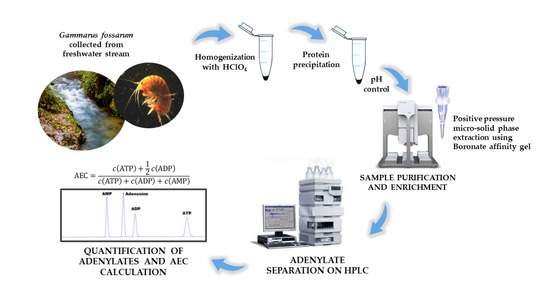
Determination of Adenylate Nucleotides in Amphipod Gammarus fossarum by Ion-Pair Reverse Phase Liquid Chromatography: Possibilities of Positive Pressure Micro-Solid Phase Extraction
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Sample Collection
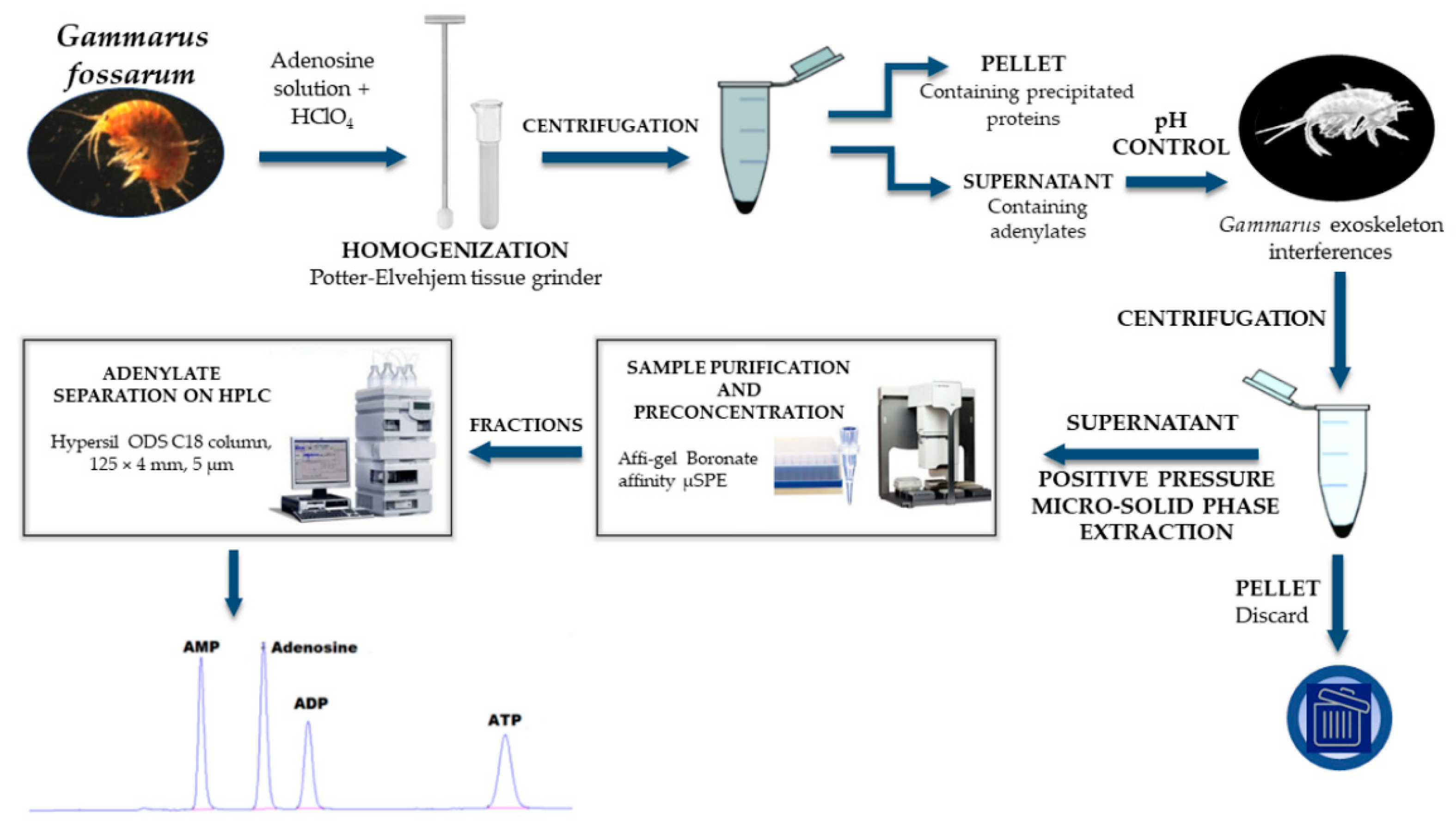
2.3. Preparation of G. fossarum Samples and Standard Solutions
