serial dilution was continued until the tenth well; lastly, 100 µL of the mixture was discarded from the tenth well. The final concentration of the extract was half of the original concentration in each well. Ten µL of each bacterial suspension was added in vertical rows individually, except for the twelfth well (sterility control). One hundred µL of antibiotic ampicillin (10 µg/50 µL) was used, except Pseudomonas aeruginosa MTCC 2453, in which neomycin (30 µg/50 µL) was added in the eleventh well of each row (positive control). After 48 hrs of incubation at 37 °C, 25 µL of resazurin (0.01%) was added to all wells and incubated at 37 °C for 75 min in the dark. Color changes were observed and recorded. The lowest concentration before the color change was considered as the minimum inhibitory concentration for that particular test organism [47,48].
2.10. Determination of the Individual and Combined Effect of Fusarium tardicrescens NFCCI 5201 Extract Containing Beauvericin and Amphotericin B on Pathogenic Fungi of Agricultural Importance
The antifungal potential of the individual and combined effect of F. tardicrescens NFCCI 5201 extract containing beauvericin and amphotericin B on agriculturally important pathogenic fungi was carried out using the poisoned food technique as described earlier [49]. Fungal pathogens Pythium sp. NFCCI 3482, Geotrichum candidum NFCCI 3744, Rhizoctonia solani NFCCI 4327, Rhizopus sp. NFCCI 2108, Sclerotium rolfsii NFCCI 4263, Bipolaris sorokiniana NFCCI 4690 and Absidia sp. NFCCI 2716 were selected for the antifungal study. These pathogenic fungi were grown on PDA and incubated at 25 ± 2 °C for 5–7 days.
Agar discs with mycelia (5 mm in diameter) were cut out from the actively growing regions of the 5–7 days old pure cultures using a sterile cork borer and aseptically inoculated at the center of four different Petri plates, as follows:
- Control plate containing PDA;
- PDA supplemented with amphotericin B (40 µg/mL);
- PDA supplemented with F. tardicrescens NFCCI 5201 extract containing beauvericin (40 µg/mL);
- PDA supplemented with amphotericin B (20 µg/mL) and F. tardicrescens NFCCI 5201 extract containing beauvericin (20 µg/mL).
The growth of fungal colonies on different media was measured between 3 and 7 days depending upon the growth of fungi. Each treatment was repeated thrice, and the plates were incubated at 25 °C. The diameter of the fungal colony was measured after 3 and 7 days of incubation. The percentage inhibition of the mycelial growth of the test fungi was calculated using the formula described earlier [50]. Inhibition of mycelial growth (%) = (dc−dt)/dc × 100, where dc is the mean diameter (in mm) of the colony in the control sample and dt is the diameter (in mm) of the colony in the treatment.
2.11. Antioxidant Activity of Extract of Fusarium tardicrescens NFCCI 5201 Containing Beauvericin
The extract was tested for in vitro antioxidant activity using the 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging method.
The concentration of the extract as well as standard solutions (control) glucose (Himedia) (negative) and ascorbic acid (Sigma) (positive) used were 1, 0.9, 0.8, 0.7, 0.6, 0.5, 0.4, 0.3, 0.2 and 0.1 mg/mL in acetonitrile. The extract or standard solution (10 µL) was added to DPPH in acetonitrile solution (200 µL, 100 µM) in a 96-well microtiter plate (Tarsons Products (P) Ltd., Kolkata, India). After incubation at 37 °C for 30 min, the absorbance of each solution was determined at 517 nm using a Synergy HT Multi-detection microplate reader (BioTek, Winooski, VT, USA).
The radical scavenging activity was calculated by the following formula:
Radical scavenging activity (%) = (OD control − OD sample)/OD control × 100 [51,52]. Three replicates were maintained for each treatment.
3. Results
3.1. Phylogenetic Analysis
The nrDNA (ITS), translation elongation factor 1-alpha (Tef1-α), 28S large subunit of the nrDNA (LSU), RNA polymerase second largest subunit (Rpb2), beta-tubulin (β-tub) and calmodulin (CaM) loci sequence alignments were together used to confirm the resolution of the isolates used in this study. The concatenated file contained sequence data of 186 taxa. Alignment contained 6944 columns, 3815 distinct patterns, 2049 parsimony-informative, 950 singleton sites and 3944 constant sites. TIM2e + R4 was found to be the best-fit model of 286 models tested and was chosen based on the Bayesian Information Criterion (BIC). The phylogeny was inferred using the Maximum Likelihood Method based on the model mentioned above. The log-likelihood of the consensus tree was –59368.419. Rate parameters: A-C: 1.27988, A-G: 2.69656, A-T: 1.27988, C-G: 1.00000, C-T: 5.18110, G-T: 1.00000; Base frequencies: A: 0.250, C: 0.250, G: 0.250, T: 0.250; Site proportion and rates: (0.587, 0.115) (0.171, 1.101) (0.200, 2.369) (0.042, 6.470) (Figure 3).
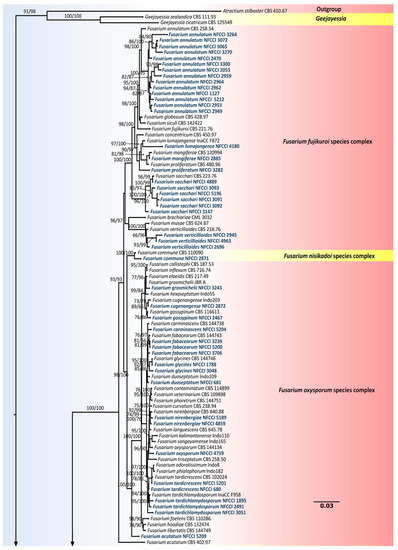
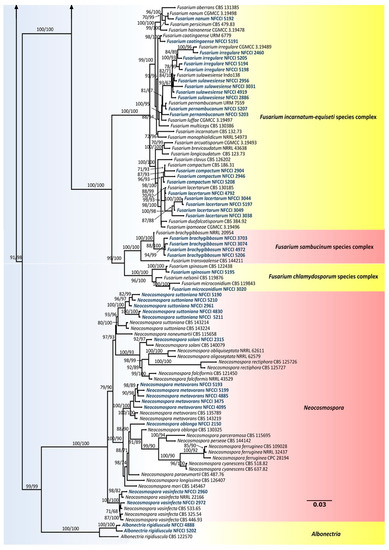
Figure 3.
Maximum-likelihood (IQ-TREE-ML) consensus tree inferred from the combined ITS, LSU, Rpb2, β-tub, CaM and Tef-1α multiple sequence alignment of genus Fusarium and related genera. Numbers at the branches indicate statistical support values (UFBS and SH-aLRT). The scale bar indicates expected changes per site. The tree is rooted to Atractium stilbaster (CBS 410.67). Isolates used in this study are shown in blue bold.
