Simple Summary
This study aimed to assess prevalence and antimicrobial resistance of Campylobacter spp. in cattle in Mongolia. Thirty-five Campylobacter spp., including 23 C. jejuni, 7 C. hyointestinalis, 4 C. fetus, and 1 C. lari, were isolated. Multilocus sequence typing of C. jejuni cattle isolates revealed substantial genetic diversity. The antimicrobial resistance patterns of the C. jejuni cattle isolates completely differed from those of previously reported chicken isolates; excluding one ciprofloxacin-resistant isolate, all C. jejuni isolates were susceptible to tetracycline and ciprofloxacin. To the best of our knowledge, this is the first report on the characterization of Campylobacter spp. in cattle in Mongolia.
Abstract
Poultry and cattle are the major reservoirs of Campylobacter infection in humans. However, no information is available on Campylobacter spp. in cattle in Mongolia. Thus, this study aimed to assess their prevalence and antimicrobial resistance. Between 2019 and 2023, rectal swabs were collected from cattle on dairy farms around Ulaanbaatar city and in total, 35 Campylobacter spp., including 23 C. jejuni, 7 C. hyointestinalis, 4 C. fetus, and 1 C. lari, were isolated. Multilocus sequence typing of C. jejuni cattle isolates revealed substantial genetic diversity and identified 7 sequence types (STs) including ST61, which is known to be associated with cattle and sheep. Interestingly, the antimicrobial resistance patterns of the C. jejuni cattle isolates completely differed from those of previously reported chicken isolates. Excluding one ciprofloxacin-resistant isolate, all isolates were susceptible to tetracycline and ciprofloxacin. This is the first report on the characterization of Campylobacter spp. in cattle in Mongolia. Although no official statistics of human campylobacteriosis are currently available in Mongolia, data on Campylobacter spp. in food-producing animals represent valuable information for investigating potential sources and infection routes to humans.
1. Introduction
Campylobacter infection is a leading cause of foodborne bacterial diseases worldwide. Campylobacter spp. colonize in the intestines of homoiothermal animals asymptomatically in most cases except for C. fetus infection in cattle, which causes bovine genital campylobacteriosis. Among the Campylobacter spp., C. jejuni and C. coli are the predominant causes of campylobacteriosis in humans. Additionally, species such as C. fetus, C. lari, and C. hyointestinalis are also known to be pathogenic to humans [1,2]. The primary clinical symptoms of Campylobacter infection are diarrhea, abdominal pain, fever, headache, nausea, and vomiting. Although most cases are self-limited within 3–6 days without any treatment, macrolides and fluoroquinolones (FQs) are prescribed to patients with severe or prolonged symptoms. Campylobacter infection is recognized as a risk factor for Guillain-Barré syndrome (GBS), an autoimmune demyelinating polyneuropathy [3]. Nearly 30% of GBS cases are associated with antecedent infection of Campylobacter [4,5], and about 0.1% of infected individuals develop GBS [6].
Livestock such as poultry and ruminants are major attributed sources of Campylobacter infection in humans, and companion animals and environments including surface water are also sources of infection [7,8]. As the route of infection can vary by region depending on differences in livestock breeding conditions, livestock hygiene management, food hygiene practice, food culture, and the coexisting environment between humans and animals, the identification of the source and infection route to humans is important for implementing effective measures against campylobacteriosis.
In Mongolia, meat is thoroughly cooked before consumption; thus, it is unlikely that undercooked meat poses a risk of infection. However, there are other risks of foodborne campylobacteriosis such as ingestion of traditional dairy products made from unpasteurized animal milk, secondary contamination during processing and along the food chain, the spread of ready-to-eat foods such as salads, the westernization of dietary habits, and the use of surface water in rural areas.
We recently reported the high antimicrobial resistance (AMR) ratios of Campylobacter spp. in chickens in Mongolia. All isolates were resistant to tetracycline (TET), and more than half were resistant to FQs. Half of the C. coli isolates exhibited muti-drug resistance, including resistance to erythromycin (EM) [9]. Because WHO assigned EM and ciprofloxacin (CPFX) as critically important antimicrobials for treating severe campylobacteriosis in humans, the presence of macrolide- and/or FQ-resistant Campylobacter spp. in food-producing animals, foods, and environments represents a public health concern. However, no data on the existence and AMR of Campylobacter spp. in other livestock and contamination of Campylobacter spp. in food chain are available. Additionally, campylobacteriosis in humans has not been officially reported to date, probably because of the insufficient diagnostic capacity.
