Abstract
Aflatoxin B1 (AFB1) and ochratoxin A (OTA) are typical contaminants of food and feed, which have serious implications for human and animal health, even at low concentrations. Therefore, a transcriptomic study was carried out to analyze gene expression changes triggered by low doses of AFB1 and OTA (100 nM; 7 days), individually and combined, in human lymphoblastic T cells. RNA-sequencing analysis showed that AFB1-exposure resulted in 99 differential gene expressions (DEGs), while 77 DEGs were obtained in OTA-exposure and 3236 DEGs in the combined one. Overall, 16% of human genome expression was altered. Gene ontology analysis revealed, for all studied conditions, biological processes and molecular functions typically associated with the immune system. PathVisio analysis pointed to ataxia telangiectasia mutated signaling as the most significantly altered pathway in AFB1-exposure, glycolysis in OTA-exposure, and ferroptosis in the mixed condition (Z-score > 1.96; adjusted p-value ≤ 0.05). Thus, the results demonstrated the potential DNA damage caused by AFB1, the possible metabolic reprogramming promoted by OTA, and the plausible cell death with oxidative stress prompted by the mixed exposure. They may be considered viable mechanisms of action to promote immune toxicity in vitro.
1. Introduction
Aflatoxin B1 (AFB1) is the most toxic compound produced by several species of Aspergillus, which contamination occurs in a broad range of food commodities []. Once ingested, AFB1 is metabolized in the liver releasing reactive metabolites that are considered the causative agents of growth suppression, malnutrition, immune system alterations, and onset of hepatocellular carcinoma []. AFB1-toxicity has been also reported in the pancreas, bladder, kidney, and central nervous system [,]. In view of this, AFB1 is classified in group 1 as carcinogenic to humans by the International Agency for Research on Cancer (IARC) [].
Another widespread mycotoxin, ochratoxin A (OTA), is mostly synthesized by Penicillium verrucosum, Aspergillus ochraceus and Aspergillus niger. Typical OTA-contaminated foods are cereals, wine, tea, coffee, cheese, meat, fruits, dried fruits, spices, and vegetables []. OTA is firstly absorbed in the kidney’s proximal tubule, and its slow excretion led to a potential accumulation in the body []. Several studies have demonstrated as OTA exposure can lead to hepatotoxicity, nephrotoxicity, and neurotoxicity [,]. According to this evidence, OTA has been classified in group 2B as a possible human carcinogen [].
AFB1 and OTA are also well-known to be immune toxic agents in vitro and in vivo [,]. Exposure to AFB1 may induce an immune and pro-inflammatory response in macrophages with reactive oxygen species (ROS) generation, autophagy, and extracellular trap formation []. In macrophages, AFB1 also aggravated swine influenza virus infection, inflammation and damage of pulmonary tissue by activating TLR4-NFκB signaling pathway []. The suppression of immune response has been also observed in rats and broilers [,,]. Similarly, OTA may disrupt the phagocytosis function of heterophils with intracellular ROS production []. In macrophages, OTA exposure was also correlated with rheumatoid arthritis progression [], while in vivo OTA administration provoked apoptosis, immune stress, inflammation, and spleen damage []. Furthermore, it has been observed that AFB1 and OTA in combination caused inflammation, oxidative stress, and apoptosis in 3D4/21 cell line via NF-κB signaling pathway []. However, very little information on AFB1 and OTA-induced toxicity in human T lymphocytes has been reported, and their underlying mechanism has rarely been investigated.
In toxicological studies for human health risk assessment, the use of low experimental doses is necessarily required to reproduce a real scenario []. In human blood, AFB1 and OTA concentrations varied according to the studied population []. For AFB1, doses ranging from 0.6 to 237 nM were observed in the serum of Gambian children, while low values between 0.5 and 4 nM were detected in serum samples from healthy Iraqi patients [,]. As regards OTA, values of 22–25 nM were observed in plasma samples from Chinese and Bolivian patients, whereas only 2 nM in the serum of Swedish adolescents [,,]. In relation to AFB1-metabolites, their detection is quite difficult []. For the main AFB1-metabolite, i.e., AFM1, extremely low values ranging from 0.01 to 0.03 nM were observed in urinary samples from Brazilian, Guinean, and Bangladeshi patients [,,]. Regarding OTA-metabolism, the European Food Safety Authority (ESFA) reported that in vivo OTA-biotransformation, especially in humans and animals, is very low []. For OTα, the main OTA-metabolite, values of 0.5 nM were detected in the plasma of Bangladeshi adolescents as well as in urinary samples from Spanish and German patients [,,]. Nevertheless, OTα has been described as a non-toxic compound and its formation has been considered an important OTA-detoxification pathway [,].
In light of the above, a transcriptomic analysis was designed to assess AFB1 and OTA toxicological effects at low doses (both at 100 nM), individually and in combination, after seven days of exposure the gene expression profile of Jurkat cells.
2. Material and Methods
2.1. Chemicals
The reagents and compounds used for cell culture, Roswell Park Memorial Institute (RPMI)-glutamax medium, fetal bovine serum (FBS), penicillin/streptomycin, and phosphate buffer saline were purchased by Sigma Chemical Co. (St. Louis, MO, USA); Dimethyl sulfoxide (DMSO) and methanol were obtained from Fisher Scientific (Madrid, Spain); Deionized water (<18, MΩcm resistivity) was obtained using Milli-QSP® Reagent Water System (Millipore, Bedford, MA, USA); AFB1 (MW: 312.28 g/mol) and OTA (MW: 403.81 g/mol) standards were acquired from Sigma-Aldrich (St. Louis, MO, USA). Stock solutions were prepared in methanol solvent at a concentration of 1000 mg/L and kept at −20 °C.
2.2. Cell Culture and Exposure Conditions
Jurkat cells (ATCC-TIB152) were maintained in RPMI-glutamax medium complemented with 100 U/mL penicillin, 100 mg/mL streptomycin and 10% FBS. Cells were incubated at pH 7.4, 5% CO2 at 37 °C and air atmosphere at a constant humidity of 95%. The medium was changed every 2–3 days. To achieve the goal of the study, Jurkat cells were plated at a density of 2.5 × 105 cells/mL in 6-well tissue culture plates and exposed for 7 days to DMSO solvent at 0.1% as control condition in maintenance medium (n = 3) as well as AFB1 and OTA at 100 nM, individually and in combination, in 0.1% DMSO (n = 3).
2.3. RNA Extraction and Next Generation Sequencing (NGS)
Firstly, RNA was extracted from Jurkat cells and purified from DNA contamination by using a ReliaPrepTM RNA Cell Miniprep System Kit (Promega, WI, USA). Quantity and quality of obtained RNA were assessed using a NanoDrop™ 2000 spectrophotometer (Thermo Scientific™, Madrid, Spain), showing 260/280 nm and 260/230 nm ratios both around 2.
Secondly, Illumina NextSeq 500, supplied by the Genomics section of the Central Service for Experimental Research (SCSIE, University of Valencia), was employed for sequencing high-quality RNA samples, being the Integrity Numbers above 8. The standard protocol of Illumina was also carried out to create RNA-seq libraries, using TruSeq-stranded mRNA. Subsequently, results were generated as one archive for each sample, 12 in total.
2.4. Data Processing
FastQC software v0.11.8 (Babraham bioinformatics, Cambridge, UK) was used to calculate the percentage of mapped reads and to ensure quality control (QC) of reads []. The trimming was carried out employing FASTX-Toolkit v0.13 [], eliminating bases from 5′- and 3′-extremes. Then, reads with low quality and identical sequences were reduced into a single sequence but maintained counts. The trimmed reads alignment was performed by Spliced Transcripts Alignment to a Reference (STAR) software v2.7 (Cold Spring Harbor Laboratory, New York, NY, USA) [], using the Genome Reference Consortium Human Build 38 version as a reference. SAM tools software v1.10 (GitHub, San Francisco, CA, USA) [] allowed to transform Sequence Alignment Map (SAM) files into their binary version (BAM). BAM files were used with STAR to generate an expression matrix in R software []. Annotation, normalization and statistical analysis were performed according to the user guide of edgeR package [] to contrast differential expression among mycotoxin exposures and control, using gene-wise negative binomial generalized linear models with quasi-likelihood tests. Differentially expressed genes (DEGs) with p-value ≤ 0.05 were considered significant. Venn diagrams and heat maps were built by using Vennerable and pheatmap packages [,] to assess coincident DEGs between conditions [,].