Of the isolates, 88 were found to belong to Fusarium and related genera, i.e., Albonectria and Neocosmospora. Strains used in this study were found to represent 35 species, including Albonectria rigidiuscula, Fusarium acutatum, F. annulatum, F. brachygibbosum, F. caatingaense, F. carminascens, F. commune, F. compactum, F. cugenangense, F. duoseptatum, F. fabacearum, F. glycines, F. gossypinum, F. grosmichelii, F. irregulare, F. lacertarum, F. lumajangense, F. mangiferae, F. microconidium, F. nanum, F. nirenbergiae, F. oxysporum, F. pernambucanum, F. proliferatum, F. sacchari, F. spinosum, F. sulawesiense, F. tardichlamydosporum, F. tardicrescens, F. verticillioides, Neocosmospora metavorans, N. oblonga, N. solani, N. suttoniana and N. vasinfecta.
Fusarium isolates were found to represent six species complexes. The species falling into a particular species complex are included in brackets, viz. Fusarium sambucinum species complex [F. brachygibbosum], Fusarium chlamydosporum species complex [F. microconidium, F. spinosum], Fusarium incarnatum-equiseti species complex [F. caatingaense, F. compactum, F. irregulare, F. lacertarum, F. nanum, F. pernambucanum, F. sulawesiense], Fusarium oxysporum species complex [F. carminascens, F. cugenangense, F. duoseptatum, F. fabacearum, F. glycines, F. gossypinum, F. grosmichelii, F. nirenbergiae, F. oxysporum, F. tardichlamydosporum, F. tardicrescens], Fusarium nisikadoi species complex [F. commune] and Fusarium fujikuroi species complex [F. acutatum, F. annulatum, F. lumajangense, F. mangiferae, F. proliferatum, F. sacchari, F. verticillioides].
Interestingly, out of 35 species reported in this study, 17 species, Fusarium caatingaense, F. carminascens, F. compactum, F. cugenangense, F. duoseptatum, F. fabacearum, F. glycines, F. gossypinum, F. grosmichelii, F. lumajangense, F. microconidium, F. nanum, F. nirenbergiae, F. sulawesiense, F. tardichlamydosporum, F. tardicrescens, Neocosmospora oblonga and N. suttoniana, were found to be new records from India [53,54], most of them being reported for the first time from a new host in this study (Table 4).

Table 4.
New records of Fusarium from India along with their host detail.
3.2. Detection of Beauvericin Produced
3.2.1. Thin Layer Chromatography
Thin layer chromatography analysis of fungal extracts along with the standard beauvericin showed spots at the same retention factor as the standard beauvericin, depicting the possible presence of beauvericin in the fungal extracts (Figure 4).
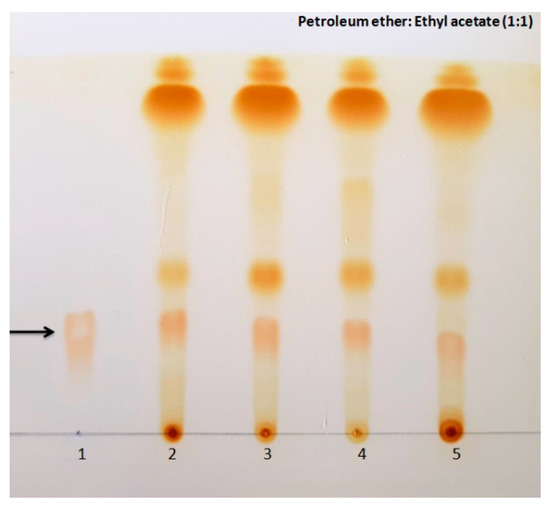
Figure 4.
Thin layer chromatography developed in petroleum ether: Ethyl acetate (1:1) showing spots of beauvericin detected using iodine vapours. Black arrow shows spots of beauvericin (1) Standard beauvericin (500 µg/mL) (2) Fusarium tardicrescens NFCCI 5201 (3) F. cugenangense NFCCI 2872 (4) Neocosmospora vasinfecta NFCCI 2960 (5) Fusarium annulatum NFCCI 2962.
3.2.2. High-Performance Liquid Chromatography
There was a linear correlation between the concentration of the standard beauvericin and the areas of the peak in HPLC chromatogram. The retention time of standard beauvericin was found to be 9.1 min (Figure 5). Biomass produced by various isolates varied from 4.41 to 14.17 g/L of FDM. Among 88, 50 isolates (56%) were found to be capable of beauvericin production which varied from 0.01 to 15.82 mg/g of biomass. Table 5 shows the mycelial biomass, BEA content of mycelial biomass and final medium pH after a week’s fermentation of different Fusarium isolates in Fusarium defined medium. F. tardicrescens NFCCI 5201 showed maximum beauvericin production of 15.82 mg/g of biomass. F. carminascens NFCCI 5204 and F. fabacearum NFCCI 5200 also produced a significant amount of beauvericin: 13.54 and 14.25 mg/g of biomass, respectively (Figure 6). The final pH of the fermented broth varied from 6.38 to 8.96 for different isolates. The correlation coefficient between biomass produced and the beauvericin produced was 0.2692; that between the pH of the fermented broth and the beauvericin produced was 0.16, indicating very weak and/or no association in both cases.
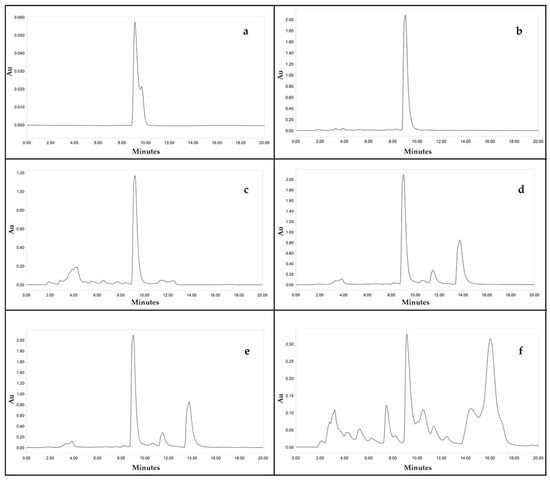
Figure 5.
HPLC chromatograms of (a) standard beauvericin (500 µg/mL) (b) Fusarium fabacearum NFCCI 5200 extract (c) F. sacchari NFCCI 4889 extract (d) F. carminascens NFCCI 5204 extract (e) F. tardicrescens NFCCI 5201 extract (f) F. cugenangense NFCCI 2872 extract.

Table 5.
Mycelial biomass and BEA content of Fusarium isolates and their pH in the Fusarium defined medium (on day 7).
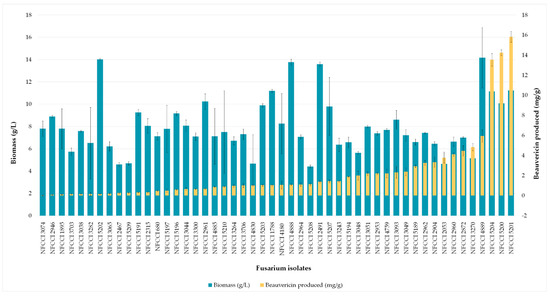
Figure 6.