More than 5 million cattle, 25 million sheep, and 23 million goats are raised in Mongolia. Meat is one of the primary food products in Mongolia, and beef and mutton are widely consumed. The annual consumption is approximately 134,000 tons for beef and 275,000 tons for sheep and goat meat [10]. Additionally, Mongolia has a unique culture of using animal milk and consumes 160,000 tons of milk per year. These facts imply that ruminants are possible sources of Campylobacter infection in humans. Therefore, in the current study, we characterized Campylobacter spp. isolated from cattle to provide scientific data for investigating the zoonotic transmission route of Campylobacter spp. in Mongolia.
2. Materials and Methods
2.1. Sampling
In total, 100 rectal swabs were collected from 10 dairy farms in Ulaanbaatar, Mongolia (Table 1). Most samples were collected from the Songinokhairkhan district, a cattle farm-concentrated area in Ulaanbaatar. Rectal swabs were collected using sterile cotton swabs. Each swab was directly placed in 5 mL of Brucella broth (Becton Dickinson, Franklin Lakes, NJ, USA) supplemented with Modified Preston Campylobacter Supplement (Oxoid, Basingstoke, UK) and 5% lysed horse blood (LHB, Nippon Bio-test Laboratories Inc., Asaka, Japan; the supplemented broth is hereafter termed Preston broth) and kept cooled during transportation to the laboratory.

Table 1.
Campylobacter spp. isolated from cattle.
2.2. Isolation and Identification of Campylobacter spp.
Preston broth containing the rectal swabs was incubated for 24 h at 42 °C under microaerobic conditions using AnaeroPacks-MicroAero (Mitsubishi Gas Chemical Co., Inc., Tokyo, Japan) to enrich thermophilic Campylobacter spp. Then, one loop of the broth was streaked on Campylobacter Blood-Free Selective Agar Base (Oxoid) with Charcoal Cefoperazone Deoxycholate Agar (CCDA) selective supplement (Oxoid; hereafter termed mCCDA), and the mCCDA plates were incubated for 48 h at 37 °C under microaerobic conditions. Typical Campylobacter colonies on the mCCDA plates were picked up using sterilized toothpicks and inoculated into Campylobacter Blood-Free Selective Agar Base without CCDA supplement to analyze bacterial growth under microaerobic and anaerobic conditions using AnaeroPacks-MicroAero and AneroPak-Anaero (Mitsubishi Gas Chemical Co., Inc.), respectively.
The suspected Campylobacter colonies were cultured in 2 mL of Brucella broth for 48 h at 37 °C under microaerobic conditions, and DNA was extracted from the cultures using CICA Geneus DNA Extraction Reagent (Kanto Chemical Co., Inc., Tokyo, Japan) according to the supplier’s instructions. Species-specific multiplex PCR targeting cdtC gene [11], which can distinguish C. jejuni, C. coli, and C. fetus, was performed as described previously [9]. Campylobacter spp. were also confirmed by matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry using the MALDI Biotyper Compass (Bruker Daltonics, Bremen, Germany).
2.3. Antimicrobial Susceptibility Test
The antimicrobial susceptibility of the isolates was analyzed using the E-test and broth dilution method. E-test strips of TET, EM, nalidixic acid (NA), and ciprofloxacin (CPFX) were purchased from BioMérieux (Marcy-l’Etoile, France), and E-test was performed as descrived previously [9]. The minimum inhibitory concentrations (MIC) for NA were also determined using the broth dilution method according to the CLSI guidelines. Antimicrobial susceptibility was determined according to CLSI M45 third edition, with resistant breakpoints of EM (≥32 μg/mL), CPFX (≥4 μg/mL), and TET (≥16 μg/mL), and according to Japanese Veterinary Antimicrobial Resistance Monitoring System (JVARM) for NA (≥16 μg/mL).
