Abstract
Green mold, caused by Penicillium digitatum, is the major cause of citrus postharvest decay. Currently, the application of sterol demethylation inhibitor (DMI) fungicide is one of the main control measures to prevent green mold. However, the fungicide-resistance problem in the pathogen P. digitatum is growing. The regulatory mechanism of DMI fungicide resistance in P. digitatum is poorly understood. Here, we first performed transcriptomic analysis of the P. digitatum strain Pdw03 treated with imazalil (IMZ) for 2 and 12 h. A total of 1338 genes were up-regulated and 1635 were down-regulated under IMZ treatment for 2 h compared to control while 1700 were up-regulated and 1661 down-regulated under IMZ treatment for 12 h. The expression of about half of the genes in the ergosterol biosynthesis pathway was affected during IMZ stress. Further analysis identified that 84 of 320 transcription factors (TFs) were differentially expressed at both conditions, making them potential regulators in DMI resistance. To confirm their roles, three differentially expressed TFs were selected to generate disruption mutants using the CRISPR/Cas9 technology. The results showed that two of them had no response to IMZ stress while ∆PdflbC was more sensitive compared with the wild type. However, disruption of PdflbC did not affect the ergosterol content. The defect in IMZ sensitivity of ∆PdflbC was restored by genetic complementation of the mutant with a functional copy of PdflbC. Taken together, our results offer a rich source of information to identify novel regulators in DMI resistance.
1. Introduction
Citrus (including all species in the Citrus genus) is one of the most important economic crops in the world, with its fruit yield ranking first. However, postharvest diseases caused by microbial pathogens during packaging, storage, transportation, and marketing can cause heavy economic losses of up to 30–50% of the total production when fruits are not timely treated with postharvest fungicides and/or placed in refrigeration []. Citrus green mold caused by Penicillium digitatum is the main cause of postharvest disease in citrus, which accounts for up to 90% of the total postharvest losses []. At present, the massive application of fungicides to marketed fruits is the major means to control the citrus green mold. In the mid-1970s, the sterol demethylation inhibitor (DMI) fungicides imazalil (IMZ) and prochloraz were introduced to control postharvest diseases of citrus fruits worldwide, owing to their good inhibitory effects on fruit decay symptoms, and these fungicides are still widely used in citrus preservation all over the world []. However, with the extensive and continuous use of DMI fungicides, the resistance problem in pathogens including P. digitatum is growing. According to previous studies, the resistance of pathogenic fungi to DMI fungicides is mainly caused by three molecular mechanisms: (a) point mutation occurs in the coding region of target gene CYP51s, and the resultant 14α-demethylase cannot bind to drugs or has a decreased drug affinity [,,,,], (b) the target gene CYP51s is overexpressed, resulting in the need for increased drug dose to inhibit the activity of 14α-demethylase [,,,,,], or (c) the expression of ATP-binding cassette (ABC) transporters or major facilitator superfamily (MFS) transporters on the cell membrane was up-regulated, which leads to accelerated elimination of intracellular drugs [,]. The reported molecular mechanisms of DMI resistance in P. digitatum are mainly caused by overexpression of drug target genes, which can be divided into R1, R2, and R3 resistance types: the presence of five tandem repeats of a 126-bp enhancer in the CYP51A gene’s promoter (R1) [], the insertion of a unique 199-bp transposable element in the CYP51A gene’s promoter (R2) [], and the insertion of the 199-bp element in the CYP51B gene’s promoter (R3) []. The R3 resistance type now exists predominantly in the DMI-resistant population of P. digitatum in the main producing areas of citrus, making the control strategies severely challenged [,].
As the ergosterol synthesis pathway is targeted by DMI fungicides, how this pathway is regulated has received widespread attention. In eukaryotes, the primary regulators of the sterol production pathway are called sterol regulatory element binding proteins, or SREBPs []. SREBPs belong to the bHLH class of transcription factors and are widely found in fungi from genera like Cryptococcus, Schizomyces, and Aspergillus []. SREBPs can recognize and bind to sterol regulatory elements (SREs) in the promoter region of sterol synthesis pathway genes, thereby regulating the expression of target genes []. However, the copy number and function of SREBP homologous protein in different species may differ. For example, two SREBP homologs, Sre1 and Sre2, are found in Schizosaccharides pombe; Sre1 is the main regulator of ergosterol synthesis and hypoxic adaptation, while Sre2 is not involved in the regulation of these two important functions []. In Aspergillus fumigatus, two SREBP homologous proteins, SrbA and SrbB, have been identified, and both show an important role in hypoxic growth and pathogenicity []. In Magnaporthe oryzae, the SREBP encoding gene deletion mutant, ΔMosre1, showed increased susceptibility to hypoxia and delayed growth in host cells. However, such defects did not result in a substantial decrease in disease severity []. In the medicinal fungus Ganoderma lingzhi, the metabolism of Ganoderic acids, lipids, and ergosterol was significantly increased when SREBP was overexpressed []. There are two SREBP homologous proteins, PdSreA and PdSreB, in P. digitatum. Deletion of PdsreA or PdsreB led to abnormal mycelia growth, decreased intracellular ergosterol content, and increased susceptibility to DMI fungicide, but the influence on virulence is very limited [].
Although some yeast fungi like S. cerevisiae, C. albicans, C. glabrata, and C. parapsilosis also contain SREBP homologous proteins, these SREBPs are not involved in the regulation of intracellular ergosterol synthesis []. Instead, these fungi utilize transcription factor Upc2 to control ergosterol synthesis, drug resistance, and hypoxic response [,,,,]. Surprisingly, both SREBP and Upc2 exist in the Fusarium graminearum genome; however, they are not associated with ergosterol regulation and sensitivity to DMI fungicides. Intriguingly, a transcriptional factor named FgSR in F. graminearum was reported to bind to the promoter region of 20 genes involved in sterol synthesis, and deletion of the FgSR gene resulted in decreased sterol synthesis, increased DMI sensitivity, less deoxynivalenol production, and reduced virulence, proving that FgSR is a novel and master regulator of sterol biosynthesis in F. graminearum [].
According to previous studies, it is speculated that there may be a variety of regulatory pathways controlling ergosterol synthesis in fungi. In addition to SREBPs, other transcription factors that may participate in the regulation of ergosterol content and DMI resistance in P. digitatum are still not known. Thus, the objective of this study is to find novel regulators of DMI fungicide resistance in the citrus postharvest pathogen P. digitatum.