Mycelial biomass and BEA content produced by different Fusarium isolates in the Fusarium defined medium (on day 7).
3.2.3. High-Resolution Mass Spectrometry (HRMS)
The HRMS results showed molecular ion peaks at 806.3999 (m/z) [M + Na]+,indicating the presence of beauvericin (C45H57N3O9) in the fungal extracts (Figure 7). Similar (m/z) [M + Na]+ 806.3956 have been reported for beauvericin (C45H57N3O9) [71]. It was observed that the fungal extract contains, majorly, beauvericin, as maximum ion intensity was observed with m/z 806.3999.
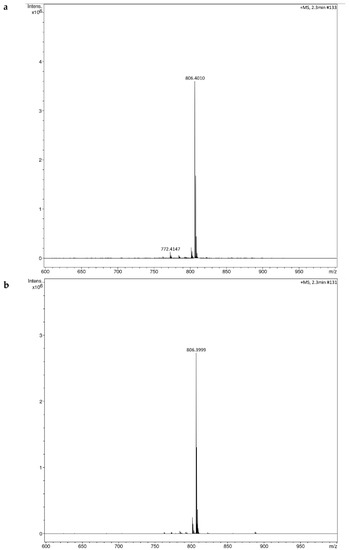
Figure 7.
HRMS spectra from higher-collision dissociation of the [M + Na]+ ions of fungal extract, (a) Fusarium sacchari NFCCI 4889 (b) F. tardicrescens NFCCI 5201.
3.3. Large Scale Fermentation
Fermentation of 3.5 liters of Fusarium defined medium resulted in 36.27 gms of biomass which, on extraction, resulted in 144 mg of crude which was dissolved in 36 mL of acetonitrile, making the final concentration 4 mg/mL. This extract was subjected to HPLC; it was found that beauvericin is the major compound in the extract, as the area covered by it was found to be nearly 64–68%. This stock was diluted as per requirement for further studies.
3.4. Antimicrobial and MIC of Crude Extract of Fusarium tardicrescens NFCCI 5201
The extract showed promising results against Staphylococcus aureus MLS16 MTCC 2940 and Micrococcus luteus MTCC 2470. The MIC of the crude extract of F. tardicrescens NFCCI 5201 was found to be 15.63 µg/mL against Micrococcus luteus MTCC 2470 and 62.5 µg/mL against Staphylococcus aureus MLS16 MTCC 2940 (Figure 8). The extract did not show any antimicrobial activity against Escherichia coli MTCC 739, Bacillus subtilis MTCC 121, Raoultella planticola MTCC 530, Pseudomonas aeruginosa MTCC 2453 and Staphylococcus aureus MTCC 96.
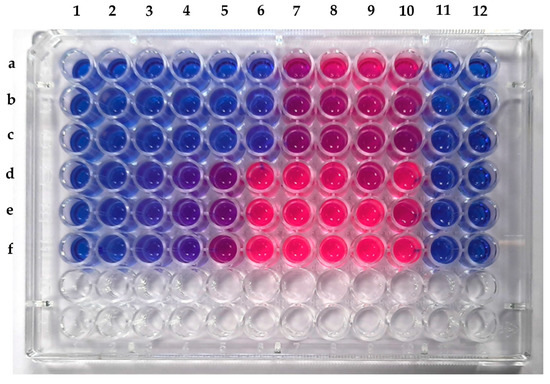
Figure 8.
Microtiter plate showing MIC of crude extract of F. tardicrescens NFCCI 5201 against Staphylococcus aureus MLS16 MTCC 2940 (Rows a–c) and Micrococcus luteus MTCC 2470 (Rows d–f).
3.5. Individual and Combined Effect of Extract of Fusarium tardicrescens NFCCI 5201 Containing Beauvericin and Amphotericin B on Pathogenic Fungi
The antifungal activity was observed as a reduction in the mycelial growth of pathogenic fungi in poisoned plates when compared to the control plates (Figure 9). The extract of F. tardicrescens NFCCI 5201 containing beauvericin (40 µg/mL) showed good antifungal activity against plant pathogenic fungi, Rhizoctonia solani NFCCI 4327, Sclerotium rolfsii NFCCI 4263, Geotrichum candidum NFCCI 3744 and Pythium sp. NFCCI 3482 showed a % inhibition of 84.31, 49.76, 38.22 and 35.13. Amphotericin B (40 µg/mL) inhibited Geotrichum candidum NFCCI 3744, Rhizoctonia solani NFCCI 4327, Sclerotium rolfsii NFCCI 4263 and Absidia sp. NFCCI 2716, showing % inhibition of 71.76, 55.72, 56.82 and 48.68. Interestingly, the results were remarkable when the extracts of F. tardicrescens NFCCI 5201 containing beauvericin (20 µg/mL) and amphotericin B (20 µg/mL) were used in combination against Rhizopus sp. NFCCI 2108, Sclerotium rolfsii NFCCI 4263, Bipolaris sorokiniana NFCCI 4690 and Absidia sp. NFCCI 2716, showing % inhibition of 50.35, 79.37, 48.07 and 76.72%, respectively. Individually, extract of F. tardicrescens NFCCI 5201 containing beauvericin (40 µg/mL) showed % inhibition of 1.68, 49.76, 2.56 and 7.9 against Rhizopus sp. NFCCI 2108, Sclerotium rolfsii NFCCI 4263, Bipolaris sorokiniana NFCCI 4690 and Absidia sp. NFCCI 2716. Individually, amphotericin B (40 µg/mL) showed % inhibition of 11.51, 56.82, 1.28 and 48.67 against Rhizopus sp. NFCCI 2108, Sclerotium rolfsii NFCCI 4263, Bipolaris sorokiniana NFCCI 4690 and Absidia sp. NFCCI 2716. The extract of F. tardicrescens NFCCI 5201 containing beauvericin (40 µg/mL) showed better results than amphotericin B in the case of Pythium sp. NFCCI 3482 and Rhizoctonia solani NFCCI 4327 (Figure 10).
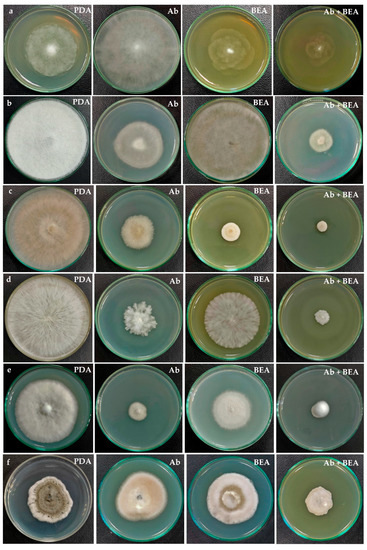
Figure 9.