2.4. Genetic Analysis
Nucleotide sequencing of the gyrA gene to analyze codon 86 amino acid substitution for estimating FQ susceptibility and 16S rDNA for species identification, was performed as described elsewhere [9]. Multi-locus sequence typing (MLST) was performed to identify sequence types (STs) and clonal complexes (CCs) of the C. jejuni isolates. Seven housekeeping genes (aspA, glnA, gltA, glyA, pgm, tkt, and uncA) were amplified by PCR using the primer sets listed in the Campylobacter MLST database [12], and the amplified DNA fragments were purified using a FastGene Gel/PCR Extraction Kit (Nippon Genetics Co., Ltd., Tokyo, Japan). Nucleotide sequences of the amplified fragments were determined using the BigDye Terminator 3.1 Cycle Sequencing Kit and 3130-Avant Genetic Analyzer (Applied Biosystems, Waltham, MA, USA). Phylogenetic analyses were conducted using Molecular Evolutionary Genetics Analysis version 11 [13]. The nucleotide sequences of aspA (409 base pairs [bp], nucleotide [nt] 669–1077), glnA (452 bp, nt 247–698), gltA (398 bp, nt 321–719), glyA (403 bp, nt 392–794), tkt (438 bp, nt 247–684), uncA (482 bp, nt 676–1157), and pgm (498 bp, nt 421–1118) were used for the phylogenetic analyses.
3. Results
3.1. Isolation and Identification of Campylobacter spp.
Between 2019 and 2023, we visited 10 dairy farms (9 farms in Songinokhairkhan district were within a 40-km radius of central Ulaanbaatar, while farm 20I in Baganuur district was 130 km away from central Ulaanbaatar), and collected 10 rectal swabs from cattle in each farm. A total of 35 Campylobacter spp. were isolated (Table 1), including 23 C. jejuni (65.7%) and 4 C. fetus (11.4%) identified by multiplex PCR. The remaining isolates were identified as C. hyointestinalis (n = 7, 20.0%) by 16S rDNA sequencing analysis and MALDI Biotyper analysis and C. lari (n = 1, 2.9%) by MALDI Biotyper analysis. No C. coli was isolated in the current study. All C. jejuni and C. fetus isolates were also confirmed by MALDI Biotyper analysis. Of the 10 farms, C. jejuni was isolated from 9 farms, whereas in farm 19C, only C. hyointestinalis was isolated. In farms 19D, 20I, and 23B, only C. jejuni was isolated, whereas more than one Campylobacter spp. including C. jejuni, C. fetus, C. hyointestinalis, or C. lari were isolated from six farms (19A, 19B, 22A, 22B, 23A, and 23C). Two different Campylobacter spp., C. jejuni and C. hyointestinalis, were isolated from cattle ID CA2 in farm 19B and cattle ID CA11 in farm 22B (Table 2).

Table 2.
ST and antimicrobial resistance of Campylobacter isolates.
3.2. AMR of Campylobacter spp. Isolates
We previously reported that C. jejuni isolates from chickens in Mongolia exhibited extremely high AMR ratios; all the isolates were resistant to TET and half of the isolates were resistant to NA and FQs [9]. Different from the antimicrobial resistance patterns of C. jejuni isolates from chickens, most of the C. jejuni isolates from cattle in Mongolia were sensitive to TET and CPFX; among the 23 C. jejuni isolates, only one isolate (22A-CA10-1) was resistant to CPFX and NA. Nucleotide sequence analysis of gyrA of isolate 22A-CA10-1 confirmed that nucleotide substitution at nt257 (ACA to ATA) resulted in a Thr-to-Ile amino acid substitution at codon 86, which is associated with FQ resistance in C. jejuni [14]. Consistent with the susceptibility to CPFX, other C. jejuni isolates carried Thr at codon 86 (Table 2). C. hyointestinalis, C. fetus, and C. lari are known to be intrinsically resistant to NA [15,16,17], and consistent with this, all three Campylobacter spp. isolated in this study were resistant to NA (Table 2).