2. Materials and Methods
2.1. Fungal Strains and Growth Conditions
This study was conducted with the wild-type P. digitatum strain Pdw03 (CBS130527), isolated from an infected citrus fruit (Quzhou of Zhejiang province, China) and resistant to IMZ []. Pdw03 and its derived mutants were kept on solid potato dextrose agar (PDA), and conidia were collected after 5–7 days of incubation. Mycelia were harvested from potato dextrose broth (PDB) after incubation at 160 rpm at 25 °C for 2 days. To prepare RNA sequencing samples, conidial suspensions (106 spores/mL) 100 μL of P. digitatum Pdw03 were cultured in PDB medium at 25 °C for 48 h, and IMZ was then added to PDB medium (6 μg/mL), and samples were collected at 0, 2, and 12 h, respectively.
2.2. Transcriptome Analysis
Each sample’s collected mycelia were ground in liquid nitrogen, and total RNA was extracted with an RNAprep Pure Kit (Tiangen Biotech Co., Ltd., Beijing, China). RNA-Seq libraries were generated with the Illumina TruSeq RNA Sample Preparation Kit and sequenced on an Illumina NovaSeq6000 platform by the Biomarker Technology Co., Ltd. (Beijing, China). The RNA-Seq was conducted for three biological replicates of each sample. Trimmomatic v0.32 was used to filter low-quality reads and remove adaptor sequences from raw reads []. Clean reads were mapped to the reference genome with HISAT2 []. The number of reads mapped to each gene was counted using featureCounts v2.0.0 []. Principal component analysis (PCA) was performed to identify whether samples clustered based on treatment using the function prcomp() in R 4.2.3 (https://rdrr.io/r/stats/prcomp.html (accessed on 10 March 2023)). The differential gene expression was analyzed using DESeq2 []. Transcripts were considered differentially expressed when their absolute value of log2 fold change (log2FC) was more than 1 and the false-discovery rate (FDR) value was less than 0.01. DEGs were analyzed for gene ontology (GO) enrichment using topGO v2.28.0 []. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted with KOBAS-I []. The transcriptome project has been deposited at NCBI BioProject under the accession PRJNA1072022.
2.3. Quantitative Real-Time PCR
To validate the reliability of the transcriptome data, we chose 8 genes to measure their expression levels through qRT-PCR. Reverse transcription was performed with 1 μg of each RNA sample using the HiScript III All-in-one RT SuperMix reagent kit (Vazyme Biotech Co., Ltd., Nanjing, China). The target genes’ relative transcript levels were determined in triplicate using a 7500 Real-Time PCR equipment (ABI, Foster City, CA, USA). The actin gene (AB030227) served as an internal reference, and the data were analyzed using the 2−ΔΔCT method.
2.4. Disruption of P. digitatum Genes and Mutant Complementation
To ensure their roles in DMI resistance, we then selected 3 differentially expressed transcription factor encoding genes, Pdw03_2654 (bZIP, down-regulated), Pdw03_3762 (Zn_clus, down-regulated), and Pdw03_3571 (zf-C2H2, up-regulated, PdflbC), for functional analyses in P. digitatum. Gene disruption was performed according to the recently developed CRISPR/Cas9 tools in P. digitatum with minor modification []. The 20-bp protospacer for each target gene was designed with the GuideMaker v0.4.0 software []. The sgRNA expression constructs were synthesized by Shangya Biotechnology Co., Ltd., Hangzhou, China. After digesting with the BglII ligase, the sgRNA expression construct was inserted into the pFC332 vector to generate the gene disruption plasmid pFC332-Gene-D. Then, this vector was transformed into the protoplasts of P. digitatum Pdw03 according to the previously described protocol []. Transformants grown on PDA with hygromycin (100 mg/L) were isolated, and mutants were confirmed by PCR and Sanger sequencing. For genetic complementation, a functional copy of PdflbC DNA fragment under its native promoter was amplified and inserted into the pA1300-NEO vector []. The resultant vector was subsequently transformed into ∆PdflbC protoplasts. Transformants were recovered from neomycin-supplemented PDA (100 mg/L) and examined by PCR and Sanger sequencing. Primers are listed in Table S1.
2.5. Mycelial Growth Tests
To know whether these transcription factors play a role in environmental adaptation, P. digitatum wild type and mutant strains were grown on PDA plates containing 4 μg/mL IMZ, 0.7 M NaCl, 0.05 mM Menadione, or 200 μg/mL Congo red (CR), 200 μg/mL Sodium dodecyl sulfate (SDS). Each plate was inoculated with a 5 mm mycelial plug, and the colony diameters were measured after 5 days of incubation at 25 °C. The percentage of the mycelial radial growth inhibition (MRGI) rate was calculated using the formula MRGI% = ((C − N)/(C − 0.5)) × 100%, where C is colony diameter of the control (PDA only) and N is that of a treatment. The experiments were repeated thrice.
2.6. Virulence Assays
To examine the fungal virulence, infection assays were performed on mandarin (Citrus reticulata Blanco) fruits using the wild type and 3 gene deletion mutants of P. digitatum. Citrus peel was first wounded (1–2 mm deep) with sterile needles and then inoculated with 10 μL conidial suspensions (1 × 106 conidia/mL) of the wild type and mutant strains of P. digitatum. The infected citrus fruits were kept at 25 °C for 4 days before being tested for lesion size. Six fruits were inoculated for each strain, and the experiment was conducted two times.
2.7. Determination of Intracellular Ergosterol Content
To extract ergosterol, fungal spores were placed in a 50 mL yeast glucose (YG) medium (15 g/L glucose, 5 g/L yeast extract), which was then shaken continuously at 25 °C for two days. Mycelia were then collected for ergosterol extraction according to the published protocol (Wang et al., 2014 []). Each sample was analyzed using the Waters e2695 high-performance liquid chromatography (HPLC) system. Ergosterol was separated at room temperature on a Discovery C18 250 nm × 4.6 nm, 5 μL analytical column using 100% methanol (chromatography pure) as mobile phase. Quantification of ergosterol was measured based on retention time and co-chromatography of a commercial standard of ergosterol (Sigma, St. Louis, MO, USA) under the detection wavelength of 282 nm. This experiment was repeated twice.