Antifungal activity of individual and combined effect of extract of F. tardicrescens NFCCI 5201 containing beauvericin and amphotericin B on various pathogenic fungi (a) Pythium sp. NFCCI 3482 (b) Rhizopus sp. NFCCI 2108 (c) Rhizoctonia solani NFCCI 4327 (d) Sclerotium rolfsii NFCCI 4263 (e) Geotrichum candidum NFCCI 3744 (f) Bipolaris sorokiniana NFCCI 4690. PDA: Potato dextrose agar (Control); Ab: Potato dextrose agar containing amphotericin B (40 µg/mL); BEA: Potato dextrose agar having extract of F. tardicrescens NFCCI 5201 containing beauvericin (40 µg/mL); Ab + BEA: Potato dextrose agar having amphotericin B (20 µg/mL) and extract of F. tardicrescens NFCCI 5201 containing beauvericin (20 µg/mL).
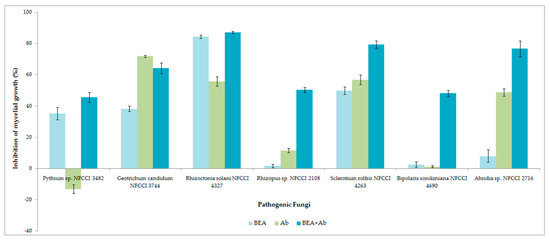
Figure 10.
Inhibition of mycelial growth (%) of individual and combined effect of extract of F. tardicrescens NFCCI 5201 containing beauvericin and amphotericin B on various pathogenic fungi using food poison technique.
3.6. Antioxidant Effect of Extract of Fusarium tardicrescens NFCCI 5201 Containing Beauvericin
Figure 11 shows the free radical scavenging activity of the extract of F. tardicrescens NFCCI 5201 containing beauvericin and the standard ascorbic acid when tested for in vitro antioxidant activity. The extract of F. tardicrescens NFCCI 5201 containing beauvericin showed good satisfactory dose-dependent DPPH radical scavenging activity with an IC50 value of 0.675 mg/mL when tested with standard ascorbic acid, which showed IC50 value of 0.146 mg/mL.
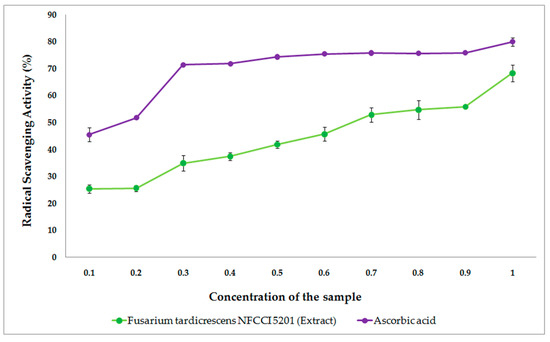
Figure 11.
The free radical scavenging activity of the extract of F. tardicrescens NFCCI 5201 containing beauvericin (mg/mL) and the standard ascorbic acid (mg/mL) when tested for in vitro antioxidant activity.
4. Discussion
One of the greatest hindrances in studying Fusarium has been the ambiguous nomenclature or incorrect species names of the isolates because of the limitations in recognition of the species based on morphology [72]. Being the world’s most important pathogens, its prophylaxis and management needs correct and rapid identification [73]. As mentioned earlier, ITS and LSU frequently fail to distinguish at the species level and, preferably, Tef1-α and Rpb2 can be used to distinguish isolates at the species level. Literature reveals that Indian Fusarium isolates have been mostly identified on the basis of morphology. Very few studies were found where the Fusarium isolates had been identified using molecular studies. Wherever molecular studies were performed, they were based on the ITS gene region; very limited studies used either Tef1-α or Rpb2 or both of these gene regions [74]. To our knowledge, this is the first study from India where Fusarium isolates have been identified on the basis of six gene regions, nrDNA (ITS), translation elongation factor 1-alpha (Tef1-α), 28S large subunit of the nrDNA (LSU), RNA polymerase second largest subunit (Rpb2), beta-tubulin (β-tub) and calmodulin (CaM). Interestingly, in this study, 17 species, Fusarium caatingaense, F. carminascens, F. compactum, F. cugenangense, F. duoseptatum, F. fabacearum, F. glycines, F. gossypinum, F. grosmichelii, F. lumajangense, F. microconidium, F. nanum, F. nirenbergiae, F. sulawesiense, F. tardichlamydosporum, F. tardicrescens, Neocosmospora oblonga and N. suttoniana were found to be new records for India, most of them being reported from new hosts [53,54].
BEA is a potent bioactive compound possessing antimicrobial, anti-insecticidal, antitumor and antiplatelet activities at extremely low concentration and with unique uncharacterized active mechanisms. A review of the literature indicates that no study was undertaken in India on BEA production from Indian species of Fusarium, except [43] who had reported BEA production from two isolates of Fusarium viz., F. anthophilum (Host: Sugarcane; 1300 µg/g) and F. nygamai (Host: Cajanus indicus; 3 µg/g) [43]. This work was undertaken in Bari, Italy. This suggests that Indian Fusarium isolates have remained unexplored for their capability of BEA production and its applicability.
Albonectria rigidiuscula, F. acutatum, F. annulatum, F. brachygibbosum, F. caatingaense, F. carminascens, F. compactum, F. cugenangense, F. fabacearum, F. glycines, F. gossypinum, F. grosmichelii, F. irregulare, F. lacertarum, F. lumajangense, F. nirenbergiae, F. oxysporum, F. pernambucanum, F. proliferatum, F. sacchari, F. tardichlamydosporum, F. tardicrescens, N. metavorans, N. solani, N. suttoniana and N. vasinfecta were found to be the positive producers of BEA in this study, many of them being reported as BEA producers for the first time in this study. Fusarium tardicrescens NFCCI 5201 showed maximum beauvericin production of 15.82 mg/g of biomass. Fusarium carminascens NFCCI 5204 and F. fabacearum NFCCI 5200 also produced a significantly ample amount of beauvericin, 13.54 and 14.25 mg/g of biomass, respectively, all three of them being reported for the first time as significant BEA producers in this study.
In earlier reports, BEA was detected in cultures of Fusarium moniliforme, F. semitectum [75], F. subglutinans, F. thapsinum [76], F. sambucinum, F. acuminatum, F. equiseti, F. longipes, F. anthophilum, F. oxysporum, F. poae, F. avenaceum, F. beomiforme, F. dlamini, F. bulbicola, F. nygamai [43], F. chlamydosporum, F. solani, F. proliferatum and F. sacchari [77].