3.3. MLST and Phylogenetic Analysis
For MLST, 17 and 10 C. jejuni isolates from cattle (this study) and chickens [9] in Mongolia, respectively, were used. The 17 cattle isolates were classified into 7 STs (ST19, ST22, ST61, ST918, ST2100, ST2217, and ST3098) (Table 2), suggesting the diversity of C. jejuni in cattle in Mongolia. Twelve C. jejuni isolates from cattle with ST19, ST22, ST61, ST918, and ST2100 belonged to CC19, CC21, CC22, CC48, and CC52, respectively. The CCs of the remaining 5 isolates with ST2217 or ST3098 were not identified. The chicken isolates 15M-B7 and 15M-B18 were assigned to ST14448 (CC21), a new ST (Figure 1).
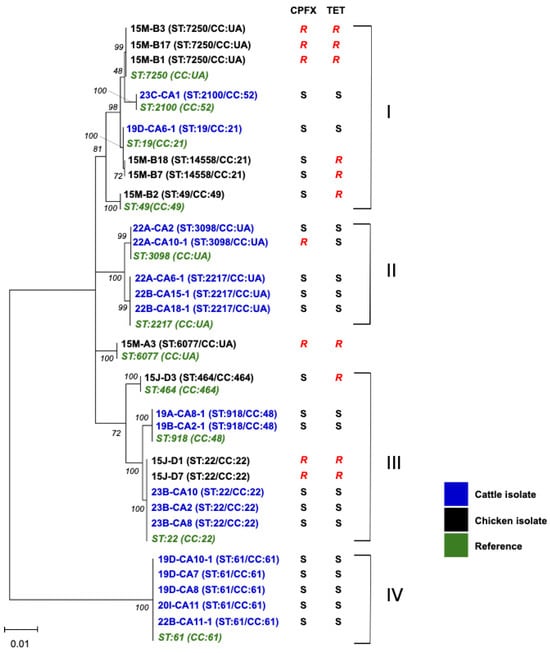
Figure 1.
Phylogenetic analysis of C. jejuni isolates from cattle and chickens in Mongolia. Nucleotide sequences of a total of 3080 nt from aspA, glnA, gltA, glyA, tkt, uncA, and pgm were used. The nucleotide sequences of ST19 (CC21), ST22 (CC22), ST49 (CC49), ST464 (CC464), ST918 (CC48), ST2100 (CC52), ST2217 (CC:UA), ST3098 (CC:UA), ST6077 (CC:UA), ST7250 (CC:UA), ST14558 (CC21), and ST61 (CC61) were obtained from the Campylobacter Sequence Typing database [12]. The percentages (50%) of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown in italic numbers next to the branches.
Figure 1 shows phylogenetic relationships of the isolates from cattle and chickens based on the seven genes used for MLST analysis. Clusters I and III comprised isolates from chickens and cattle, whereas clusters II and IV solely contained cattle isolates. Within cluster II, ST3098 and ST2217 have only been reported in cattle or beef, with the exception of a single human blood isolate, suggesting the association of these STs with cattle [12]. Cluster III included ST22 isolates from both chickens and cattle; however, the cattle isolates were susceptible to TET and CPFX but chicken isolates were resistant to these antibiotics. Cluster IV only included ST61, which is known as a cattle/sheep-associated ST worldwide [8]. ST61 C. jejuni were isolated from different farms, suggesting a wide distribution among cattle in Mongolia.