2.5. DEGs Analysis
DEGs were submitted to gene ontology (GO) analysis by ConsensusPathDB []. Pathway assignments were carried out through PathVisio software (University of Maastricht, Maastricht, The Netherlands) by using WikiPathways as biological pathways database [,]. Z-score > 1.96 and adjusted p ≤ 0.05 were used as thresholds to identify significant pathways.
2.6. Gene Selection and Primer Design
Specific primers for each gene were designed by Primer-BLAST establishing the default software settings with PCR products of amplification ranging from 97 to 145 bp and melting temperature of 60 °C. qPCR analysis was performed by StepOne Plus Real-time PCR instrument (Applied Biosystems, Foster City, CA, USA). The reliability of primer amplification was determined from standard curves of each gene, by measuring linearity (R2 values) and efficiency of Ct mean values against the log cDNA dilution factor. Table 1 reports primers employed in the present study.

Table 1.
Gene names, forward (FS) and reverse (RS) primer sequences, PCR efficiency (E%) and R2 value for the target genes and the reference gene 18S rRNA.
2.7. Reverse Transcription and qPCR
Amplification solutions were prepared in 96 well plates using SYBR Green as fluorescent dye. Reactions mixes consisted of 100 ng template, 500 nM of each primer, and the required amount of 2× Fast SYBR Green in a reaction volume of 10 μL. The PCR temperature cycling conditions for cystatin A (CSTA) and DNA nucleotidylexotransferase (DNTT) were as follows: initial denaturation step at 95 °C for 10 min to activate Taq DNA polymerase, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 58 °C for 30 s, and elongation at 72 °C for 15 s. The melting curve was generated by heating the amplicon from 60 to 90 °C. Threshold cycles (Ct) were generated by StepOne Plus Software version 2.3 and relative gene expressions were assessed using the 2−ΔΔCT method []. The relative quantification values were transformed to log2 (Log2RQ) for normalization. Samples were run in triplicate according to Minimum Information for Publication of Quantitative Real-Time PCR Experiments guidelines [].
2.8. Statistical Analysis
Statistical analysis was carried out by ΔCt values (experimental Ct and housekeeping Ct mean) obtained using the StepOne Plus Software version 2.3 (Applied Biosystems, Foster City, CA, USA). Variances among groups were evaluated by Levene’s test and all the group variances were equal. T-Student defined differences between controls and treated groups. A p value ≤ 0.05 was considered for statistically significant differences.
3. Results
3.1. DEGs Profile
Gene expression of lymphocytes T exposed for 7 days to AFB1 and OTA (100 nM), individually or in combination, significantly differs from the expression of untreated cells. Considering a p-value ≤ 0.05 and log2FC |0.5|, AFB1-exposure resulted in 99 DEGs with 61 up- and 38 down-regulated. A total of 77 DEGs (23 up- and 54 down-regulated) were obtained in OTA exposure and 3236 DEGs (2255 up- and 981 down-regulated) in the combined AFB1-OTA exposure. Overall, the total DEGs number was 3412 with 68.5% up-regulation and 31.5% down-regulation by compromising the usual expression levels up to 16% of the human genome, containing 21,000 protein-coding genes (HGNC database). Interestingly, 42 DEGs (1.3%) overlapped in AFB1 and combined exposure, 8 DEGs (0.2%) among AFB1 and OTA individually, and 32 DEGs (1%) between OTA and mixed exposure. Solely 3 DEGs (0.1%): DNTT, adhesion G-protein-coupled receptor E1 (ADGRE1), and voltage-dependent T-type calcium channel subunit alpha-1I (CACNA1l) were found by overlapping all conditions (Figure 1). Although DNTT was up-regulated for each exposure, ADGRE1 and CACNA1l were down-regulated in AFB1 and OTA individually and over-expressed in the combined exposure.
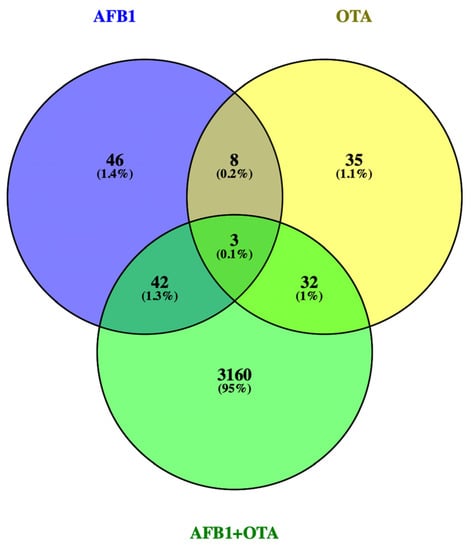
Figure 1.
Venn diagram for the DEGs found by RNA-seq in Jurkat cells treated with AFB1, OTA, AFB1 + OTA at 100 nM in 0.1% DMSO compared to 0.1% DMSO control exposure. DEGs p-value ≤ 0.05 and log2FC |0.5|.
3.2. GO and Pathway Identification
The characterization of biological processes (BP) and molecular functions (MF) in which DEGs are involved is a major step in this transcriptomics study. It includes the comparison between the DEGs list and the rest of the genome for over-represented functions and gene set enrichment analysis [,]. Table 2 shows relevant GOs with their relative categories obtained by the over-representation analysis in ConsensusPathDB.

Table 2.
Relevant GOs resulting from the over-representation analysis of the selected DEGs obtained by AFB1, OTA, AFB1 + OTA exposures (100 nM in all cases) in ConsensusPathDB.
3.2.1. AFB1-Exposure
Regarding the AFB1 condition, the over-representation analysis by ConsesusPathDB provided a list of GO terms in which several BP linked to system development, cell adhesion, and regulation of the immune system process were some of the most over-represented. MF associated with cation, steroid, and GTPase binding, DNA and RNA polymerase transcription factor activity, and cytokine receptor function were significant in the DEGs set at levels 3 and 4. Plasma membrane for cellular components (CC), was significantly represented in the individual AFB1-exposure (Table 2).
PathVisio analysis reported a number of 3691 data points (N), among them 753 met criterion (R). (N) denotes the total number of genes measured in the dataset where (R) indicates the filter analysis criterion []. Several pathways were statistically significant after AFB1 exposure (Z-score > 1.96; adjusted p-value ≤ 0.05). According to the number of genes affected, the most significant pathways were as follows: ApoE and miRNA-146 in inflammation and atherosclerosis (80%), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) actives NLRP3 inflammasome (67%), miRNA-124 interaction with cell cycle and differentiation (67%), and SARS-CoV-2 antagonizes innate immune activation (62%). The analysis also showed a significant alteration in the mitochondrial immune response to SARS-CoV-2 and ataxia telangiectasia mutated (ATM) signaling in development and disease, with 45% of genes affected in each pathway (Table 3).

Table 3.
Pathways overlapped in AFB1-exposure (100 nM) by PathVisio.
The genes involved in the ATM signaling pathway (Z-score: 4.15) for Homo sapiens are shown in Figure 2, indicating in red the over-expressed genes and in green the down-regulated genes ones. A slight alteration in ATM-related gene expression was obtained.
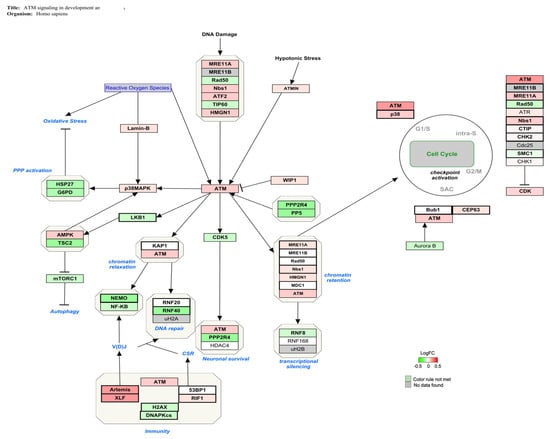
Figure 2.
Genes involved in ATM signaling for Homo sapiens are shown in red and green. The up-regulated genes are represented in red and the down-regulated ones after AFB1 exposure (100 nM) are represented in green.
3.2.2. OTA Exposure
The consensusPathDB analysis pointed to blood coagulation, secretion, hypoxia, hemostasis, and regulation of the immune and inflammatory response as some of the most over-represented BP upon OTA exposure. The activity of dioxygenase and ubiquitin proteins was the most significant MF in the DEGs set at levels 3 and 4, whereas CC such as basolateral plasma membrane and its integral components were significantly affected (Table 2).