The crude extract of F. tardicrescens NFCCI 5201 showed promising results against Staphylococcus aureus MLS16 MTCC 2940 and Micrococcus luteus MTCC 2470, with MIC values of 15.63 µg/mL against Micrococcus luteus MTCC 2470 and 62.5 µg/mL against Staphylococcus aureus MLS16 MTCC 2940. BEA from F. oxysporum had a potent inhibitory effect on the growth of pathogenic Staphylococcus aureus with a MIC of 3.91 µM [23]. Earlier reports reveal that inhibitory concentration (IC50) values of BEA against Bacillus subtilis, Staphylococcus haemolyticus, Pseudomonas lachrymans, Agrobacterium tumefaciens, Escherichia coli and Xanthomo vesicatoria by a 96-well microplate broth dilution–MTT assay ranged between 18.45 and 70.41 µg/mL [78]. BEA has been known to inhibit bacteria, including Bacillus pumilus (0.1 µg of BEA per disk), several other species of Bacillus and Paenibacillus (1 µg of BEA per disk), P. validus, Bifidobacterium adolescentis, Clostridium perfringens, Eubacterium biforme, Peptostreptococcus anaerobius and P. productus (25 µg of BEA per disk) [18]. Strong antimicrobial activity has been reported against Staphylococcus aureus, Salmonella typhimurium and Bacillus cereus with MIC values of 3.91 µM, 6.25 and 3.12 µg/mL, respectively [19,23].
With BEA being a high score ABC inhibitor, it has been reported to show strong synergy with some azole compounds (miconazole, ketoconazole) against Candida albicans and Candida parapsilosis (in vitro as well as in vivo). It has been reported that BEA individually fails to show antifungal activity, but it was found to synergize the effect of ketoconazole [25].
In this study, the antifungal activity was observed as a reduction in the mycelial growth of pathogenic fungi of agricultural importance in poisoned plates when compared to the control plates. The extract of F. tardicrescens NFCCI 5201 containing beauvericin (40 µg/mL) showed good antifungal activity against pathogenic fungi Rhizoctonia solani NFCCI 4327, Sclerotium rolfsii NFCCI 4263, Geotrichum candidum NFCCI 3744 and Pythium sp. NFCCI 3482 showing % inhibition of 84.31, 49.76, 38.22 and 35.13. Interestingly, when the extract of F. tardicrescens NFCCI 5201 containing beauvericin (20 µg/mL) and amphotericin B (20 µg/mL) were used in combination, they produced a synergized antifungal effect against Rhizopus sp. NFCCI 2108, Sclerotium rolfsii NFCCI 4263, Bipolaris sorokiniana NFCCI 4690 and Absidia sp. NFCCI 2716, showing % inhibition of 50.35, 79.37, 48.07 and 76.72%, respectively. Individually, the extract of F. tardicrescens NFCCI 5201 containing beauvericin (40 µg/mL) showed a % inhibition of 1.68, 49.76, 2.56 and 7.9 against Rhizopus sp. NFCCI 2108, Sclerotium rolfsii NFCCI 4263, Bipolaris sorokiniana NFCCI 4690 and Absidia sp. NFCCI 2716, and amphotericin B (40 µg/mL) showed a % inhibition of 11.51, 56.82, 1.28 and 48.67 against Rhizopus sp. NFCCI 2108, Sclerotium rolfsii NFCCI 4263, Bipolaris sorokiniana NFCCI 4690 and Absidia sp. NFCCI 2716. The extract of F. tardicrescens NFCCI 5201 containing beauvericin (40 µg/mL) showed better results than amphotericin B in the case of Pythium sp. NFCCI 3482, Rhizoctonia solani NFCCI 4327. This is the first report on the testing of BEA against filamentous, agriculturally important fungal pathogens. The synergistic effect of BEA along with amphotericin B against filamentous agriculturally important pathogens has also been studied for the first time. Earlier studies focused on the synergistic effect of BEA and azoles (miconazole, ketoconazole) on yeast (Candida albicans and Candida parapsilosis). This study focuses on the synergistic effect of fungal extracts and polyene (amphotericin B) on agriculturally important filamentous fungal pathogens. Moreover, the extract of F. tardicrescens NFCCI 5201 containing beauvericin showed good dose-dependent DPPH radical scavenging activity with an IC50 value of 0.675 mg/mL. Antioxidant activity has been reported from metabolites produced by Fusarium oxysporum [79]. The DPPH-radicals’ scavenging activity has been reported from F. solani with IC50 value of 24 µg/mL [80].
5. Conclusions
Fusarium spp. have been known globally for centuries for their pathogenic behavior; however, they can prove to be a treasure trove because of their capability to produce other interesting metabolites of great applicability in various sectors such as food, agriculture and medicine. As shown in this study, the antifungal activity became synergized when a half dosage of the fungal extract and amphotericin B was used. Therefore, this study indicates that concerted efforts are required, in which high-throughput screening needs to be carried out with dosage of the BEA and the other antifungal compound, which can possibly be used at a large scale for calculating the optimum dosage required. Similar studies can be carried out in the field of medicine where the dosage is optimized and the delivery of the drug combinations is also targeted.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/jof8070662/s1, Figure S1: Fermentation of different isolates in Fusarium defined media after a week’s fermentation (a) Control (Uninnoculated FDM medium) (b) Fusarium fabacearum NFCCI 5200 (c) F. annulatum NFCCI 3300 (d) F. sulawesiense NFCCI 4919 (e) F. sacchari NFCCI 4889 (f) F. annulatum NFCCI 5212 (g) F. nirenbergiae NFCCI 5189 (h) F. tardicrescens NFCCI 5201 (i) F. lacertarum NFCCI 4792 (j) F. lumajangense NFCCI 4180 (k) F. pernambucanum NFCCI 5203 (l) F. duoseptatum NFCCI 681 (m) F. tardichlamydosporum NFCCI 1895 (n) Neocosmospora solani NFCCI 2315 (o) F. mangiferae NFCCI 2885 (p) F. annulatum NFCCI 2964 (q) F. sacchari NFCCI 3147 (r) F. grosmichelii NFCCI 3243; Table S1: GenBank Accession No. and other details of the isolates used in the phylogenetic study.