4. Discussion
AMR monitoring of C. jejuni in humans and food-producing animals conducted by member states of the European Unions revealed extremely high ratios of TET and CPFX resistance in three animal species with sufficient isolate numbers including broilers (ranging from 24.1 to 84.6% and 38.0 to 97.3%, respectively), fattening turkeys (18.8–76.3% and 34.1–93.2%, respectively), and cattle under 1-year old (35.3–92.6% and 45.7–79.3%, respectively), across all nations excluding Nordic countries [18]. Increased trends of CPFX-resistant C. jejuni both in human and livestock have been reported in European countries and globally [19,20]. Because EM and CPFX are assigned as critically important antimicrobials for treating severe campylobacteriosis in humans [21], the presence of macrolide- and/or FQ-resistant Campylobacter spp. in food-producing animals, foods, and environments represents a public health concern. We previously reported that C. jejuni isolated from chickens in Mongolia showed extremely high AMR ratios; all isolates were resistant to TET and nearly half were resistant to FQs [9]. However, this trend was not observed in C.jejuni isolates from cattle in this study, as all isolates were susceptible to TET and CPFX, excluding one isolate with CPFX-resistance (22A-CA10-1) carried a Thr-to-Ile substitution at codon 86 in gyrA. This unique difference in TET and FQ resistance between chickens and cattle suggests that the transmission of Campylobacter spp. between the two livestock species is rare in Mongolia. In the questionnaire survey, we could not obtain any evidence of FQ use on chicken and dairy farms. It has been noted that no fitness burden is required for FQ-resistant C. jejuni to colonize the chicken intestine [22,23]. Similarly, it is reported that FQ-resistant C. jejuni might have a fitness advantage over FQ-sensitive isolates in colonizing the cattle intestine [24,25]. Thus, although the current prevalence of FQ-resistant C jejuni is low in cattle, AMR monitoring, including that of Campylobacter spp. in cattle, should be strengthened to detect trends as quickly as possible.
ST61 C. jejuni is known to be associated with cattle and sheep [8] and associated with CPFX sensitivity significantly [26]. Consistent with these findings, ST61 was only found in cattle isolates, all of which were sensitive to CPFX. Although undercooked beef is not consumed often because of the Mongolian food culture, C. jejuni in cattle can spread to humans via cross-contaminated foods during processing and along the food chain, surface water contaminated with cattle feces, and the consumption of raw milk and dairy products. Indeed, Campylobacter spp. are considered as one of the major hazards associated with raw drinking milk [27]. In Japan, where undercooked poultry is the main attribution source of food-borne campylobacteriosis, ST61 C. jejuni caused an outbreak by consumption of raw cow milk [28]. Raw ruminant milk is a potential risk of Campylobacter infection in humans in Mongolia because it is used for a variety of traditional dairy products such as cream and cheese.
One of the limitations in this study is that most of the samples were collected in the Songinokhairkhan district of Ulaanbaatar, a region where many dairy farms reside. Genetic diversity was observed among the C. jejuni isolates from this district; however, continued analysis of a larger number of Campylobacter spp. isolated from different regions will be required to clarify the characteristics of Campylobacter spp. in cattle in Mongolia.
Although there are no official statistics on campylobacteriosis in humans in Mongolia, a genetic fragment of Campylobacter spp. has been detected by PCR in human diarrhea samples of unknown cause (personal communication, Dr. Tundev Odgerel at National Center for Communicable Disease, Mongolia). At present, no information on the attribution source of human campylobacteriosis is available. Future molecular genetic studies of isolates from humans, animals, foods, and environments under the One Health concept will uncover the exposure route of Campylobacter spp. to humans.
5. Conclusions
We recently reported the high antimicrobial resistance ratios of C. jejuni in chickens in Mongolia; all isolates were resistant to TET, and nearly half were resistant to FQs. However, this study revealed that the antimicrobial resistance patterns of the C. jejuni cattle isolates completely differed from those of chicken isolates. This is the first report on the characterization of Campylobacter spp. in cattle in Mongolia. Data on Campylobacter spp. in food-producing animals will be valuable for investigating potential sources and infection routes to humans.
Author Contributions
Conceptualization, project administration, funding acquisition, M.H. and P.N.-O.; methodology and investigation, E.B. and Z.B.; resources, B.N., B.C., T.B., B.S. and P.N.-O., methodology, E.K., A.S., J.T.-H. and T.S.; Writing—Original Draft Preparation, E.B.; Writing—Review & Editing, M.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by International Collaborative Research (JSPS KAKENHI Grant Number 24KK0134) and the World-leading Innovative and Smart Education (WISE) Program (1801) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan. E.B. is supported by JICA Agriculture Studies Networks for Food Security (Agri-Net) program (D2202684).
Institutional Review Board Statement
This research was approved by the Ethical Committee of Veterinary Science and Bio-Medical Research of the Mongolian University of Life Sciences (approval no. 22/01–02).
Informed Consent Statement
All farms participating in the study have agreed to sample collection and authorized the authors to use the analyzed results, including publication. We have obtained the written informed consent from all farms.