PathVisio analysis reported a total number of 3662 data points, of which only 193 met the criterion. Several pathways were statistically significant upon OTA exposure (Z-score > 1.96; adjusted p-value ≤ 0.05). Based on the number of genes affected, the most significant pathways were glycolysis (43%) and the Cori cycle (34%). The analysis indicated a significant alteration in HIF1α and PPAR-γ regulated glycolysis (29%) and innate immune response to SARS-CoV-2 with 25% of genes affected (Table 4).

Table 4.
Pathways overlapped in OTA exposure (100 nM) by PathVisio.
Figure 3 displays the genes involved in the glycolysis signaling pathway (Z-score: 4.45) for Homo sapiens. In this case, low values of fold changes were observed.
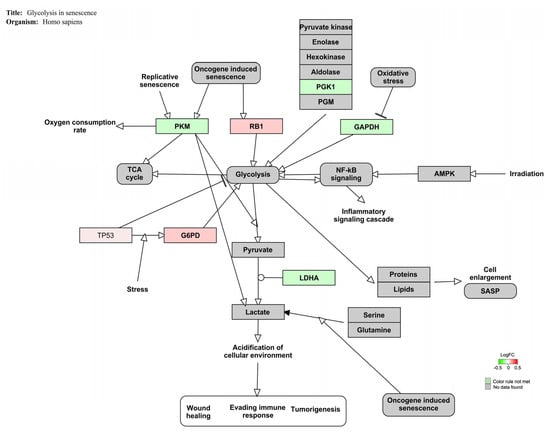
Figure 3.
Genes implicated in glycolysis senescence pathway for Homo sapiens are represented in red and green. The up-regulated genes are represented in red and the down-regulated ones upon OTA exposure (100 nM) are represented in green.
3.2.3. AFB1-OTA Combined Exposure
The analysis conducted by ConsensusPathDB showed that in AFB1-OTA mixed exposure, the most over-represented BP were the regulation of immune system process, cytokine production, cell adhesion, lymphocyte differentiation, and T cell cytotoxicity, among others. MF linked to G protein, cytokine receptor activity and C-C chemokine binding were statistically significant in the DEGs set at levels 4 and 5. CC such as cell cortex and membrane, chromatin, blood microparticle, and MHC class I protein complex were significantly affected to a greater extent (Table 2).
PathVisio analysis showed 3744 data points, of which a large number (2489) matched the criterion. Solely 9 of the pathways were statistically relevant following the combined exposure (Z-score > 1.96; adjusted p-value ≤ 0.05). In this case, all pathways found showed more than 84% of genes affected. The 100% of genes involved in the tricarboxylic acid cycle (TCA), dual hijack model of HIV infection, and hyperlipidemia were statistically altered (Table 5).

Table 5.
Pathways overlapped in AFB1-OTA-exposure (100 nM) by PathVisio.
The genes involved in the ferroptosis signaling pathway (Z-score: 2.41) for Homo sapiens are shown in Figure 4, and the majority of them were down-regulated.
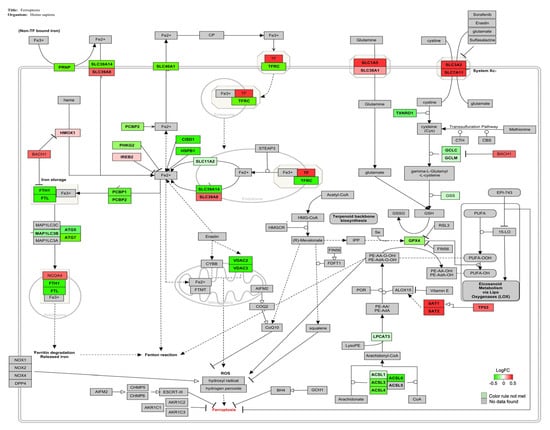
Figure 4.
Genes involved in ferroptosis pathway for Homo sapiens are represented in red and green. The up-regulated genes are represented in red and the down-regulated ones following AFB1-OTA exposure (100 nM for both toxins) are represented in green.
3.3. Validation of NGS Results by qPCR
The expression of CSTA, a cysteine protease inhibitor gene, was assessed after exposure to AFB1 whereas DNTT expression, an independent DNA polymerase gene, was evaluated upon exposure to AFB1, OTA, and mycotoxins mixture. The experimental conditions used in the qPCR analysis were the same as those used for the NGS assay. Additionally, CSTA and DNTT were chosen because they were the most affected genes in the sequencing analysis. Moreover, qPCR assay confirmed NGS results with the strong up-regulation of CSTA and DNTT when compared to the control (Figure 5A,B). A ribosomal protein 18S was employed as endogenous gene control.
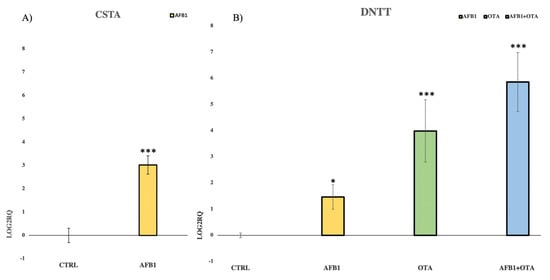
Figure 5.
Bar plots showing the relative expression of (A) CSTA and (B) DNTT when compared to the control (Log2RQ = 0) after 7 days of exposure in Jurkat cells to AFB1 (100 nM) for CSTA and to AFB1, OTA, AFB1 + OTA (100 nM in all cases) for DNTT by qPCR. RQ: relative quantification; CTRL: control; CSTA: cystatin A; DNTT: DNA nucleotidylexotransferase. p ≤ 0.05 (*), p ≤ 0.001 (***).
4. Discussion
In the present study, the possible mechanism of action (MoA) by which low doses of AFB1 and OTA, comparable with those found in human blood, promoted immune toxicity was investigated after seven days in vitro exposure. The choice of this exposure time reflects the peak of the immune response in vivo, which precisely occurs after the seventh day of infection. Thereafter, the number of T lymphocytes undergoes a programmed contraction as well as the immune system gradually tends to deactivate [,]. Reproducing a realistic scenario, it is interesting to observe as the number of DEGs was significantly increased in the combined exposure (3236) when compared to the individual ones (AFB1 DEGs = 99 and OTA DEGs = 77). This finding is relevant to human health risk assessment, as the human population is constantly exposed to multiple mycotoxins contaminating food and our findings may suggest that AFB1 and OTA had an additive in vitro effect []. Similar results were obtained by [], exposing HepG2 liver cells with high concentrations of AFB1 (1.5–150 μM; 3 and 24 h) and OTA (50–800 μM; 3 and 24 h). It has been observed more prominent toxic effects in the combined exposure compared to individual ones, suggesting a synergism between mycotoxins. In Vero kidney cells, AFB1 and OTA (1–50 μM; 24 h) not only showed an additive cytotoxic effect but also synergism to promote genotoxicity with increased DNA fragmentation []. It has been also shown that OTA significantly increased AFB1 mutagenicity with a higher percentage of mutations than AFB1 alone, indicating the potential risk of mycotoxins co-occurrence []. On the contrary, different findings were obtained by [], treating chicken LMH liver cells with AFB1 (0–3 μM; 48 h) and OTA (0–20 μM; 48 h). The cytotoxicity assay and transcriptome analysis revealed the antagonist effect between mycotoxins to induce chicken liver toxicity. The discrepancy in results was probably associated with the use of experimental conditions (doses, exposure time, and non-human cell line) less realistic compared to those employed in the present study.
4.1. AFB1 Exposure
ATM signaling was the most affected pathway after AFB1 exposure (Table 3). It is well known that ATM plays a major role in the cellular response to DNA damage, by involving ATM protein kinase as the main downstream signal effector []. Several studies showed that in vitro and in vivo exposure to AFB1-induced DNA double-strand breaks (DSBs), activation of ATM signaling, and the up-regulation of all ATM kinases related to chromatin relaxation, cell cycle regulation, and immunity [,,]. It has been also demonstrated that low AFB1 doses (5–80 nM; 24 h) were sufficient to promote DNA damage and ATM up-regulation in BEAS-2B cells with possible genome alterations in the human respiratory system [].
ATM signaling is also a core component of the DNA repair system by activating the ubiquitin ligase RNF40, which in turn covers a key role in chromatin reorganization and timely DNA-DSB repair [,]. The reduced expression of RNF40 was associated with replicative stress, chromosomal instability, cell cycle checkpoint inactivation, and inhibition of DNA repair mechanism in vitro [,,]. In turkey embryos, exposure to AFB1 (1 μg/for injection; 24 h) has been shown to downregulate the activity of ubiquitin ligase and impair the cellular response to genotoxic damage [,]. Therefore, the activation of ATM signaling with up-regulation of ATM kinases and RNF40 down-regulation pointed to the induction of DNA damage and the disruption of the repair mechanism as a possible MoA by which AFB1 induces immune toxicity in vitro (Figure 2).