Author Contributions
Conceptualization, S.R. and S.K.S.; methodology, S.R. and S.K.S.; formal analysis, S.R. and S.K.S.; investigation, S.R.; resources, S.K.S.; writing—original draft preparation, S.R.; writing—review and editing, S.R., S.K.S. and L.D.; supervision, S.K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Prashant Dhakephalkar, Director MACS-Agharkar Research Institute, Pune for providing the necessary facilities and encouragement to carry out the research work. Shiwali Rana acknowledges the University Grants Commission (U.G.C.), New Delhi for granting a Senior Research Fellowship (SRF), and S.P. Pune University, Pune for registering for the Ph.D. degree. We acknowledge technical support and help by Deepak Kumar Maurya and Subhash Gaikwad of Agharkar Research Institute, Pune. Laurent Dufossé deeply acknowledges the strong financial support from Conseil Régional de Bretagne and Conseil Régional de La Réunion for research activities dedicated to microbial biotechnology.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gupta, A.K.; Baran, R.; Summerbell, R.C. Fusarium infections of the skin. Curr. Opin. Infect. Dis. 2000, 13, 121–128. [Google Scholar] [CrossRef]
- Summerell, B.A. Resolving Fusarium: Current status of the genus. Annu. Rev. Phytopathol. 2019, 57, 323–339. [Google Scholar] [CrossRef]
- Taylor, J.W.; Jacobson, D.J.; Kroken, S.; Kasuga, T.; Geiser, D.M.; Hibbett, D.S.; Fisher, M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000, 31, 21–32. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Bolchacova, E.; Voigt, K.; Crous, P.W.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Odonnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Biol. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E.; Nirenberg, H.I. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 1998, 90, 465–493. [Google Scholar] [CrossRef]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Gueidan, C.; Crous, P.W.; Geiser, D.M. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J. Clin. Microbiol. 2009, 47, 3851–3861. [Google Scholar] [CrossRef]
- O’Donnell, K. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 2000, 92, 919–938. [Google Scholar] [CrossRef]
- Nalim, F.A.; Samuels, G.J.; Wijesundera, R.L.; Geiser, D.M. New species from the Fusarium solani species complex derived from perithecia and soil in the Old-World tropics. Mycologia 2011, 103, 1302–1330. [Google Scholar] [CrossRef]
- Herron, D.A.; Wingfield, M.J.; Wingfield, B.D.; Rodas, C.A.; Marincowitz, S.; Steenkamp, E.T. Novel taxa in the Fusarium fujikuroi species complex from Pinus spp. Stud. Mycol. 2015, 80, 131–150. [Google Scholar] [CrossRef]
- Crous, P.W.; Lombard, L.; Sandoval-Denis, M.; Seifert, K.A.; Schroers, H.J.; Chaverri, P.; Gene, J.; Guarro, J.; Hirooka, Y.; Bensch, K.; et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021, 98, 100116. [Google Scholar] [CrossRef]
- Zhang, T.; Jia, X.P.; Zhuo, Y.; Liu, M.; Gao, H.; Liu, J.T.; Zhang, L.X. Cloning and characterization of a novel 2-ketoisovalerate reductase from the beauvericin producer Fusarium proliferatum LF061. BMC Biotechnol. 2012, 12, 55. [Google Scholar] [CrossRef]
- Sood, S.; Sandhu, S.S.; Mukherjee, T.K. Pharmacological and therapeutic potential of beauvericin: A Short Review. J. Proteom. Bioinform. 2017, 10, 18–23. [Google Scholar] [CrossRef]
- Hilgenfeld, R.; Saenger, W. Structural chemistry of natural and synthetic ionophores and their complexes with cations. Top. Curr. Chem. 1982, 101, 1–82. [Google Scholar]
- Ovchinnikov, Y.A.; Ivanov, V.T.; Evstratov, A.E.; Mikhaleva, I.I.; Bystrov, V.F.; Portnova, S.L.; Balashova, T.A.; Meshcheryakova, E.N.; Tulchinsky, V.M. The enniatins ionophores, conformation and ion binding properties. Int. J. Pept. Protein Res. 1974, 6, 465–498. [Google Scholar] [CrossRef]
- Steinrauf, L.K. Beauvericin and the other enniatins. Met. Ions Biol. Syst. 1985, 19, 139–171. [Google Scholar]
- Lin, H.I.; Lee, Y.J.; Chen, B.F.; Tsai, M.C.; Lu, J.L.; Chou, C.J.; Jow, G.M. Involvement of Bcl-2 family, cytochrome c and caspase 3 in induction of apoptosis by beauvericin in human non-small cell lung cancer cells. Cancer Lett. 2005, 230, 248–259. [Google Scholar] [CrossRef]
- Castlebury, L.A.; Sutherland, J.B.; Tanner, L.A.; Henderson, A.L.; Cerniglia, C.E. Short Communication: Use of a bioassay to evaluate the toxicity of beauvericin to bacteria. World J. Microbiol. Biotechnol. 1999, 15, 131–133. [Google Scholar] [CrossRef]
- Dzoyem, J.P.; Melong, R.; Tsamo, A.T.; Maffo, T.; Kapche, D.; Ngadjui, B.T.; McGaw, L.J.; Eloff, J.N. Cytotoxicity, antioxidant and antibacterial activity of four compounds produced by an endophytic fungus Epicoccum nigrum associated with Entada abyssinica. Rev. Bras. Farmacogn. 2017, 27, 251–253. [Google Scholar] [CrossRef]
- Fotso, J.; Smith, J.S. Evaluation of beauvericin toxicity with the bacterial bioluminescence assay and the Ames mutagenicity bioassay. J. Food Sci. 2003, 68, 1938–1941. [Google Scholar] [CrossRef]
- Meca, G.; Sospedra, I.; Soriano, J.M.; Ritieni, A.; Moretti, A.; Manes, J. Antibacterial effect of the bioactive compound beauvericin produced by Fusarium proliferatum on solid medium of wheat. Toxicon 2010, 56, 349–354. [Google Scholar] [CrossRef]
- Nilanonta, C.; Isaka, M.; Kittakoop, P.; Trakulnaleamsai, S.; Tanticharoen, M.; Thebtaranonth, Y. Precursor-directed biosynthesis of beauvericin analogs by the insect pathogenic fungus Paecilomyces tenuipes BCC 1614. Tetrahedron 2002, 58, 3355–3360. [Google Scholar] [CrossRef]
- Zhang, H.W.; Ruan, C.F.; Bai, X.L.; Zhang, M.; Zhu, S.S.; Jiang, Y.Y. Isolation and identification of the antimicrobial agent beauvericin from the endophytic Fusarium oxysporum 5-19 with NMR and ESI-MS/MS. Biomed Res. Int. 2016, 2016, 1084670. [Google Scholar] [CrossRef]
- Wu, X.F.; Xu, R.; Ouyang, Z.J.; Qian, C.; Shen, Y.; Wu, X.D.; Gu, Y.H.; Xu, Q.; Sun, Y. Beauvericin ameliorates experimental colitis by inhibiting activated T Cells via downregulation of the PI3K/Akt signaling pathway. PLoS ONE 2013, 8, e83013. [Google Scholar] [CrossRef]