Data Availability Statement
The original contributions presented in this study are included in the article. Nucleotide sequences of representative isolates of each ST, ST19: 19D-CA6-1; ST22: 23B-CA10; ST61: 19D-CA10-1; ST918: 19A-CA8-1; ST2100: 23C-CA1; ST2217: 22A-CA6-1; ST-3098: 22A-CA2; ST14558: 15MA-B18, have been deposited to DDBJ. Accession numbers assigned are from LC888096 to LC888151. Further inquiries can be directed to the corresponding author(s).
Acknowledgments
We thank the Japan International Cooperation Agency (JICA) technical cooperation project “Strengthening the Capacity for Human Resource Development in the Field of Veterinary and Animal Husbandry” for continuous support.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Jansen, A.; Stark, K.; Kunkel, J.; Schreier, E.; Ignatius, R.; Liesenfeld, O.; Werber, D.; Göbel, U.B.; Zeitz, M.; Schneider, T. Aetiology of community-acquired, acute gastroenteritis in hospitalised adults: A prospective cohort study. BMC Infect. Dis. 2008, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global Epidemiology of Campylobacter Infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Engberg, J. Clinical Aspects of Campylobacter jejuni and Campylobacter coli Infection, 3rd ed.; Nachamkin, I., Szymanski, C.M., Blaser, M.J., Eds.; ASM Press: Washington, DC, USA, 2008. [Google Scholar]
- Poropatich, K.O.; Walker, C.L.; Black, R.E. Quantifying the association between Campylobacter infection and Guillain-Barré syndrome: A systematic review. J. Health Popul. Nutr. 2010, 28, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Rees, J.H.; Soudain, S.E.; Gregson, N.A.; Hughes, R.A. Campylobacter jejuni infection and Guillain-Barré syndrome. N. Engl. J. Med. 1995, 333, 1374–1379. [Google Scholar] [CrossRef]
- Keithlin, J.; Sargeant, J.; Thomas, M.K.; Fazil, A. Systematic review and meta-analysis of the proportion of Campylobacter cases that develop chronic sequelae. BMC Public Health 2014, 14, 1203. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Pijnacker, R.; Coipan, C.; Mulder, A.C.; Fernandes Veludo, A.; de Rijk, S.; van Hoek, A.; Buij, R.; Muskens, G.; Koene, M.; et al. Sources and transmission routes of campylobacteriosis: A combined analysis of genome and exposure data. J. Infect. 2021, 82, 216–226. [Google Scholar] [CrossRef]
- Sheppard, S.K.; Dallas, J.F.; Strachan, N.J.; MacRae, M.; McCarthy, N.D.; Wilson, D.J.; Gormley, F.J.; Falush, D.; Ogden, I.D.; Maiden, M.C.; et al. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 2009, 48, 1072–1078. [Google Scholar] [CrossRef]
- Byambajav, Z.; Bulgan, E.; Hirai, Y.; Nakayama, M.; Tanaka, M.; Nitta, Y.; Suzuki, A.; Umemura, T.; Altankhuu, B.; Tsagaan, A.; et al. Research Note: Antimicrobial resistance of Campylobacter species isolated from chickens near Ulaanbaatar city, Mongolia. Poult. Sci. 2021, 100, 100916. [Google Scholar] [CrossRef]
- Ministry of Food, Agriculture and Light Industry. Available online: https://mofa.gov.mn/branch/huns/616fa7c073bc4a5fc70f2228 (accessed on 14 August 2025).