The central position of ATM signaling to maintain genomic stability is demonstrated by its involvement in the G2/M cell cycle transition []. It has been observed as AFB1-exposure prompted the up-regulation of ATM kinase, checkpoint kinase 2 (chk2) and Ataxia telangiectasia Rad3-related protein (ATR), which in turn caused G2/M cell cycle arrest in bronchial epithelial cells []. Similar findings were also observed in the chicken bursa of Fabricius, spleen, and jejunum, thus confirming the correlation between in vivo AFB1 administration and the disruption of cell cycle machinery at G2/M stage by ATM signaling activation [,,]. In line with these findings, 7 days of exposure to AFB1 can induce a potential alteration in cell cycle distribution (Figure 2). However, gene expression changes (i.e., ATR, chk2) are very mild and further research is needed to confirm this hypothesis.
Moreover, ATM signaling is involved in T cell development by controlling V(D)J recombination process, which is indispensable to constitute the variable domain of T cell receptor [,]. Alterations in ATM signaling and its main effectors can disrupt V(D)J process and predispose human T cells to leukemia and lymphoma []. In this context, Artemis protein, a well-known ATM substrate, has a critical role in V(D)J recombination [,]. It has been also reported the association between in vitro deleterious effects such as immunodeficiency, cell cycle arrest, DNA damage, and Artemis over-expression [,,]. Thus, ATM signaling activation with ATM kinase and Artemis upregulation may suggest the possible interference of AFB1 in T cell recombination and functionality (Figure 2).
4.2. OTA Exposure
Mycotoxins can induce in vitro toxicity by reprogramming cell metabolism []. For instance, OTA (1.25–5 μM; 24 h) promoted mitochondrial toxicity by reprogramming energy metabolism from oxidative phosphorylation (OxPhos) to glycolysis in human gastric cells []. Similar metabolic disturbances were obtained by [] in human esophageal cells after 24 h exposure to OTA (2.5–10 μM; 24 h) and by [], analyzing the liver of rats treated with moderate doses of AFB1 in contaminated feed (1.6 mg/kg; 12 days). In line with this evidence, PathVisio analysis revealed the promotion of metabolic reprogramming and pointed to glycolysis as the most affected pathway after 7 days of in vitro exposure to OTA (Table 4).
Figure 3 showed that all genes related to the glycolysis pathway were slightly affected by mycotoxins exposure. In detail, retinoblastoma 1 (RB1) is a tumor suppressor involved in cell cycle regulation, cell differentiation, proliferation, and death []. It has been demonstrated that RB1 over-expression was associated with metabolic changes and glycolysis promotion in human breast cells []. Similarly, the up-regulation of glucose-6-phosphate dehydrogenase (G6PD), a pivotal enzyme in NADPH production and cell redox balance, enhanced glycolysis activation in acute myeloid leukemia cells []. G6PD over-expression has been also observed in beauvericin and enniatin, B promoted mitochondrial disturbance and reduced ATP production in vitro []. Phosphoglycerate kinase 1 (PGK1) catalyzes the first reaction of anaerobic glycolysis. The microarray analysis performed in HepG2 cells after OTA exposure (2.5–10 μM; 24 h) showed the downregulation of PGK1 and its involvement in OTA-disrupted liver metabolism []. Likewise, the reduced expression and activity of pyruvate kinase M2 (PKM2), which converts phosphoenolpyruvate to pyruvate with ATP production, has been correlated with OTA-induced metabolic disturbance and glycolysis in human gastric cells [,]. Additionally, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), previously considered a simple housekeeping gene, has been shown to be involved in many cellular processes such as glycolysis and cell metabolism []. Indeed, GAPDH down-regulation was related to significant alterations in energy metabolism after exposure to ZEA (10 μM; 24 h) in human breast MCF10F cells []. Lastly, lactate dehydrogenase A (LDHA) catalyzes the last step of anaerobic glycolysis by converting pyruvate to lactate []. Interestingly, Fusaric acid (4 and 256 μM; 6 h), a neglected foodborne mycotoxin, downregulated LDHA gene expression, among others, and induced bioenergetic adaptations by switching energy metabolism from mitochondrial OxPhos to glycolysis in HepG2 cells []. The metabolic disturbance promoted by OTA exposure in Jurkat cells is further supported by the alteration of HIF1α, PPAR-γ, and Cori cycle signaling pathways (Table 4). HIF1a and PPAR-γ pathways are key regulators of metabolic reprogramming and glycolysis in vitro whereas the Cori cycle is a typical metabolic route activated with high lactate concentrations [,,].
4.3. AFB1-OTA Combined Exposure
Ferroptosis was the main signaling pathway altered by the combined AFB1-OTA exposure with 85% of genes affected (Table 5). Ferroptosis is a type of regulated necrosis triggered by the combination of excessive intracellular iron overload, induction of lipid peroxidation with plasma membrane damage and inhibition of glutathione peroxidase 4 (GPX4) activity [].
The intracellular iron accumulation can be mediated by several factors. Firstly, transferrin (TF), a strong inducer of the process, is a carrier protein that binds extracellular ferric iron (Fe3+) and transports it into cells through its transporter (TFR) by endocytosis []. It has been demonstrated the strong correlation between TF over-expression and ferroptosis induction in broiler hearts upon AFB1 administration in contaminated feed (1 mg/kg; 21 days). The down-regulation of Heat Shock Protein Family B1 (HSPB1), a negative regulator of intracellular iron uptake and accumulation, also confirmed the imbalance of iron levels in AFB1-promoted ferroptosis []. Conversely, TFR1 downregulation may lead to iron accumulation, oxidative stress, and ROS generation in vitro []. Indeed, RNA-seq analysis performed on DON-treated IPEC-J2 cells (0.5 μg/mL; 48 h) revealed the alteration of several genes related to iron homeostasis, including the down-regulation of TFR1, which was considered a key initial signal of ferroptosis []. Another pivotal driver of ferroptosis is the transcription factor BTB domain and CNC homolog 1 (BACH1), a regulator of iron metabolism. In vitro upregulation of BACH1 can induce the repression of several genes involved in iron storage (such as ferritin, a protein complex consisting of heavy, FTH1, and light chains, FTL), thereby causing the release of unstable iron into the cytoplasm and worsening the ferroptotic condition [,].
High concentrations of free iron in the cytoplasm can increase the production of endogenous hydrogen peroxide and hydroxyl radical by the Fenton reaction, by promoting lipid peroxidation [,]. During ferroptosis, lipid oxidation occurs as an intermediate event, in which ASCL4 and ASCL3 are the main regulators [,]. For instance, T-2 toxin (2.5–10 nM for 6–20 h) promoted ferroptosis, lipid peroxidation, and intracellular ROS generation through the alteration of several genes expression, including the slight downregulation of ASCL4 []. Likewise, the downregulation of ASCL3 has been related to ferroptosis and oxidative stress in vitro and in vivo [].
GPX4, a selenium-dependent glutathione peroxidase, protects cells from lipid hydroperoxides formed during oxidative stress, using reduced glutathione as an enzyme co-substrate []. In human T cells, the reduced GPX4 activity can lead to lipid peroxidation and ferroptotic cell death, altered cellular homeostasis, and increased susceptibility to acute infections [,]. It has been also reported that exposure to AFB1 and OTA downregulated GPX4 gene expression inducing lipid peroxidation, ROS generation, and ferroptosis in several in vitro and in vivo models [,,]. Based on this evidence, AFB1-OTA combined exposure may induce ferroptosis in Jurkat cells by altering the expression of several key genes involved in intracellular iron overload (TF, TFR1, BACH1, FTH1, FTL, and NCO4), lipid peroxidation (ASCL4, ASCL3, SAT1) and antioxidant cell defense (GPX4) (Figure 5). Interestingly, AFB1 and OTA also altered the PPAR-α signaling pathway, a novel route associated with ferroptosis due to its active function in lipid remodeling and peroxidation (Table 5) [].
In the present study, AFB1 and OTA, individually and in combination, resulted in DNTT over-expression. Moreover, qPCR analysis confirmed not only NGS results but also the synergism between mycotoxins, as their combined transcriptional effect on DNTT was more pronounced than individual exposures. It is well known that DNTT plays a key role in T cell recombination and proliferation, showing a strong anti-apoptotic function. Consequently, DNTT up-regulation has been reported to confer resistance to tumor cells against chemotherapeutic agents []. Since the increased DNTT activity has been also found in peripheral blood cells derived from patients with acute lymphoblastic leukemia, it cannot be excluded as exposure to AFB1 and OTA may worsen the leukemic condition [,,].