- Fukuda, T.; Arai, M.; Tomoda, H.; Omura, S. New beauvericins, potentiators of antifungal miconazole activity, produced by Beauveria sp FKI-1366-II. Structure elucidation. J. Antibiot. 2004, 57, 117–124. [Google Scholar] [CrossRef]
- Shekhar-Guturja, T.; Tebung, W.A.; Mount, H.; Liu, N.N.; Kohler, J.R.; Whiteway, M.; Cowen, L.E. Beauvericin potentiates azole activity via inhibition of multidrug efflux, blocks Candida albicans morphogenesis, and is effluxed via Yor1 and circuitry controlled by Zcf29. Antimicrob. Agents Chemother. 2016, 60, 7468–7480. [Google Scholar] [CrossRef]
- Zhang, L.X.; Yan, K.Z.; Zhang, Y.; Huang, R.; Bian, J.; Zheng, C.S.; Sun, H.X.; Chen, Z.H.; Sun, N.; An, R.; et al. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc. Natl. Acad. Sci. USA 2007, 104, 4606–4611. [Google Scholar] [CrossRef]
- Campos, F.F.; Sales, P.A.; Romanha, A.J.; Araujo, M.S.S.; Siqueira, E.P.; Resende, J.M.; Alves, T.M.A.; Martins, O.A.; dos Santos, V.L.; Rosa, C.A.; et al. Bioactive endophytic fungi isolated from Caesalpinia echinata Lam. (Brazilwood) and identification of beauvericin as a trypanocidal metabolite from Fusarium sp. Mem. Inst. Oswaldo Cruz 2015, 110, 65–74. [Google Scholar] [CrossRef]
- Aamir, S.; Sutar, S.; Singh, S.K.; Baghela, A. A rapid and efficient method of fungal genomic DNA extraction, suitable for PCR based molecular methods. Plant Pathol. Quar. 2015, 5, 74–81. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innes, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Vilgalys, R.; Sun, B.L. Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA-sequences. Proc. Natl. Acad. Sci. USA 1994, 91, 7832. [Google Scholar] [CrossRef] [PubMed]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.J.; Whelen, S.; Benjamin, D.H. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Yonezawa, T.; Lee, K.; Kumagai, S.; Sugita-Konishi, Y.; Goto, K.; Hara-Kudo, Y. Molecular phylogeny of the higher and lower taxonomy of the Fusarium genus and differences in the evolutionary histories of multiple genes. BMC Evol. Biol. 2011, 11, 322. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.W. Multilocus DNA sequence analysis shows that Penicillium biourgeianum is a distinct species closely related to P. brevicompactum and P. olsonii. Mycol. Res. 2004, 108, 434–440. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Aberkane, A.; Cuenca-Estrella, M.; Gomez-Lopez, A.; Petrikkou, E.; Mellado, E.; Monzón, A.; Rodriguez-Tudela, J.L. Comparative evaluation of two different methods of inoculum preparation for antifungal susceptibility testing of filamentous fungi. J. Antimicrob. Chemother. 2002, 50, 719–722. [Google Scholar] [CrossRef]
- Lee, H.; Kang, J.; Kim, B.; Park, S.; Lee, C. Statistical optimization of culture conditions for the production of enniatins H, I, and MK1688 by Fusarium oxysporum KFCC 11363P. J. Biosci. Bioeng. 2011, 111, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Logrieco, A.; Moretti, A.; Castella, G.; Kostecki, M.; Golinski, P.; Ritieni, A.; Chelkowski, J. Beauvericin production by Fusarium species. Appl. Environ. Microbiol. 1998, 64, 3084–3088. [Google Scholar] [CrossRef] [PubMed]
- Touchstone, J.C. Practice of Thin Layer Chromatography, 3rd ed.; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 1992; pp. 1–14. [Google Scholar]
- Fessenden, R.J.; Fessenden, J.S.; Feist, P. Organic Laboratory Techniques, 3rd ed.; Cengage Learning: Boston, MA, USA; Brooks/Cole Thomson Leaning, Inc.: Belmont, CA, USA, 2001; pp. 133–138. [Google Scholar]
- Xu, L.J.; Liu, Y.S.; Zhou, L.G.; Wu, J.Y. Optimization of a liquid medium for beauvericin production in Fusarium redolens Dzf2 mycelial culture. Biotechnol. Bioprocess Eng. 2010, 15, 460–466. [Google Scholar] [CrossRef]
- Teh, C.H.; Nazni, W.A.; Nurulhusna, A.; Norazah, A.; Lee, H.L. Determination of antibacterial activity and minimum inhibitory concentration of larval extract of fly via resazurin-based turbidometric assay. BMC Microbiol. 2017, 17, 36. [Google Scholar] [CrossRef]
- Foerster, S.; Desilvestro, V.; Hathaway, L.J.; Althaus, C.L.; Unemo, M. A new rapid resazurin-based microdilution assay for antimicrobial susceptibility testing of Neisseria gonorrhoeae. J. Antimicrob. Chemother. 2017, 72, 1961–1968. [Google Scholar] [CrossRef]
- Adjou, E.S.; Kouton, S.; Dahouenon-Ahoussi, E.; Sohounhloue, C.K.; Soumanou, M.M. Antifungal activity of Ocimum canum essential oil against toxinogenic fungi isolated from peanut seeds in post-harvest in Benin. Int. Res. J. Biol. 2012, 1, 20–26. [Google Scholar]
- Philippe, S.; Soua¨ıbou, F.; Guy, A.; Sébastien, D.T.; Boniface, Y.; Paulin, A.; Issaka, Y.; Dominique, S. Chemical composition and antifungal activity of essential oil of fresh leaves of Ocimum gratissimum from Benin against six mycotoxigenic fungi isolated from traditional cheese wagashi. Res. J. Biol. Sci. 2012, 1, 22–27. [Google Scholar]
- Jinesh, V.K.; Jaishree, V.; Badami, S.; Shyam, W. Comparative evaluation of antioxidant properties of edible and non-edible leaves of Anethum graveolens Linn. Indian J. Nat. Prod. Reso. 2010, 1, 168–173. [Google Scholar]
- Elizabeth, K.; Rao, M.W.A. Oxygen radical scavenging activity of Curcumin. Int. J. Pharmaceu. 1990, 58, 237–240. [Google Scholar]
- Jamaluddin; Goswami, M.S.; Ojha, B.M. Fungi of India 1989–2001; Scientific Publishers: Jodhpur, India, 2004; pp. 1–326. [Google Scholar]
- Manoharachary, C.; Atri, N.S.; Devi, T.P.; Kamil, D.; Singh, S.K.; Singh, A.P. Bilgrami’s Fungi of India List and References (1988–2020); Today & Tomorrow’s Printers and Publishers: New Delhi, India, 2022; pp. 1–475. [Google Scholar]
- Santos, A.C.D.; Trindade, J.V.C.; Lima, C.S.; Barbosa, R.D.; da Costa, A.F.; Tiago, P.V.; Oliveira, N. Morphology, phylogeny, and sexual stage of Fusarium caatingaense and Fusarium pernambucanum, new species of the Fusarium incarnatum-equiseti species complex associated with insects in Brazil. Mycologia 2019, 111, 244–259. [Google Scholar] [CrossRef]