3. Results
3.1. Transcriptomic Analysis of P. digitatum in Response to IMZ
The number of clean reads ranged from 20,838,987 to 29,875,237, and the percentage of Q30 bases in all samples was no less than 89% (Table S2). The proportion of reads that aligned to the genome ranged between 86.9 and 93.8% (Table S2). As demonstrated in Figure 1, principal component 1 (PC1) accounts for 49.2% of the sample variability, whereas principal component 2 (PC2) accounts for 34.2%. Each condition was clearly distinguished from the other, and samples in the same condition were more similar (Figure 1A). These results indicate the good quality of the transcriptome data.
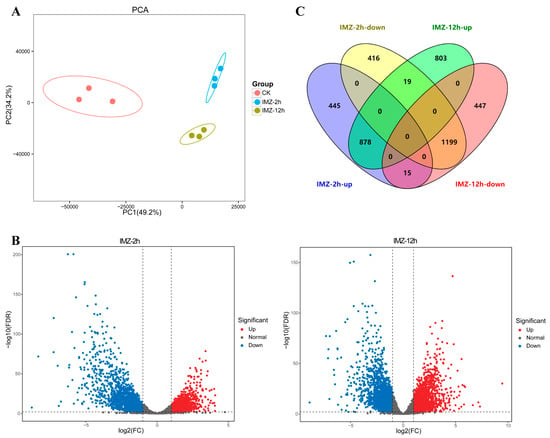
Figure 1.
Overview of the differentially expressed genes. (A) PCA plot of RNA-sequencing samples. Each dot represents a sample. The distance between dots represents the similarity of samples. The horizontal axis and the vertical axis represent the contribution of PC1 and PC2 to the identification of distinct samples. (B) Volcano plots of global gene expression pattern. Each dot represents a gene. Blue dots are down-regulated genes, while red dots are up-regulated ones, and black dots are genes without significant difference. (C) Venn diagram displaying the up-regulated and down-regulated genes after IMZ treatment for 2 h or 12 h compared to that of the control 0 h.
Gene expression comparisons were then performed between the IMZ treatment and control samples, i.e., 2 h vs. 0 h and 12 h vs. 0 h. In total, 2973 differentially expressed genes, including 1338 up-regulated genes and 1635 down-regulated genes, were identified in P. digitatum under IMZ treatment for 2 h compared to the control (Figure 1B,C). GO enrichment analysis revealed that the up-regulated genes were significantly enriched for the biological progress categories “nucleic acid metabolic process”, “RNA metabolic process”, “organic cyclic compound metabolic process”, “nucleobase-containing compound metabolic process”, “cellular aromatic compound metabolic process”, “heterocycle metabolic process”, “ribosome biogenesis”, “cellular process”, “ribonucleoprotein complex biogenesis”, “ncRNA processing”, and “DNA-templated transcription”. Nevertheless, GO analyses did not show any significant annotation in the down-regulated gene set (Figure 2). KEGG analysis showed that the up-regulated genes were significantly enriched in “Cell cycle”, “Meiosis “, and “Ribosome biogenesis in eukaryotes”. However, the down-regulated genes were not significantly enriched in any pathway (Figure 3).
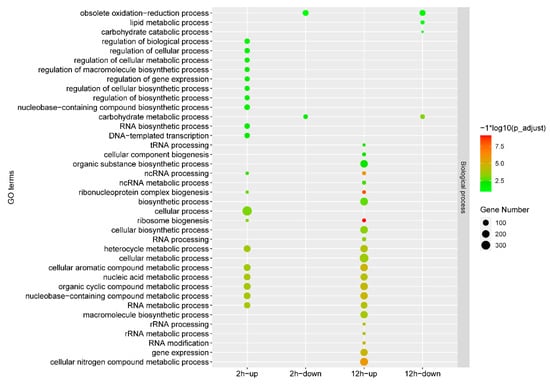
Figure 2.
GO analysis of the differentially expressed genes under different conditions. The x-axis indicates different gene sets. The y-axis indicates the GO term.
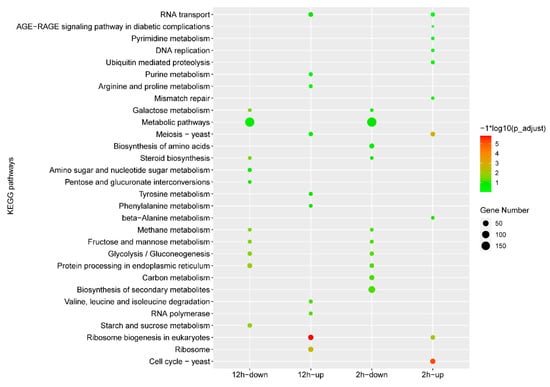
Figure 3.
KEGG analysis of the differentially expressed genes under different conditions. The x-axis indicates different gene sets. The y-axis indicates the pathway name.
Overall, 3361 genes were differentially expressed in P. digitatum under IMZ treatment for 12 h compared to control, comprising 1700 up-regulated and 1661 down-regulated genes (Figure 1B,C). GO enrichment analysis revealed that the up-regulated genes were significantly enriched for the biological progress categories “ribosome biogenesis”, “ribonucleoprotein complex biogenesis”, “cellular nitrogen compound metabolic process”, “cellular aromatic compound metabolic process”, “gene expression”, “organic cyclic compound metabolic process”, “nucleobase-containing compound metabolic process”, “nucleic acid metabolic process”, “heterocycle metabolic process”, “macromolecule biosynthetic process”, “cellular metabolic process”, “cellular biosynthetic process”, “organic substance biosynthetic process”, “cellular component biogenesis”, and some RNA related categories such as “RNA modification“ and “RNA processing”, while those down-regulated genes were only significantly enriched for the category “carbohydrate metabolic process” (Figure 2). KEGG pathway enrichment analysis showed the up-regulated genes were significantly enriched in “Ribosome” and “Ribosome biogenesis in eukaryotes”, while the down-regulated genes were significantly enriched in “Starch and sucrose metabolism”, “Protein processing in endoplasmic reticulum”, “Glycolysis/Gluconeogenesis”, “Methane metabolism”, “Fructose and mannose metabolism”, “Steroid biosynthesis”, and “Galactose metabolism” (Figure 3).