5. Conclusions
These results contribute to a better understanding of the main cellular pathways involved in AFB1 and OTA-induced immune toxicity in Jurkat cells. RNA-seq analysis revealed that ATM signaling pathway was the most altered in AFB1 exposure. In detail, potential alterations in G2/M cell cycle checkpoint, T cell recombination, and induction of DNA damage with impairment of repair mechanisms have been mainly related to AFB1 toxicity. Regarding OTA exposure, the glycolysis signaling pathway was the most affected. Therefore, energy metabolism reprogramming by glycolysis activation may be considered the main OTA-MoA in Jurkat cells. In combination, AFB1 and OTA mainly affected the ferroptosis signaling pathway. In this case, the potential accumulation of intracellular iron with lipid peroxidation and oxidative stress accompanied by the disruption of antioxidant cell defense may explain the intrinsic MoA by which the mixture of mycotoxins promoted immune toxicity in vitro. Although the transcriptome analysis identified the main pathways altered by low doses of AFB1 and OTA in a human T cell line, further investigations are required to confirm these hypotheses and to better explore the underlying mechanism by which AFB1 and OTA may weaken the immune system, by rendering it more susceptible to infections.
Author Contributions
Conceptualization, L.M.; methodology, L.M.; software, M.F. and M.L.; formal analysis, M.F., M.L. and L.M.; investigation, M.F., M.L. and A.C.; data curation, M.F., L.M. and A.C.; writing—original draft preparation, M.F., M.L., L.M. and A.C.; writing—review and editing, M.F., L.M. and G.F.; visualization, M.F., L.M. and G.F; supervision, L.M. and G.F.; project administration, L.M. and G.F.; funding acquisition, G.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Spanish Ministry of Science and Innovation (PID2019-108070RB-I00-ALI).
Data Availability Statement
All related data and methods are presented in this paper. Additional inquiries should be addressed to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Umesha, S.; Manukumar, H.M.G.; Chandrasekhar, B.; Shivakumara, P.; Shiva Kumar, J.; Raghava, S.; Prakash, H.S. Aflatoxins and food pathogens: Impact of biologically active aflatoxins and their control strategies. J. Sci. Food Agric. 2017, 97, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Manyes, L.; Font, G. Mycotoxins: Toxicity, Occurrence, Risk Assessment and Prevention. In Encyclopedia of Human Nutrition, 4th ed.; Elsevier: New York, NY, USA, 2022. [Google Scholar] [CrossRef]
- Frangiamone, M.; Alonso-Garrido, M.; Font, G.; Cimbalo, A.; Manyes, L. Pumpkin extract and fermented whey individually and in combination alleviated AFB1-and OTA-induced alterations on neuronal differentiation in vitro. Food Chem. Toxicol. 2022, 164, 113011. [Google Scholar] [CrossRef] [PubMed]
- Rushing, B.R.; Selim, M.I. Aflatoxin B1: A review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem. Toxicol. 2019, 124, 81–100. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Chemical agents and related occupations. IARC Monogr. Eval. Carcinog. Risks Hum. 2012, 100(Pt. F), 9. [Google Scholar]
- Chen, W.; Li, C.; Zhang, B.; Zhou, Z.; Shen, Y.; Liao, X.; Yang, J.; Wang, Y.; Li, X.; Li, Y.; et al. Advances in biodetoxification of ochratoxin A: A review of the past five decades. Front. Microbiol. 2018, 9, 1386. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, B.; Dai, Y.; Li, H.; Xu, W. A review: Epigenetic mechanism in ochratoxin A toxicity studies. Toxins 2017, 9, 113. [Google Scholar] [CrossRef]
- Cimbalo, A.; Frangiamone, M.; Font, G.; Manyes, L. The importance of transcriptomics and proteomics for studying molecular mechanisms of mycotoxin exposure: A review. Food Chem. Toxicol. 2022, 169, 113396. [Google Scholar] [CrossRef]
- Tao, Y.; Xie, S.; Xu, F.; Liu, A.; Wang, Y.; Chen, D.; Pan, Y.; Huang, L.; Peng, D.; Wang, X.; et al. Ochratoxin A: Toxicity, oxidative stress and metabolism. Food Chem. Toxicol. 2018, 112, 320–331. [Google Scholar] [CrossRef]
- Cimbalo, A.; Frangiamone, M.; Lozano, M.; Escrivá, L.; Vila-Donat, P.; Manyes, L. Protective role of fermented whey and pumpkin extract against aflatoxin B1 and ochratoxin A toxicity in Jurkat T-cells. World Mycotoxin J. 2022, 1–14. [Google Scholar] [CrossRef]
- Frangiamone, M.; Cimbalo, A.; Alonso-Garrido, M.; Vila-Donat, P.; Manyes, L. In vitro and in vivo evaluation of AFB1 and OTA-toxicity through immunofluorescence and flow cytometry techniques: A systematic review. Food Chem. Toxicol. 2022, 160, 112798. [Google Scholar] [CrossRef]
- An, Y.; Shi, X.; Tang, X.; Wang, Y.; Shen, F.; Zhang, Q.; Yu, L. Aflatoxin B1 induces reactive oxygen species-mediated autophagy and extracellular trap formation in macrophages. Front. Cell Inf. Microbiol. 2017, 7, 53. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Z.; Liu, D.; Chen, J.; Gan, F.; Huang, K. Low-level aflatoxin B1 promotes influenza infection and modulates a switch in macrophage polarization from M1 to M2. Cell. Physiol. Biochem. 2018, 49, 1151–1167. [Google Scholar] [CrossRef]
- Bakheet, S.A.; Attia, S.M.; Alwetaid, M.Y.; Ansari, M.A.; Zoheir, K.M.; Nadeem, A.; Ahmad, S.F. β-1, 3-Glucan reverses aflatoxin B1-mediated suppression of immune responses in mice. Life Sci. 2016, 152, 1–13. [Google Scholar] [CrossRef]
- Ben Salah-Abbes, J.; Jebali, R.; Sharafi, H.; Akbari Noghabi, K.; Oueslati, R.; Abbès, S. Immuno-physiological alterations from AFB1 in rats counteracted by treatments with Lactobacillus paracasei BEJ01 and montmorillonite clay mixture. J. Immunotoxicol. 2016, 13, 628–637. [Google Scholar] [CrossRef]
- Guan, K.; Li, H.; Zuo, Z.; Wang, F.; Hu, P.; Peng, X.; Ouyang, P. The Molecular Mechanisms of Protective Role of Se on the G0/G1 Phase Arrest Caused by AFB1 in Broiler’s Thymocytes. Biol. Trace Elem. Res. 2019, 189, 556–566. [Google Scholar] [CrossRef]
- Han, Z.; Zhang, Y.; Wang, C.; Liu, X.; Jiang, A.; Liu, Z.; Wei, Z. Ochratoxin A-triggered chicken heterophil extracellular traps release through reactive oxygen species production dependent on activation of NADPH oxidase, ERK, and p38 MAPK signaling pathways. J. Agric. Food Chem. 2019, 67, 11230–11235. [Google Scholar] [CrossRef]
- Jahreis, S.; Kuhn, S.; Madaj, A.M.; Bauer, M.; Polte, T. Mold metabolites drive rheumatoid arthritis in mice via promotion of IFN-gamma-and IL-17-producing T cells. Food Chem. Toxicol. 2017, 109, 405–413. [Google Scholar] [CrossRef]
- Liu, D.; Su, J.; Lin, J.; Qian, G.; Chen, X.; Song, S.; Huang, K. Activation of AMPK-dependent SIRT-1 by astragalus polysaccharide protects against ochratoxin A-induced immune stress in vitro and in vivo. Int. J. Biol. Macromol. 2018, 120, 683–692. [Google Scholar] [CrossRef]
- Hou, L.; Gan, F.; Zhou, X.; Zhou, Y.; Qian, G.; Liu, Z.; Huang, K. Immunotoxicity of ochratoxin A and aflatoxin B1 in combination is associated with the nuclear factor kappa B signaling pathway in 3D4/21 cells. Chemosphere 2018, 199, 718–727. [Google Scholar] [CrossRef]