- Lombard, L.; Sandoval-Denis, M.; Lamprecht, S.C.; Crous, P.W. Epitypification of Fusarium oxysporum—Clearing the taxonomic chaos. Persoonia 2019, 43, 1–47. [Google Scholar] [CrossRef]
- Vanwyk, P.S.; LOS, O.; Palterl, G.D.C.; Marasas, W.F.O. Geographic distribution and pathogenicity of Fusarium species associated with crown rot of wheat in the orange free state, South Africa. Phytophylactica 1987, 19, 271–274. [Google Scholar]
- Matic, S.; Tabone, G.; Gullino, M.L.; Garibaldi, A.; Guarnaccia, V. Emerging leafy vegetable crop diseases caused by the Fusarium incarnatum-equiseti species complex. Phytopathol. Mediterr. 2020, 59, 303–317. [Google Scholar] [CrossRef]
- Sever, Z.; Ivić, D.; Kos, T.; Miličević, T. Fusarium rot in stored apples in Croatia Arh. Hig. Rada. Toksikol. 2012, 63, 463–470. [Google Scholar] [CrossRef]
- Frisullo, S.; Logrieco, A.; Moretti, A.; Grammatikaki, G.; Bottalico, A. Banana Corm and Root Rot by Fusarium compactum, in Crete. Phytopathol. Mediterr. 1994, 33, 78–82. [Google Scholar]
- Mazhar, M.A.; Jahangir, A.Z.; Banihashemi, Z.; Izadpanah, K. First report of isolation and pathogenicity of Fusarium compactum on safflower. Iran. J. Plant Pathol. 2013, 49, 277. [Google Scholar]
- Brizuela, A.M.; De la Lastra, E.; Marin-Guirao, J.I.; Galvez, L.; de Cara-Garcia, M.; Capote, N.; Palmero, D. Fusarium consortium populations associated with Asparagus crop in Spain and their role on field decline syndrome. J. Fungi 2020, 6, 336. [Google Scholar] [CrossRef]
- Maryani, N.; Lombard, L.; Poerba, Y.S.; Subandiyah, S.; Crous, P.W.; Kema, G.H.J. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud. Mycol. 2019, 92, 155–194. [Google Scholar] [CrossRef]
- Ujat, A.; Vadamalai, G.; Hattori, Y.; Nakashima, C.; Wong, C.; Zulperi, D. Current classification and diversity of Fusarium species complex, the causal pathogen of Fusarium wilt disease of banana in Malaysia. Agronomy 2021, 11, 1955. [Google Scholar] [CrossRef]
- Maryani, N.; Sandoval-Denis, M.; Lombard, L.; Crous, P.W.; Kema, G.H.J. New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Persoonia 2019, 43, 48–69. [Google Scholar] [CrossRef]
- Wang, M.M.; Chen, Q.; Diao, Y.Z.; Duan, W.J.; Cai, L. Fusarium incarnatum-equiseti complex from China. Persoonia 2019, 43, 70–89. [Google Scholar] [CrossRef]
- Lu, Y.A.; Qiu, J.B.; Wang, S.F.; Xu, J.H.; Ma, G.Z.; Shi, J.R.; Bao, Z.H. Species diversity and toxigenic potential of Fusarium incarnatum-equiseti species complex isolates from rice and soybean in China. Plant Dis. 2021, 105, 2628–2636. [Google Scholar] [CrossRef]
- Yi, R.H.; Lian, T.; Su, J.J.; Chen, J. First report of internal black rot on Carica papaya fruit caused by Fusarium sulawesiense in China. Plant Dis. 2022, 106, 319. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, Y.; Li, Q.; Huang, S.; Tang, L.; Chen, X.; Guo, T.; Mo, J.; Ma, L. First report of leaf blight caused by Fusarium pernambucanum and Fusarium sulawesiense on Plum in Sichuan, China. Plant Dis. 2022. [Google Scholar] [CrossRef]
- Araújo, M.B.M.; Moreira, G.M.; Nascimento, L.V.; Nogueira, G.D.; Nascimento, S.R.D.; Pfenning, L.H.; Ambrosio, M.M.D. Fusarium rot of melon is caused by several Fusarium species. Plant Pathol. 2021, 70, 712–721. [Google Scholar] [CrossRef]
- Urbaniak, M.; Stepien, L.; Uhlig, S. Evidence for naturally produced beauvericins containing N-Methyl-Tyrosine in Hypocreales fungi. Toxins 2019, 11, 182. [Google Scholar] [CrossRef]
- Geiser, D.M.; Jimenez-Gasco, M.D.; Kang, S.C.; Makalowska, I.; Veeraraghavan, N.; Ward, T.J.; Zhang, N.; Kuldau, G.A.; O’Donnell, K. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004, 110, 473–479. [Google Scholar] [CrossRef]
- Wingfield, M.J.; De Beer, Z.W.; Slippers, B.; Wingfield, B.D.; Groenewald, J.Z.; Lombard, L.; Crous, P.W. One fungus, one name promotes progressive plant pathology. Mol. Plant Pathol. 2012, 13, 604–613. [Google Scholar] [CrossRef]
- Aiyaz, M.; Thimmappa Divakara, S.; Mudili, V.; George Moore, G.; Kumar Gupta, V.; i Yli-Mattila, T.; Chandra Nayaka, S.; Ramachandrappa Niranjana, S. Molecular diversity of seed-borne Fusarium species associated with maize in India. Curr. Genom. 2016, 17, 132–144. [Google Scholar] [CrossRef][Green Version]
- Gupta, S.; Krasnoff, S.B.; Underwood, N.L.; Renwick, J.A.A.; Roberts, D.W. Isolation of beauvericin as an insect toxin from Fusarium semitectum and Fusarium moniliforme var. subglutinans. Mycopathologia 1991, 155, 185–189. [Google Scholar] [CrossRef]
- Moretti, A.; Logrieco, A.; Bottalico, A.; Ritieni, A.; Fogliano, V.; Randazzo, G. Diversity in beauvericin and fusaproliferin production by different populations of Gibberella fujikuroi (Fusarium section Liseola). Sydowia 1997, 48, 44–56. [Google Scholar]
- Azliza, I.N.; Hafizi, R.; Nurhazrati, M.; Salleh, B. Production of major mycotoxins by Fusarium species isolated from wild grasses in peninsular Malaysia. Sains Malays. 2014, 43, 89–94. [Google Scholar]
- Xu, L.; Wang, J.; Zhao, J.; Li, P.; Shan, T.; Wang, J.; Li, X.; Zhou, L. Beauvericin from the endophytic fungus, Fusarium redolens, isolated from Dioscorea zingiberensis and its antibacterial activity. Nat. Prod. Commun. 2010, 5, 811–814. [Google Scholar] [CrossRef]
- Caicedo, N.H.; Davalos, A.F.; Puente, P.A.; Rodriguez, A.Y.; Caicedo, P.A. Antioxidant activity of exo-metabolites produced by Fusarium oxysporum: An endophytic fungus isolated from leaves of Otoba gracilipes. Open Microbiol. J. 2019, 8, e903. [Google Scholar] [CrossRef]
- Menezes, B.S.; Solidade, L.S.; Conceição, A.A.; Santos Junior, M.N.; Leal, P.L.; de Brito, E.S.; Canuto, K.M.; Mendonça, S.; de Siqueira, F.G.; Marques, L.M. Pigment production by Fusarium solani BRM054066 and determination of antioxidant and anti-inflammatory properties. AMB Express 2020, 10, 117. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).