The results of qRT-PCR revealed that though the level of fold changes between the two techniques for several of the genes in the two conditions varied, both generally displayed comparable patterns in transcript accumulation (Figure S1).
3.2. Expression of the Ergosterol Biosynthesis Pathway
The results showed that the expression of about half of the genes in this pathway was affected after IMZ treatment. All DEGs involved in ergosterol biosynthesis were down-regulated except for three genes encoding Mvd1 (diphosphomevalonate decarboxylase), ERG7 (lanosterol cyclase), and ERG27 (3-ketosteroid reductase), which up-regulated their expression level by 2~3 folds. Reduced expression of the genes encoding Hmg2 (HMG-CoA reductase), ERG1 (Squalene epoxidase), ERG26 (C-3 sterol dehydrogenase), ERG3A (C-5 sterol desaturase), Hyd1 (cholesterol delta-isomerase), ERG25 (C-4 methylsterol oxidase), and ERG11A/B (Sterol 14-alpha demethylase) was detected in both conditions. The transcript levels of left DEGs, namely smt1, ERG24, ERG2, and ERG5, were significantly decreased in only one condition (2 h or 12 h post IMZ treatment) (Figure 4).
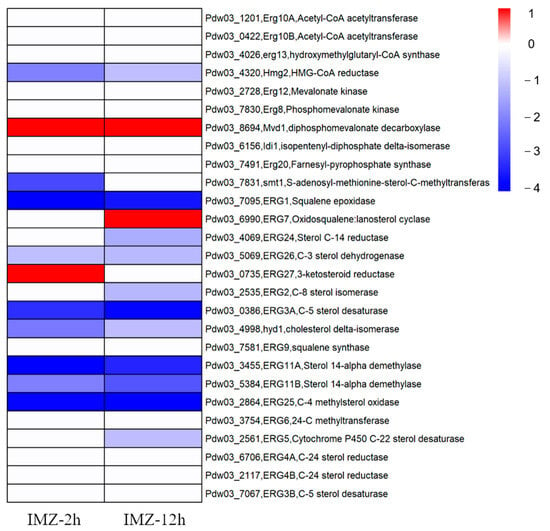
Figure 4.
Heat map visualization of gene expressions associated with the ergosterol biosynthesis pathway under different conditions.
3.3. Expression of Potential Regulators
After comparing two sets of DEGs associated with IMZ treatment, 878 and 1199 DEGs were up-regulated and down-regulated in both conditions, respectively (Figure 1C). We then searched for potential regulators of ergosterol biosynthesis and DMI resistance from these core DEGs. Finally, a total of 84 transcription factors were discovered to be differentially expressed (26.3% of all transcription factors in the genome), with the expressions of 54 being up-regulated and 30 being down-regulated (Table S3).
3.4. Disruption of Three Transcription Factors in P. digitatum
We obtained two mutants for Pdw03_2654, two for Pdw03_3762, and one for Pdw03_3571 (Figure S2). The growth assay showed that the average colony diameter of the ∆PdflbC mutant was slightly larger than that of the wild type, while the others showed a similar growth rate with the wild type (Figure 5). However, the vegetative growth of ∆PdflbC was strongly retarded on medium supplemented with 4 μg/mL IMZ fungicide, while the other mutants and the complemented CPPdflbC mutant showed wild-type levels of growth (Figure 5 and Figure S3A). These results demonstrated that PdflbC is a crucial regulator in DMI resistance in P. digitatum.
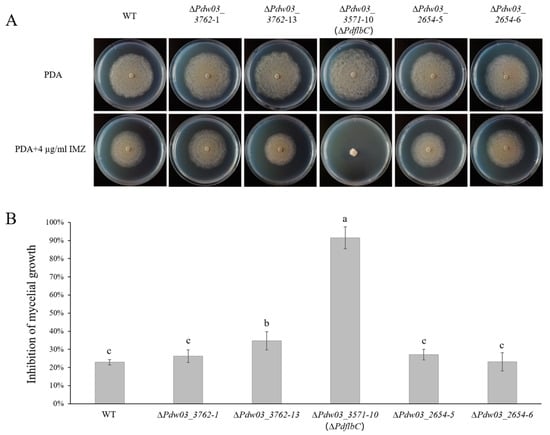
Figure 5.
Sensitivity of the wild type and gene deletion mutants to IMZ. (A) Colony morphology of the wild type and gene deletion mutants grown on potato dextrose agar (PDA) supplemented with or without IMZ. All plants were incubated at 25 °C for 5 days. (B) Growth inhibition of gene deletion mutants grown on PDA supplemented with or without IMZ. Different lowercase letters indicate significant difference estimated by Duncan’s test (p ≤ 0.05).
The stress assays showed that ∆Pdw03_3762 exhibited similar phenotypes to the wild-type strain, ∆Pdw03_2654 was more sensitive to salt stress, and the ∆PdflbC mutant exhibited significantly increased sensitivity to osmotic stress, menadione, and cell wall interfering agents CR and SDS (Figure 6). All phenotypic defects of ∆PdflbC excluding the elevated sensitivity to NaCl were restored by genetic complementation of the mutant with the wild-type PdflbC (Figure S3B). These results indicated that PdflbC is essential for the adaptation to environmental stresses in P. digitatum.
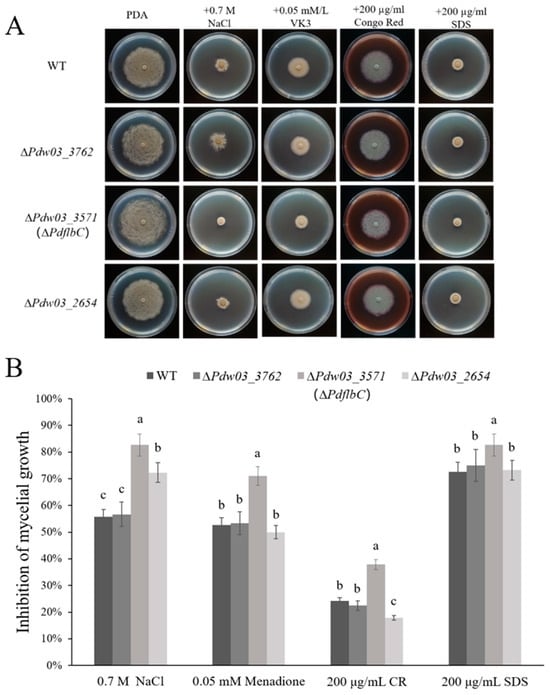
Figure 6.