- Hernández, A.F.; Tsatsakis, A.M. Human exposure to chemical mixtures: Challenges for the integration of toxicology with epidemiology data in risk assessment. Food Chem. Toxicol. 2017, 103, 188–193. [Google Scholar] [CrossRef]
- Arce-López, B.; Lizarraga, E.; Vettorazzi, A.; González-Peñas, E. Human biomonitoring of mycotoxins in blood, plasma and serum in recent years: A review. Toxins 2020, 12, 147. [Google Scholar] [CrossRef] [PubMed]
- Kareem, S.A.; Hameed, R.M.; AL-Redha, S.A.; Al-Khatt, H. Aflatoxin B1 as a Threshold Concept of Uncertain Etiology of Chronic Kidney Diseases. Indian J. Forensic Med. Toxicol. 2021, 15, 737–744. [Google Scholar] [CrossRef]
- Leong, Y.H.; Latiff, A.A.; Ahmad, N.I.; Rosma, A. Exposure measurement of aflatoxins and aflatoxin metabolites in human body fluids. A short review. Mycotoxin Res. 2012, 28, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Xu, J.; Jiang, K.; Liu, X.; Meng, J.; Di Mavungu, J.D.; Han, Z. Determination of multiple mycotoxins in paired plasma and urine samples to assess human exposure in Nanjing, China. Environ. Pollut. 2019, 248, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Ferrufino-Guardia, E.; Chavez-Rico, V.; Larondelle, Y. Ochratoxin a in human breast milk, maternal and placental blood from Cochabamba-Bolivia. Rev. Toxicol. 2019, 36, 116–125. [Google Scholar]
- Warensjö Lemming, E.; Montano Montes, A.; Schmidt, J.; Cramer, B.; Humpf, H.U.; Moraeus, L.; Olsen, M. Mycotoxins in blood and urine of Swedish adolescents—Possible associations to food intake and other background characteristics. Mycotoxin Res. 2020, 36, 193–206. [Google Scholar] [CrossRef]
- Ali, N.; Blaszkewicz, M.; Hossain, K.; Degen, G.H. Determination of aflatoxin M1 in urine samples indicates frequent dietary exposure to aflatoxin B1 in the Bangladeshi population. Int. J. Hyg. Environ. Health 2017, 220, 271–281. [Google Scholar] [CrossRef]
- Ali, N.; Manirujjaman, M.; Rana, S.; Degen, G.H. Determination of aflatoxin M1 and deoxynivalenol biomarkers in infants and children urines from Bangladesh. Arch. Toxicol. 2020, 94, 3775–3786. [Google Scholar] [CrossRef]
- De Cássia Romero, A.; Ferreira, T.R.B.; dos Santos Dias, C.T.; Calori-Domingues, M.A.; da Gloria, E.M. Occurrence of AFM1 in urine samples of a Brazilian population and association with food consumption. Food Control 2010, 21, 554–558. [Google Scholar] [CrossRef]
- EFSA Panel on Contaminants in the Food Chain (CONTAM); Schrenk, D.; Bodin, L.; Chipman, J.K.; del Mazo, J.; Grasl-Kraupp, B.; Bignami, M. Risk assessment of ochratoxin A in food. EFSA J. 2020, 18, e06113. [Google Scholar] [CrossRef]
- Ali, N.; Hossain, K.; Degen, G.H. Blood plasma biomarkers of citrinin and ochratoxin A exposure in young adults in Bangladesh. Mycotoxin Res. 2018, 34, 59–67. [Google Scholar] [CrossRef]
- Coronel, M.B.; Marin, S.; Tarragó, M.; Cano-Sancho, G.; Ramos, A.J.; Sanchis, V. Ochratoxin A and its metabolite ochratoxin alpha in urine and assessment of the exposure of inhabitants of Lleida, Spain. Food Chem. Toxicol. 2011, 49, 1436–1442. [Google Scholar] [CrossRef]
- Muñoz, K.; Blaszkewicz, M.; Degen, G.H. Simultaneous analysis of ochratoxin A and its major metabolite ochratoxin alpha in plasma and urine for an advanced biomonitoring of the mycotoxin. J. Chromatogr. B 2010, 878, 2623–2629. [Google Scholar] [CrossRef]
- Wu, Q.; Dohnal, V.; Huang, L.; Kuca, K.; Wang, X.; Chen, G.; Yuan, Z. Metabolic pathways of ochratoxin A. Curr. Drug Metab. 2011, 12, 1–10. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 25 July 2021).
- Hannon, G.J. FASTX-Toolkit. 2010. Available online: http://hannonlab.cshl.edu/fastx_toolkit (accessed on 30 July 2021).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing: Vienna, Austria, 2018. Available online: https://www.R-project.org (accessed on 21 September 2021).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps, R package Version 1.0.12; CRAN R-Project: Vienna, Austria, 2019. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 21 September 2021).
- Swinton, J. Vennerable Package. 2013. Available online: https://github.com/js229/Vennerable (accessed on 21 September 2021).
- Alonso-Garrido, M.; Escrivá, L.; Manyes, L.; Font, G. Enniatin B induces expression changes in the electron transport chain pathway related genes in lymphoblastic T-cell line. Food Chem. Toxicol. 2018, 121, 437–443. [Google Scholar] [CrossRef]
- Escrivá, L.; Jennen, D.; Caiment, F.; Manyes, L. Transcriptomic study of the toxic mechanism triggered by beauvericin in Jurkat cells. Toxicol. Lett. 2018, 284, 213–221. [Google Scholar] [CrossRef]
- Kamburov, A.; Herwig, R. ConsensusPathDB 2022: Molecular interactions update as a resource for network biology. Nucleic Acids Res. 2022, 50, D587–D595. [Google Scholar] [CrossRef]
- Kutmon, M.; van Iersel, M.P.; Bohler, A.; Kelder, T.; Nunes, N.; Pico, A.R.; Evelo, C.T. PathVisio 3: An extendable pathway analysis toolbox. PLoS Comput. Biol. 2015, 11, e1004085. [Google Scholar] [CrossRef] [PubMed]
- Martens, M.; Ammar, A.; Riutta, A.; Waagmeester, A.; Slenter, D.N.; Hanspers, K.; Kutmon, M. WikiPathways: Connecting communities. Nucleic Acids Res. 2021, 49, D613–D621. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Wittwer, C.T. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Alexeyenko, A.; Lee, W.; Pernemalm, M.; Guegan, J.; Dessen, P.; Lazar, V.; Pawitan, Y. Network enrichment analysis: Extension of gene-set enrichment analysis to gene networks. BMC Bioinform. 2012, 13, 226. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Mortazavi, A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Badovinac, V.P.; Porter, B.B.; Harty, J.T. Programmed contraction of CD8+ T cells after infection. Nat. Immunol. 2002, 3, 619–626. [Google Scholar] [CrossRef]
- Lieberman, N.A.; Peddu, V.; Xie, H.; Shrestha, L.; Huang, M.L.; Mears, M.C.; Greninger, A.L. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020, 18, e3000849. [Google Scholar] [CrossRef]
- Corcuera, L.A.; Arbillaga, L.; Vettorazzi, A.; Azqueta, A.; De Cerain, A.L. Ochratoxin A reduces aflatoxin B1 induced DNA damage detected by the comet assay in Hep G2 cells. Food Chem. Toxicol. 2011, 49, 2883–2889. [Google Scholar] [CrossRef]
- Golli-Bennour, E.E.; Kouidhi, B.; Bouslimi, A.; Abid-Essefi, S.; Hassen, W.; Bacha, H. Cytotoxicity and genotoxicity induced by aflatoxin B1, ochratoxin A, and their combination in cultured Vero cells. J. Biochem. Mol. Toxicol. 2010, 24, 42–50. [Google Scholar] [CrossRef]
- Sedmikova, M.; Reisnerova, H.; Dufkova, Z.; Barta, I.; Jilek, F. Potential hazard of simultaneous occurrence of aflatoxin B~ 1 and ochratoxin A. Vet. Med. Praha 2001, 46, 169–174. [Google Scholar] [CrossRef]
- Choi, S.Y.; Kim, T.H.; Hong, M.W.; Park, T.S.; Lee, H.; Lee, S.J. Transcriptomic alterations induced by aflatoxin B1 and ochratoxin A in LMH cell line. Poult. Sci. 2020, 99, 5265–5274. [Google Scholar] [CrossRef]