- Asakura, M.; Samosornsuk, W.; Hinenoya, A.; Misawa, N.; Nishimura, K.; Matsuhisa, A.; Yamasaki, S. Development of a cytolethal distending toxin (cdt) gene-based species-specific multiplex PCR assay for the detection and identification of Campylobacter jejuni, Campylobacter coli and Campylobacter fetus. FEMS Immunol. Med. Microbiol. 2008, 52, 260–266. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Gibreel, A.; Sjögren, E.; Kaijser, B.; Wretlind, B.; Sköld, O. Rapid emergence of high-level resistance to quinolones in Campylobacter jejuni associated with mutational changes in gyrA and parC. Antimicrob. Agents Chemother. 1998, 42, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Laatu, M.; Rautelin, H.; Hänninen, M.L. Susceptibility of Campylobacter hyointestinalis subsp. hyointestinalis to antimicrobial agents and characterization of quinolone-resistant strains. J. Antimicrob. Chemother. 2005, 55, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Leatherbarrow, A.J.; Griffiths, R.; Hart, C.A.; Kemp, R.; Williams, N.J.; Diggle, P.J.; Wright, E.J.; Sutherst, J.; Houghton, P.; French, N.P. Campylobacter lari: Genotype and antibiotic resistance of isolates from cattle, wildlife and water in an area of mixed dairy farmland in the United Kingdom. Environ. Microbiol. 2007, 9, 1772–1779. [Google Scholar] [CrossRef]
- van der Graaf-van Bloois, L.; Duim, B.; Looft, T.; Veldman, K.T.; Zomer, A.L.; Wagenaar, J.A. Antimicrobial resistance in Campylobacter fetus: Emergence and genomic evolution. Microb. Genom. 2023, 9, 934. [Google Scholar] [CrossRef]
- European Food Safety Authority/European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2022–2023. EFSA J. 2025, 23, e9237. [Google Scholar] [CrossRef]
- Mohan, V.; Strepis, N.; Mitsakakis, K.; Becker, K.; Chindelevitch, L.; Shivaperumal, N.; Swe-Han, K.S.; Hays, J.P. Antimicrobial resistance in Campylobacter spp. focussing on C. jejuni and C. coli—A narrative review. J. Glob. Antimicrob. Resist. 2025, 43, 372–389. [Google Scholar] [CrossRef]
- Sproston, E.L.; Wimalarathna, H.M.L.; Sheppard, S.K. Trends in fluoroquinolone resistance in Campylobacter. Microb. Genom. 2018, 4, e000198. [Google Scholar] [CrossRef]
- World Health Organization. WHO’s List of Medically Important Antimicrobials: A Risk Management Tool for Mitigating Antimicrobial Resistance Due to Non-Human Use; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Luo, N.; Pereira, S.; Sahin, O.; Lin, J.; Huang, S.; Michel, L.; Zhang, Q. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. USA 2005, 102, 541–546. [Google Scholar] [CrossRef]
- Zhang, Q.; Sahin, O.; McDermott, P.F.; Payot, S. Fitness of antimicrobial-resistant Campylobacter and Salmonella. Microbes Infect. 2006, 8, 1972–1978. [Google Scholar] [CrossRef]
- Goulart, D.B.; Zhang, Q.; Sahin, O. Growth kinetics and fitness of fluoroquinolone resistant and susceptible Campylobacter jejuni strains of cattle origin. Front. Vet. Sci. 2023, 10, 1117975. [Google Scholar] [CrossRef]
- Smith, A.B.; Renter, D.G.; Cernicchiaro, N.; Shi, X.; Nickell, J.S.; Keil, D.J.; Nagaraja, T.G. A Randomized Trial to Assess the Effect of Fluoroquinolone Metaphylaxis on the Fecal Prevalence and Quinolone Susceptibilities of Salmonella and Campylobacter in Feedlot Cattle. Foodborne Pathog. Dis. 2017, 14, 600–607. [Google Scholar] [CrossRef]
- Cody, A.J.; McCarthy, N.M.; Wimalarathna, H.L.; Colles, F.M.; Clark, L.; Bowler, I.C.; Maiden, M.C.; Dingle, K.E. A longitudinal 6-year study of the molecular epidemiology of clinical campylobacter isolates in Oxfordshire, United kingdom. J. Clin. Microbiol. 2012, 50, 3193–3201. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hasards. Scientific Opinion on the public health risks related to the consumption of raw drinking milk. EFSA J. 2015, 13, 3940. [Google Scholar] [CrossRef]
- Ohno, Y.; Sekizuka, T.; Kuroda, M.; Ikeda, T. Outbreaks of Campylobacteriosis Caused by Drinking Raw Milk in Japan: Evidence of Relationship Between Milk and Patients by Using Whole Genome Sequencing. Foodborne Pathog. Dis. 2023, 20, 375–380. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).