Sensitivity of the wild type and gene deletion mutants to environmental stresses. (A) Mycelial plugs of different strains were placed on PDA supplemented with NaCl, Menadione, Congo red (CR), and Sodium dodecyl sulfate (SDS) at the concentrations indicated in the figure. All plates were incubated at 25 °C for 5 days. (B) Growth inhibition of gene deletion mutants grown on PDA supplemented with different chemicals. Different lowercase letters indicate significant difference estimated by Duncan’s test (p ≤ 0.05).
Both ∆Pdw03_3762 and ∆Pdw03_2654 generated slightly increased spores compared with the wild-type strain, while ∆PdflbC produced only half the spores of the wild type (Figure 7). The wild-type sporulation level was restored in CPPdflbC (Figure S3C). However, deletion of these three genes did not affect germination time and rate of conidia compared with WT.
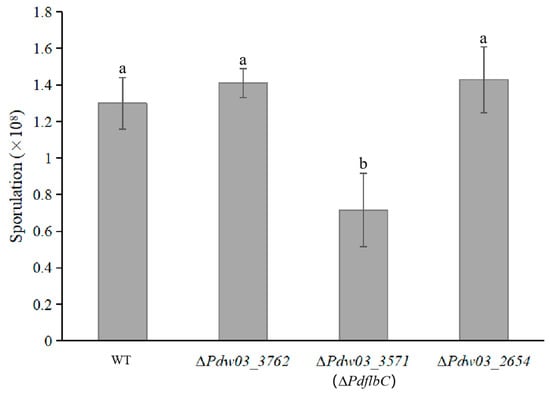
Figure 7.
Comparison of sporulation of the wild type and gene deletion mutants. Strains were inoculated on PDA at 25 °C for 5 days, and conidia were then collected and counted in a hemocytometer. Different lowercase letters indicate significant difference estimated by Duncan’s test (p ≤ 0.05).
The virulence assay revealed that all gene deletion mutants caused similar maceration lesions with the wild type, suggesting that these genes are dispensable for the pathogenicity of P. digitatum (Figure 8).

Figure 8.
Decay symptoms of the wile type and gene deletion mutants on citrus fruits. Mandarin (Citrus reticulata Blanco) fruits inoculated with the indicated strains were incubated at 25 °C for 4 days.
3.5. Roles of PdflbC in Regulating Ergosterol Biosynthesis
Ergosterol content of the wild type and the ∆PdflbC mutant were quantified using the HPLC method, and the results showed that no significant differences were found between them (Figure 9).
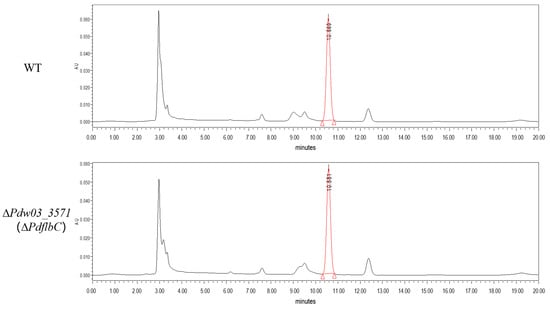
Figure 9.
High-performance liquid chromatography (HPLC) analysis of ergosterol content from the wild type and the ∆PdflbC mutant.
4. Discussion
P. digitatum is the most common cause of postharvest decay in citrus fruits around the world. At present, the problem of DMI fungicide resistance in the P. digitatum population is rising due to the extensive and continuous use of fungicides for decades. Several mechanisms underlying DMI resistance have been documented, such as enhanced gene expression because of insertions in the promoter or alterations in the coding area [,,]. The regulation of DMI fungicide resistance can be mediated by many regulators in fungi; nevertheless, related studies were poorly reported in P. digitatum. To gain further insight into the regulatory mechanism of DMI fungicide resistance in P. digitatum, we performed transcriptomic analysis of the P. digitatum strain Pdw03 before and after IMZ treatment at different time points and we showed how this strain responded to IMZ stress and identified potential regulators in this process; we also characterized the functions of three candidate genes using the CRISPR/Cas9 technology to show their relationship with IMZ sensitivity.
Our RNA-Seq data of the P. digitatum Pdw03 strain after being treated with IMZ for 2 and 12 h identified 2973 and 3361 differentially expressed genes, respectively (Figure 1). Function annotation analysis revealed that P. digitatum showed many similar responses to these two treatment times. The genes expressed at a higher degree were enriched in ribosome-related biological processes, such as ribonucleoprotein complex biogenesis, ribosome biogenesis, and nucleic acid metabolism, while the down-regulated genes were more related to carbohydrate metabolism like starch/sucrose metabolism and gluconeogenesis (Figure 2 and Figure 3). Previously, whole transcriptome analysis of P. digitatum HS-F6 strain treated with prochloraz was performed, and 1100 DEGs were identified after prochloraz treatment, with 402 up-regulated and 698 down-regulated. The top five functional categories of the DEGs of HS-F6 before and after prochloraz treatment included cation binding, ion binding, transition metal ion binding, metal ion binding, and tetrapyrrole binding []. These results revealed that, although both prochloraz and IMZ belong to DMI fungicide, their respective resistant strain showed different response patterns when fungicides were applied. This inconsistency can be explained by the phenomenon that the IMZ-resistant strain Pdw03 was more sensitive to prochloraz []. However, the underlying mechanism is not clear so far.
As the biosynthesis of ergosterol was mainly targeted by the DMI fungicides, it is expected that the expression of genes in the ergosterol biosynthesis pathway was significantly changed after DMI treatment. Usually, sterol biosynthetic genes up-regulated their expression after a DMI fungicide treatment, as maintenance or reinforcement of cell membrane integrity is a universal response when exposed to DMI fungicide. In C. beticola, 18 genes in the ergosterol biosynthesis pathway were induced in DMI-resistant strain after tetraconazole exposure []. Both P. italicum and P. digitatum showed overexpression of CYP51 genes when treated with prochloraz [,]. However, in our transcriptome data, 12 ergosterol biosynthetic genes were underexpressed after IMZ treatment, while only 3 were overexpressed (Figure 4). This may be caused by the large dose of fungicide used in our experiment, and the vegetative growth of P. digitatum Pdw03 might be strongly inhibited under the concentration of 6 μg/mL used in this study.