- Zhang, T.; Penicud, K.; Bruhn, C.; Loizou, J.I.; Kanu, N.; Wang, Z.Q.; Behrens, A. Competition between NBS1 and ATMIN controls ATM signaling pathway choice. Cell Rep. 2012, 2, 1498–1504. [Google Scholar] [CrossRef][Green Version]
- Sancar, A.; Lindsey-Boltz, L.A.; Ünsal-Kaçmaz, K.; Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [Google Scholar] [CrossRef]
- Stracker, T.H.; Roig, I.; Knobel, P.A.; Marjanovic, M. The ATM signaling network in development and disease. Front. Genet. 2013, 4, 37. [Google Scholar] [CrossRef]
- Yin, H.; Jiang, M.; Peng, X.; Cui, H.; Zhou, Y.; He, M.; Fang, J. The molecular mechanism of G2/M cell cycle arrest induced by AFB1 in the jejunum. Oncotarget 2016, 7, 35592. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Z.; Wang, X.; Wang, Y.; Zhang, X.; Lu, H.; Wang, S.L. Cytochrome P450 2A13 enhances the sensitivity of human bronchial epithelial cells to aflatoxin B1-induced DNA damage. Toxicol. Appl. Pharmacol. 2013, 270, 114–121. [Google Scholar] [CrossRef]
- Jin, M.H.; Oh, D.Y. ATM in DNA repair in cancer. Pharmacol. Ther. 2019, 203, 107391. [Google Scholar] [CrossRef]
- Moyal, L.; Lerenthal, Y.; Gana-Weisz, M.; Mass, G.; So, S.; Wang, S.Y.; Shiloh, Y. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell 2011, 41, 529–542. [Google Scholar] [CrossRef]
- Chernikova, S.B.; Razorenova, O.V.; Higgins, J.P.; Sishc, B.J.; Nicolau, M.; Dorth, J.A.; Brown, J.M. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 2012, 72, 2111–2119. [Google Scholar] [CrossRef]
- Kari, V.; Shchebet, A.; Neumann, H.; Johnsen, S.A. The H2B ubiquitin ligase RNF40 cooperates with SUPT16H to induce dynamic changes in chromatin structure during DNA double-strand break repair. Cell Cycle 2011, 10, 3495–3504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Paramasivam, M.; Cai, Q.; Dai, X.; Wang, P.; Lin, K.; Wang, Y. Arsenite binds to the RING finger domains of RNF20-RNF40 histone E3 ubiquitin ligase and inhibits DNA double-strand break repair. J. Am. Chem. Soc. 2014, 136, 12884–12887. [Google Scholar] [CrossRef] [PubMed]
- Engin, A.B.; Engin, A. DNA damage checkpoint response to aflatoxin B1. Environ. Toxicol. Pharmacol. 2019, 65, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S.; Cardona, C.J.; Coulombe, R.A.; Reed, K.M. Hepatic transcriptome responses of domesticated and wild turkey embryos to aflatoxin B1. Toxins 2016, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Lavin, M.F.; Kozlov, S. ATM activation and DNA damage response. Cell Cycle 2007, 6, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Zuo, Z.; Wang, F.; Peng, X.; Guan, K.; Li, H.; Zhou, Y. The protective role of selenium in AFB1-Induced tissue damage and cell cycle arrest in chicken’s bursa of fabricius. Biol. Trace Elem. Res. 2018, 185, 486–496. [Google Scholar] [CrossRef]
- Li, H.; Guan, K.; Zuo, Z.; Wang, F.; Peng, X.; Fang, J.; Chen, Z. Effects of aflatoxin B1 on the cell cycle distribution of splenocytes in chickens. J. Toxicol. Pathol. 2019, 32, 27–36. [Google Scholar] [CrossRef]
- Helmink, B.A.; Sleckman, B.P. The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev. Immunol. 2012, 30, 175–202. [Google Scholar] [CrossRef]
- Nussenzweig, A.; Nussenzweig, M.C. Origin of chromosomal translocations in lymphoid cancer. Cell 2010, 141, 27–38. [Google Scholar] [CrossRef]
- Matei, I.R.; Guidos, C.J.; Danska, J.S. ATM-dependent DNA damage surveillance in T-cell development and leukemogenesis: The DSB connection. Immunol. Rev. 2006, 209, 142–158. [Google Scholar] [CrossRef]
- Jeggo, P.A.; Lobrich, M. Artemis links ATM to double strand break rejoining. Cell Cycle 2005, 4, 359–362. [Google Scholar] [CrossRef]
- Rivera-Munoz, P.; Abramowski, V.; Jacquot, S.; André, P.; Charrier, S.; Lipson-Ruffert, K.; De Villartay, J. Lymphopoiesis in transgenic mice over-expressing Artemis. Gene Ther. 2016, 23, 176–186. [Google Scholar] [CrossRef]
- Multhaup, M.; Karlen, A.D.; Swanson, D.L.; Wilber, A.; Somia, N.V.; Cowan, M.J.; McIvor, R.S. Cytotoxicity associated with artemis overexpression after lentiviral vector-mediated gene transfer. Hum. Gene Ther. 2010, 21, 865–875. [Google Scholar] [CrossRef]
- Sridharan, D.M.; Whalen, M.K.; Almendrala, D.; Cucinotta, F.A.; Kawahara, M.; Yannone, S.M.; Pluth, J.M. Increased Artemis levels confer radioresistance to both high and low LET radiation exposures. Radiat. Oncol. 2012, 7, 96. [Google Scholar] [CrossRef]
- Ulus-Senguloglu, G.; Arlett, C.F.; Plowman, P.N.; Parnell, J.; Patel, N.; Bourton, E.C.; Parris, C.N. Elevated expression of artemis in human fibroblast cells is associated with cellular radiosensitivity and increased apoptosis. Br. J. Cancer 2012, 107, 1506–1513. [Google Scholar] [CrossRef]
- Prusinkiewicz, M.A.; Mymryk, J.S. Metabolic reprogramming of the host cell by human adenovirus infection. Viruses 2019, 11, 141. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, M.; Cui, J.; Wu, X.; Li, Y.; Wu, W.; Zhang, X. Ochratoxin A induces reprogramming of glucose metabolism by switching energy metabolism from oxidative phosphorylation to glycolysis in human gastric epithelium GES-1 cells in vitro. Toxicol. Lett. 2020, 333, 232–241. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, Y.; Jia, X.; Liu, W.; Zhang, X.; Cui, J. The effect of ochratoxin A on cytotoxicity and glucose metabolism in human esophageal epithelium Het-1A cells. Toxicon 2021, 198, 80–92. [Google Scholar] [CrossRef]
- Zhang, L.; Ye, Y.; An, Y.; Tian, Y.; Wang, Y.; Tang, H. Systems responses of rats to aflatoxin B1 exposure revealed with metabonomic changes in multiple biological matrices. J. Proteome Res. 2011, 10, 614–623. [Google Scholar] [CrossRef]
- Lai, P.S.; Cheah, P.Y.; Kadam, P.; Chua, C.L.M.; Lie, D.K.H.; Li, H.H.; Lee, A.S.G. Overexpression of RB1 transcript is significantly correlated with 13q14 allelic imbalance in colorectal carcinomas. Int. J. Cancer 2006, 119, 1061–1066. [Google Scholar] [CrossRef]
- Wu, Q.; Deblois, G.; Cruickshank, J.; Duan, S.; Lima-Fernandes, E.; Haight, J.; Arrowsmith, C.H. GLUT1 inhibition blocks growth of RB1-positive triple negative breast cancer. Nat. Commun. 2020, 11, 4205. [Google Scholar] [CrossRef] [PubMed]
- Poulain, L.; Sujobert, P.; Zylbersztejn, F.; Barreau, S.; Stuani, L.; Lambert, M.; Chapuis, N. High mTORC1 activity drives glycolysis addiction and sensitivity to G6PD inhibition in acute myeloid leukemia cells. Leukemia 2017, 31, 2326–2335. [Google Scholar] [CrossRef] [PubMed]
- Søderstrøm, S.; Lie, K.K.; Lundebye, A.K.; Søfteland, L. Beauvericin (BEA) and enniatin B (ENNB)-induced impairment of mitochondria and lysosomes-Potential sources of intracellular reactive iron triggering ferroptosis in Atlantic salmon primary hepatocytes. Food Chem. Toxicol. 2022, 161, 112819. [Google Scholar] [CrossRef] [PubMed]
- Hundhausen, C.; Boesch-Saadatmandi, C.; Matzner, N.; Lang, F.; Blank, R.; Wolffram, S.; Rimbach, G. Ochratoxin a lowers mRNA levels of genes encoding for key proteins of liver cell metabolism. Cancer Genom. Proteom. 2008, 5, 319–332. [Google Scholar]