Previous studies showed that transcription factors, such as SREBP, UPC2, and SR, are critical regulators of ergosterol synthesis in fungi [,,]. In P. digitatum, two SREBP homologous proteins, PdSreA and PdSreB, were previously reported to mediate ergosterol biosynthesis and DMI resistance by regulating the expression of many related genes, for example, Erg5, Erg10B, Erg7, Erg13, Erg3B, Erg1, and Cyp51A/Erg11A []. To find more regulators in DMI resistance, we identified 54 up-regulated and 30 down-regulated transcription factors, which might be good candidates for functional analyses (Table S3). Of them, functions of two transcription factors have been characterized in fungi. One is the flbC gene, which is crucial for the activation of conidiation and the co-ordination of vegetative growth and development in A. nidulans [], and the other is the pH-responsive transcription factor PacC, which is essential for pH sensing and pathogenicity in some plant pathogenic fungi [,]. However, the functions of all these genes in mediating ergosterol content and DMI resistance are largely unknown, and whether they can play as critical regulators needs further investigation.
To help establish a role in DMI resistance, three genes were selected for functional characterization based on their expression levels. To our surprise, disruption of the conidiation-related transcription factor PdflbC led to markedly increased sensitivity of the mutant to IMZ (Figure 5). FlbC contains two C2H2 zinc fingers at the C-terminus and a putative activation domain at the N-terminus []. In A. nidulans, the lack of flbC causes delayed and reduced conidiation, brlA and vosA expression, and conidial germination, while overexpression of flbC inhibits radical growth and activates expression of brlA, abaA, and vosA []. In P. oxalicum, the deletion of PoFlbC led to reduced and delayed conidiation and down-regulated the expression of brlA, while overexpression of flbC slightly affected conidiation, impaired vegetative growth, and decreased the brlA expression []. In addition, the synthesis of cellulase and hemicellulase was significantly decreased after PoflbC gene deletion. Down-regulation of genes producing cellulases, hemicellulases, and other proteins with roles in lignocellulose breakdown was discovered in the PoflbC deletion mutant []. In P. digitatum, the PdflbC gene deletion mutant also showed reduced conidiation (Figure 7), indicating that flbC is a conserved upstream developmental activator in Penicillium and Aspergillus. However, whether flbC is required for DMI fungicide resistance in other fungi needs further investigation. Although the PdflbC gene is essential for DMI resistance, the ergosterol content of the PdflbC gene deletion mutant was not changed compared with that of the wild type (Figure 9), suggesting that PdflbC-mediated DMI fungicide resistance is in a way irrelevant to the ergosterol biosynthesis pathway, and the underlying mechanism remains elusive.
5. Conclusions
In this study, we performed transcriptomic analysis of the P. digitatum strain Pdw03 after IMZ treatment and identified a large number of differentially expressed genes. Functional analysis demonstrated that one transcriptional factor, PdflbC, is crucial for the tolerance of P. digitatum to IMZ. However, functions of the majority of candidate genes are largely unknown, and their roles in DMI resistance need further validation. Taken together, our results provide new insights into the mode of action of IMZ response in P. digitatum and offer a rich source of information to identify novel regulators in DMI resistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof10050360/s1, Figure S1. qRT-PCR validation of the RNA-Seq data. Figure S2. Sequence alignment of gene locus in the selected mutants. Protospacers were marked in red. Protospacer-adjacent motifs (PAM) were underlined. Nucleotide alterations are highlighted in yellow. Figure S3. Phenotypes of the complemented strain CPPdflbC. (A) Sensitivity of the wild type, the gene deletion mutant ∆PdflbC and the complemented strain CPPdflbC to IMZ. (B) Sensitivity of the wild type, the gene deletion mutant ∆PdflbC and the complemented strain CPPdflbC to environmental stresses. (C) Sporulation of the wild type and the complemented strain CPPdflbC. Table S1. Primers used in this study. Table S2. Statistics of sequencing data. Table S3. Differentially expressed transcription factors after IMZ treatment.
Author Contributions
Y.X.: Methodology, Investigation, Data curation, Formal analysis, Visualization, Funding acquisition. J.Z.: Methodology, Investigation. B.F.: Methodology, Investigation. M.S.: Investigation. W.C.: Investigation. X.L.: Investigation. Y.G.: Software, Formal analysis, Writing. C.S.: Methodology, Supervision. H.W.: Methodology, Supervision. M.W.: Conceptualization, Supervision, Funding acquisition, Writing and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31901913) and the Graduate Research Innovation Promotion Project of Hangzhou Normal University (2022HSDYJSKY216).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Acknowledgments
We thank Qin Zhu and Cong Dang at Hangzhou Normal University for providing us with the equipment to perform the experiments.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Maria, C.S.; Giuseppe, A.; Naouel, A.; Francesco, G.; Giovanni, C.D.R. Advance in Citrus Postharvest Management: Diseases, Cold Storage and Quality Evaluation. In Citrus Pathology; Harsimran, G., Harsh, G., Eds.; IntechOpen: Rijeka, Croatia, 2017; Chapter 7. [Google Scholar]
- Kanetis, L.; Förster, H.; Adaskaveg, J.E. Comparative efficacy of the new postharvest fungicides azoxystrobin, fludioxonil, and pyrimethanil for managing citrus green mold. Plant Dis. 2007, 91, 1502–1511. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Torres, P. Molecular Mechanisms Underlying Fungicide Resistance in Citrus Postharvest Green Mold. J. Fungi 2021, 7, 783. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, Y.; Schnabel, G.; Peng, C.A.; Lagishetty, S.; Smith, K.; Luo, C.; Yuan, H. Inherent Resistance to 14α-Demethylation Inhibitor Fungicides in Colletotrichum truncatum Is Likely Linked to CYP51A and/or CYP51B Gene Variants. Phytopathology 2018, 108, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Carter, H.E.; Fraaije, B.A.; West, J.S.; Kelly, S.L.; Mehl, A.; Shaw, M.W.; Cools, H.J. Alterations in the predicted regulatory and coding regions of the sterol 14α-demethylase gene (CYP51) confer decreased azole sensitivity in the oilseed rape pathogen Pyrenopeziza brassicae. Mol. Plant Pathol. 2014, 15, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, T.M.; Berg, G.; Semaškienė, R.; Mehl, A.; Sierotzki, H.; Stammler, G.; Justesen, A.F.; Jørgensen, L.N. Impact of DMI and SDHI fungicides on disease control and CYP51 mutations in populations of Zymoseptoria tritici from Northern Europe. Eur. J. Plant Pathol. 2015, 143, 861–871. [Google Scholar] [CrossRef]
- Mair, W.J.; Deng, W.; Mullins, J.G.; West, S.; Wang, P.; Besharat, N.; Ellwood, S.R.; Oliver, R.P.; Lopez-Ruiz, F.J. Demethylase inhibitor fungicide resistance in Pyrenophora teres f. sp. teres associated with target site modification and inducible overexpression of Cyp51. Front. Microbiol. 2016, 7, 1279. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Du, J.; Chi, M.; Sun, X.; Liang, W.; Huang, J.; Li, B. The Y137H mutation in the cytochrome P450 FgCYP51B protein confers reduced sensitivity to tebuconazole in Fusarium graminearum. Pest Manag. Sci. 2018, 74, 1472–1477. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, G.; Jones, A.L. The 14α-demethylasse (CYP51A1) gene is overexpressed in Venturia inaequalis strains resistant to myclobutanil. Phytopathology 2001, 91, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Proffer, T.J.; Jacobs, J.L.; Sundin, G.W. Overexpression of the 14α-demethylase target gene (CYP51) mediates fungicide resistance in Blumeriella jaapii. Appl. Environ. Microbiol. 2006, 72, 2581–2585. [Google Scholar] [CrossRef]
- Luo, C.-X.; Cox, K.D.; Amiri, A.; Schnabel, G. Occurrence and detection of the DMI resistance-associated genetic element ‘Mona’ in Monilinia fructicola. Plant Dis. 2008, 92, 1099–1103. [Google Scholar] [CrossRef]
- Diaz-Trujillo, C.; Chong, P.; Stergiopoulos, I.; Cordovez, V.; Guzman, M.; De Wit, P.J.; Meijer, H.J.; Scalliet, G.; Sierotzki, H.; Lilia Peralta, E. A new mechanism for reduced sensitivity to demethylation-inhibitor fungicides in the fungal banana black Sigatoka pathogen Pseudocercospora fijiensis. Mol. Plant Pathol. 2018, 19, 1491–1503. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Verweij, P.E.; Melchers, W.J.G.; Moye-Rowley, W.S. Differential Functions of Individual Transcription Factor Binding Sites in the Tandem Repeats Found in Clinically Relevant cyp51A Promoters in Aspergillus fumigatus. mBio 2022, 13, e00702–e00722. [Google Scholar] [CrossRef] [PubMed]
- Mernke, D.; Dahm, S.; Walker, A.-S.; Lalève, A.; Fillinger, S.; Leroch, M.; Hahn, M. Two promoter rearrangements in a drug efflux transporter gene are responsible for the appearance and spread of multidrug resistance phenotype MDR2 in Botrytis cinerea isolates in French and German vineyards. Phytopathology 2011, 101, 1176–1183. [Google Scholar] [CrossRef] [PubMed]
- Omrane, S.; Sghyer, H.; Audéon, C.; Lanen, C.; Duplaix, C.; Walker, A.S.; Fillinger, S. Fungicide efflux and the MgMFS 1 transporter contribute to the multidrug resistance phenotype in Zymoseptoria tritici field isolates. Environ. Microbiol. 2015, 17, 2805–2823. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, H.; Hasegawa, K.; Nakaune, R.; Lee, Y.J.; Makizumi, Y.; Akutsu, K.; Hibi, T. Tandem Repeat of a Transcriptional Enhancer Upstream of the Sterol 14α-Demethylase Gene (CYP51) in Penicillium digitatum. Appl. Environ. Microbiol. 2000, 66, 3421–3426. [Google Scholar] [CrossRef] [PubMed]
- Ghosoph, J.M.; Schmidt, L.S.; Margosan, D.A.; Smilanick, J.L. Imazalil resistance linked to a unique insertion sequence in the PdCYP51 promoter region of Penicillium digitatum. Postharvest Biol. Technol. 2007, 44, 9–18. [Google Scholar] [CrossRef]
- Sun, X.; Wang, J.; Feng, D.; Ma, Z.; Li, H. PdCYP51B, a new putative sterol 14alpha-demethylase gene of Penicillium digitatum involved in resistance to imazalil and other fungicides inhibiting ergosterol synthesis. Appl. Microbiol. Biotechnol. 2011, 91, 1107–1119. [Google Scholar] [CrossRef] [PubMed]
- de Ramón-Carbonell, M.; Sánchez-Torres, P. Significance of 195 bp-enhancer of PdCYP51B in the acquisition of Penicillium digitatum DMI resistance and increase of fungal virulence. Pestic. Biochem. Physiol. 2020, 165, 104522. [Google Scholar] [CrossRef] [PubMed]
- Espenshade, P.J.; Hughes, A.L. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 2007, 41, 401–427. [Google Scholar] [CrossRef]
- Bien, C.M.; Espenshade, P.J. Sterol regulatory element binding proteins in fungi: Hypoxic transcription factors linked to pathogenesis. Eukaryot. Cell 2010, 9, 352–359. [Google Scholar] [CrossRef]