- Ferguson, E.C.; Rathmell, J.C. New roles for pyruvate kinase M2: Working out the Warburg effect. Trends biochem. Sci. 2008, 33, 359–362. [Google Scholar] [CrossRef]
- Tristan, C.; Shahani, N.; Sedlak, T.W.; Sawa, A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell Signal. 2011, 23, 317–323. [Google Scholar] [CrossRef]
- Karaman, E.F.; Ozden, S. Alterations in global DNA methylation and metabolism-related genes caused by zearalenone in MCF7 and MCF10F cells. Mycotoxin Res. 2019, 35, 309–320. [Google Scholar] [CrossRef]
- Xian, Z.Y.; Liu, J.M.; Chen, Q.K.; Chen, H.Z.; Ye, C.J.; Xue, J.; Kuang, S.J. Inhibition of LDHA suppresses tumor progression in prostate cancer. Tumor Biol. 2015, 36, 8093–8100. [Google Scholar] [CrossRef]
- Abdul, N.S.; Nagiah, S.; Chuturgoon, A.A. The neglected foodborne mycotoxin Fusaric acid induces bioenergetic adaptations by switching energy metabolism from mitochondrial processes to glycolysis in a human liver (HepG2) cell line. Toxicol. Lett. 2020, 318, 74–85. [Google Scholar] [CrossRef]
- Feng, J.; Dai, W.; Mao, Y.; Wu, L.; Li, J.; Chen, K.; Guo, C. Simvastatin re-sensitizes hepatocellular carcinoma cells to sorafenib by inhibiting HIF-1α/PPAR-γ/PKM2-mediated glycolysis. J. Exp. Clin. Cancer Res. 2020, 39, 24. [Google Scholar] [CrossRef]
- Prigione, A.; Rohwer, N.; Hoffmann, S.; Mlody, B.; Drews, K.; Bukowiecki, R.; Adjaye, J. HIF1 α Modulates Cell Fate Reprogramming Through Early Glycolytic Shift and Upregulation of PDK1–3 and PKM2. Stem Cells 2014, 32, 364–376. [Google Scholar] [CrossRef]
- Suhara, T.; Hishiki, T.; Kasahara, M.; Hayakawa, N.; Oyaizu, T.; Nakanishi, T.; Minamishima, Y.A. Inhibition of the oxygen sensor PHD2 in the liver improves survival in lactic acidosis by activating the Cori cycle. Proc. Natl. Acad. Sci. USA 2015, 112, 11642–11647. [Google Scholar] [CrossRef]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Ferroptosis in infection, inflammation, and immunity. J. Exp. Med. 2021, 218, e20210518. [Google Scholar] [CrossRef]
- Shi, Z.Z.; Fan, Z.W.; Chen, Y.X.; Xie, X.F.; Jiang, W.; Wang, W.J.; Bai, J. Ferroptosis in carcinoma: Regulatory mechanisms and new method for cancer therapy. OncoTargets Ther. 2019, 12, 11291. [Google Scholar] [CrossRef]
- Zhao, L.; Feng, Y.; Xu, Z.J.; Zhang, N.Y.; Zhang, W.P.; Zuo, G.; Sun, L.H. Selenium mitigated aflatoxin B1-induced cardiotoxicity with potential regulation of 4 selenoproteins and ferroptosis signaling in chicks. Food Chem. Toxicol. 2021, 154, 112320. [Google Scholar] [CrossRef]
- Ding, H.; Chen, S.; Pan, X.; Dai, X.; Pan, G.; Li, Z.; Xie, L. Transferrin receptor 1 ablation in satellite cells impedes skeletal muscle regeneration through activation of ferroptosis. J. Cachexia Sarcopenia Muscle 2021, 12, 746–768. [Google Scholar] [CrossRef]
- Lin, J.; Huang, F.; Liang, T.; Qin, Q.; Xu, Q.; Huang, X.; Liu, Y. EPA and DHA confer protection against deoxynivalenol-induced endoplasmic reticulum stress and iron imbalance in IPEC-1 cells. Br. J. Nutr. 2021, 128, 161–171. [Google Scholar] [CrossRef]
- Nishizawa, H.; Matsumoto, M.; Shindo, T.; Saigusa, D.; Kato, H.; Suzuki, K.; Igarashi, K. Ferroptosis is controlled by the coordinated transcriptional regulation of glutathione and labile iron metabolism by the transcription factor BACH1. J. Biol. Chem. 2020, 295, 69–82. [Google Scholar] [CrossRef]
- Xie, Y.; Hou, W.; Song, X.; Yu, Y.; Huang, J.; Sun, X.; Tang, D. Ferroptosis: Process and function. Cell Death Differ. 2016, 23, 369–379. [Google Scholar] [CrossRef]
- Au, C.; Benedetto, A.; Aschner, M. Manganese transport in eukaryotes: The role of DMT1. Neurotoxicology 2008, 29, 569–576. [Google Scholar] [CrossRef]
- He, Y.J.; Liu, X.Y.; Xing, L.; Wan, X.; Chang, X.; Jiang, H.L. Fenton reaction-independent ferroptosis therapy via glutathione and iron redox couple sequentially triggered lipid peroxide generator. Biomaterials 2020, 241, 119911. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, W.K.; Bae, K.H.; Lee, S.C.; Lee, E.W. Lipid metabolism and ferroptosis. Biology 2021, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Magtanong, L.; Ko, P.J.; To, M.; Cao, J.Y.; Forcina, G.C.; Tarangelo, A.; Dixon, S.J. Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem. Biol. 2019, 26, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Qin, S.; Zheng, Y.; Xia, C.; Zhang, P.; Zhang, L.; Deng, L. T-2 Toxin Induces Ferroptosis by Increasing Lipid Reactive Oxygen Species (ROS) and Downregulating Solute Carrier Family 7 Member 11 (SLC7A11). J. Agric. Food Chem. 2021, 69, 15716–15727. [Google Scholar] [CrossRef] [PubMed]
- Ubellacker, J.M.; Tasdogan, A.; Ramesh, V.; Shen, B.; Mitchell, E.C.; Martin-Sandoval, M.S.; Morrison, S.J. Lymph protects metastasizing melanoma cells from ferroptosis. Nature 2020, 585, 113–118. [Google Scholar] [CrossRef]
- Dixon, S.J.; Stockwell, B.R. The hallmarks of ferroptosis. Annu. Rev. Cancer Biol. 2019, 3, 35–54. [Google Scholar] [CrossRef]
- Matsushita, M.; Freigang, S.; Schneider, C.; Conrad, M.; Bornkamm, G.W.; Kopf, M. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J. Exp. Med. 2015, 212, 555–568. [Google Scholar] [CrossRef]
- Gan, F.; Xue, H.; Huang, Y.; Pan, C.; Huang, K. Selenium alleviates porcine nephrotoxicity of ochratoxin A by improving selenoenzyme expression in vitro. PLoS ONE 2015, 10, e0119808. [Google Scholar] [CrossRef]
- Vettorazzi, A.; Pastor, L.; Guruceaga, E.; de Cerain, A.L. Sex-dependent gene expression after ochratoxin A insult in F344 rat kidney. Food Chem. Toxicol. 2019, 123, 337–348. [Google Scholar] [CrossRef]
- Ren, Z.; He, H.; Fan, Y.; Chen, C.; Zuo, Z.; Deng, J. Research progress on the toxic antagonism of selenium against mycotoxins. Biol. Trace Elem. Res. 2019, 190, 273–280. [Google Scholar] [CrossRef]
- Venkatesh, D.; O’Brien, N.A.; Zandkarimi, F.; Tong, D.R.; Stokes, M.E.; Dunn, D.E.; Stockwell, B.R. MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling. Genes Dev. 2020, 34, 526–543. [Google Scholar] [CrossRef]
- Wernicke, C.M.; Richter, G.H.; Beinvogl, B.C.; Plehm, S.; Schlitter, A.M.; Bandapalli, O.R.; Grunewald, T.G. MondoA is highly overexpressed in acute lymphoblastic leukemia cells and modulates their metabolism, differentiation and survival. Leuk. Res. 2012, 36, 1185–1192. [Google Scholar] [CrossRef]
- Martinez Jimenez, L.A.; Organista Nava, J.; Illades_Aguiar, B.; Leyva Vazquez, M.A.; Gomez Gomez, Y. Terminal Deoxynucleotidyl Transferase in type B Acute Lymphoblastic Leukemia. J. Hum. Anat. Physiol. 2019, 4, 4. [Google Scholar]
- Ülger, T.G.; Uçar, A.; Çakıroğlu, F.P.; Yilmaz, S. Genotoxic effects of mycotoxins. Toxicon 2020, 185, 104–113. [Google Scholar] [CrossRef]
- Yasmineh, W.G.; Smith, B.M.; Bloomfield, C.D. DNA nucleotidylexotransferase of normal persons and leukemic patients. Clin. Chem. 1980, 26, 891–895. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).