- Goldstein, J.L.; DeBose-Boyd, R.A.; Brown, M.S. Protein sensors for membrane sterols. Cell 2006, 124, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Todd, B.L.; Espenshade, P.J. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 2005, 120, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.; Barker, B.M.; Carey, C.C.; Merriman, B.; Werner, E.R.; Lechner, B.E.; Dhingra, S.; Cheng, C.; Xu, W.; Blosser, S.J. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Path. 2014, 10, e1004487. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Chung, H.; Lee, G.-W.; Koh, S.-K.; Chae, S.-K.; Lee, Y.-H. Genome-wide analysis of hypoxia-responsive genes in the rice blast fungus, Magnaporthe oryzae. PLoS ONE 2015, 10, e0134939. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-N.; Wu, F.-Y.; Tian, R.-Y.; Shi, Y.-X.; Xu, Z.-Q.; Liu, J.-Y.; Huang, J.; Xue, F.-F.; Liu, B.-Y.; Liu, G.-Q. The bHLH-zip transcription factor SREBP regulates triterpenoid and lipid metabolisms in the medicinal fungus Ganoderma lingzhi. Commun. Biol. 2023, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Ruan, R.; Wang, M.; Liu, X.; Sun, X.; Chung, K.-R.; Li, H. Functional analysis of two sterol regulatory element binding proteins in Penicillium digitatum. PLoS ONE 2017, 12, e0176485. [Google Scholar] [CrossRef]
- Guida, A.; Lindstädt, C.; Maguire, S.L.; Ding, C.; Higgins, D.G.; Corton, N.J.; Berriman, M.; Butler, G. Using RNA-seq to determine the transcriptional landscape and the hypoxic response of the pathogenic yeast Candida parapsilosis. BMC Genom. 2011, 12, 628. [Google Scholar] [CrossRef] [PubMed]
- Synnott, J.M.; Guida, A.; Mulhern-Haughey, S.; Higgins, D.G.; Butler, G. Regulation of the hypoxic response in Candida albicans. Eukaryot. Cell 2010, 9, 1734–1746. [Google Scholar] [CrossRef] [PubMed]
- Zavrel, M.; Hoot, S.J.; White, T.C. Comparison of sterol import under aerobic and anaerobic conditions in three fungal species: Candida albicans, Candida glabrata and Saccharomyces cerevisiae. Eukaryot. Cell 2013, 12, 725–738. [Google Scholar] [CrossRef]
- Rogers, P.D. Sterol homeostasis in yeast. Nat. Chem. Biol. 2022, 18, 1170–1171. [Google Scholar] [CrossRef]
- Tan, L.; Chen, L.; Yang, H.; Jin, B.; Kim, G.; Im, Y.J. Structural basis for activation of fungal sterol receptor Upc2 and azole resistance. Nat. Chem. Biol. 2022, 18, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jian, Y.; Chen, Y.; Kistler, H.C.; He, P.; Ma, Z.; Yin, Y. A phosphorylated transcription factor regulates sterol biosynthesis in Fusarium graminearum. Nat. Commun. 2019, 10, 1228. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ruan, R.; Li, H. The completed genome sequence of the pathogenic ascomycete fungus Penicillium digitatum. Genomics 2021, 113, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenführer, J. Gene set enrichment analysis with topGO. Bioconductor Improv. 2009, 27, 1–26. [Google Scholar]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Garrigues, S.; Manzanares, P.; Marcos, J.F. Application of recyclable CRISPR/Cas9 tools for targeted genome editing in the postharvest pathogenic fungi Penicillium digitatum and Penicillium expansum. Curr. Genet. 2022, 68, 515–529. [Google Scholar] [CrossRef]
- Poudel, R.; Rodriguez, L.T.; Reisch, C.R.; Rivers, A.R. GuideMaker: Software to design CRISPR-Cas guide RNA pools in non-model genomes. GigaScience 2022, 11, giac007. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yang, X.; Ruan, R.; Fu, H.; Li, H. Csn5 Is Required for the Conidiogenesis and Pathogenesis of the Alternaria alternata Tangerine Pathotype. Front. Microbiol. 2018, 9, 508. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, C.; Zhu, C.; Sun, X.; Ruan, R.; Li, H. Os2 MAP kinase-mediated osmostress tolerance in Penicillium digitatum is associated with its positive regulation on glycerol synthesis and negative regulation on ergosterol synthesis. Microbiol. Res. 2014, 169, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, J.; Liu, J.; Yuan, Y.; Li, N.; He, M.; Qi, T.; Hui, G.; Xiong, L.; Liu, D. Novel mutations in CYP51B from Penicillium digitatum involved in prochloraz resistance. J. Microbiol. 2014, 52, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, S.; Qin, T.; Li, N.; Niu, Y.; Li, D.; Yuan, Y.; Geng, H.; Xiong, L.; Liu, D. Whole transcriptome analysis of Penicillium digitatum strains treatmented with prochloraz reveals their drug-resistant mechanisms. BMC Genom. 2015, 16, 855. [Google Scholar] [CrossRef] [PubMed]
- Bolton, M.D.; Ebert, M.K.; Faino, L.; Rivera-Varas, V.; de Jonge, R.; Van de Peer, Y.; Thomma, B.P.H.J.; Secor, G.A. RNA-sequencing of Cercospora beticola DMI-sensitive and -resistant isolates after treatment with tetraconazole identifies common and contrasting pathway induction. Fungal Genet. Biol. 2016, 92, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cao, Q.; Li, N.; Liu, D.; Yuan, Y. Transcriptome analysis of fungicide-responsive gene expression profiles in two Penicillium italicum strains with different response to the sterol demethylation inhibitor (DMI) fungicide prochloraz. BMC Genom. 2020, 21, 156. [Google Scholar] [CrossRef] [PubMed]
- Kwon, N.-J.; Garzia, A.; Espeso, E.A.; Ugalde, U.; Yu, J.-H. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol 2010, 77, 1203–1219. [Google Scholar] [CrossRef]
- Zhang, T.; Sun, X.; Xu, Q.; Candelas, L.G.; Li, H. The pH signaling transcription factor PacC is required for full virulence in Penicillium digitatum. Appl. Microbiol. Biotechnol. 2013, 97, 9087–9098. [Google Scholar] [CrossRef]
- Li, B.; Chen, Y.; Tian, S. Function of pH-dependent transcription factor PacC in regulating development, pathogenicity, and mycotoxin biosynthesis of phytopathogenic fungi. FEBS J. 2022, 289, 1723–1730. [Google Scholar] [CrossRef]
- Park, H.-S.; Yu, J.-H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Li, Z.; Wu, R.; Qin, Y.; Liu, G.; Qu, Y. Penicillium oxalicum PoFlbC regulates fungal asexual development and is important for cellulase gene expression. Fungal Genet. Biol. 2016, 86, 91–102. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).