2.4. Instrumentation, HPLC Conditions, and Adenylate Quantification
2.5. Method Validation
2.6. Positive Pressure Micro-Solid Phase Extraction (PP µSPE)
3. Results and Discussion
3.1. Nucleotide IP-RP-HPLC Method Validation
3.2. Analysis of G. fossarum Samples
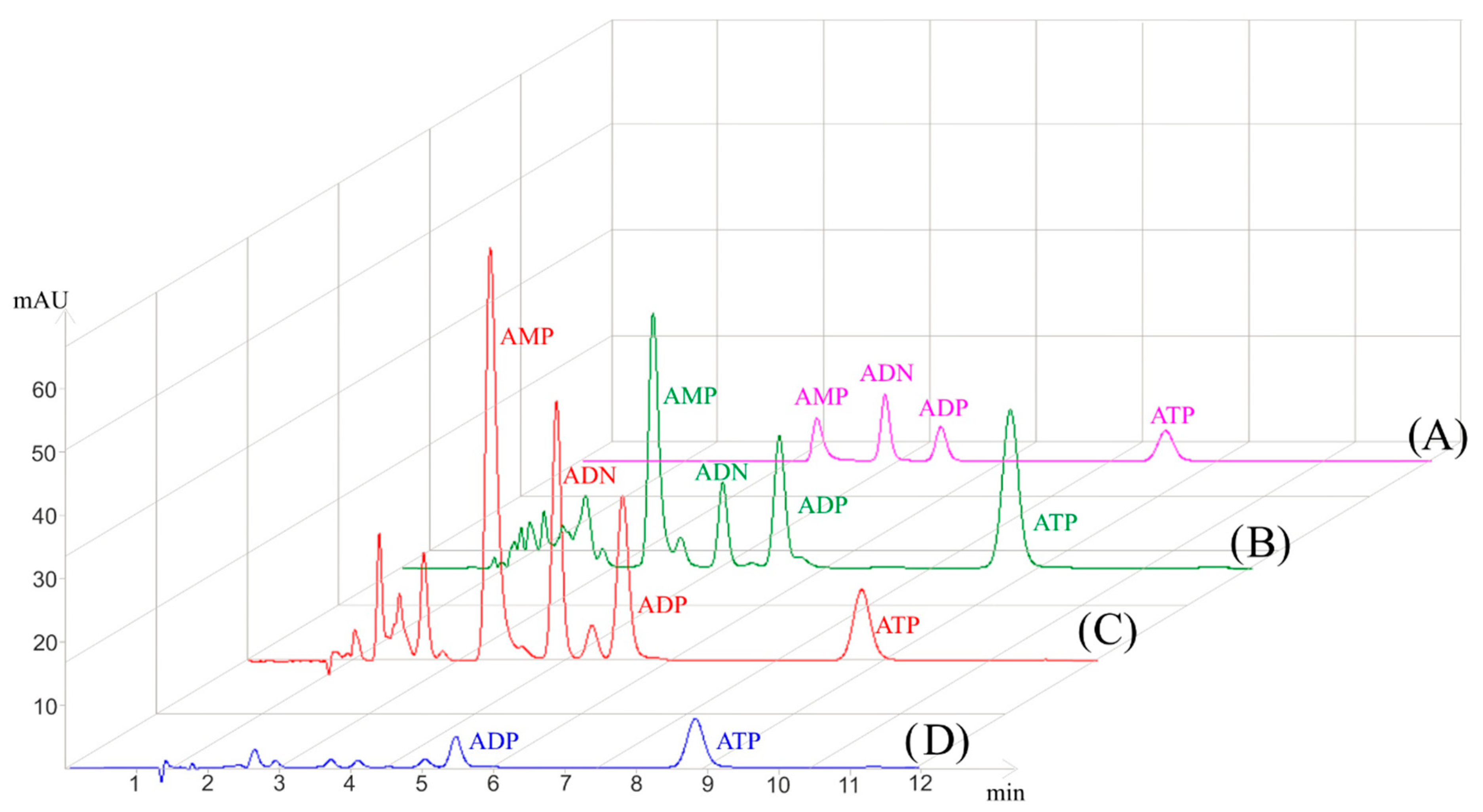
3.2.1. Influence of Homogenization Method on Adenylate Extraction
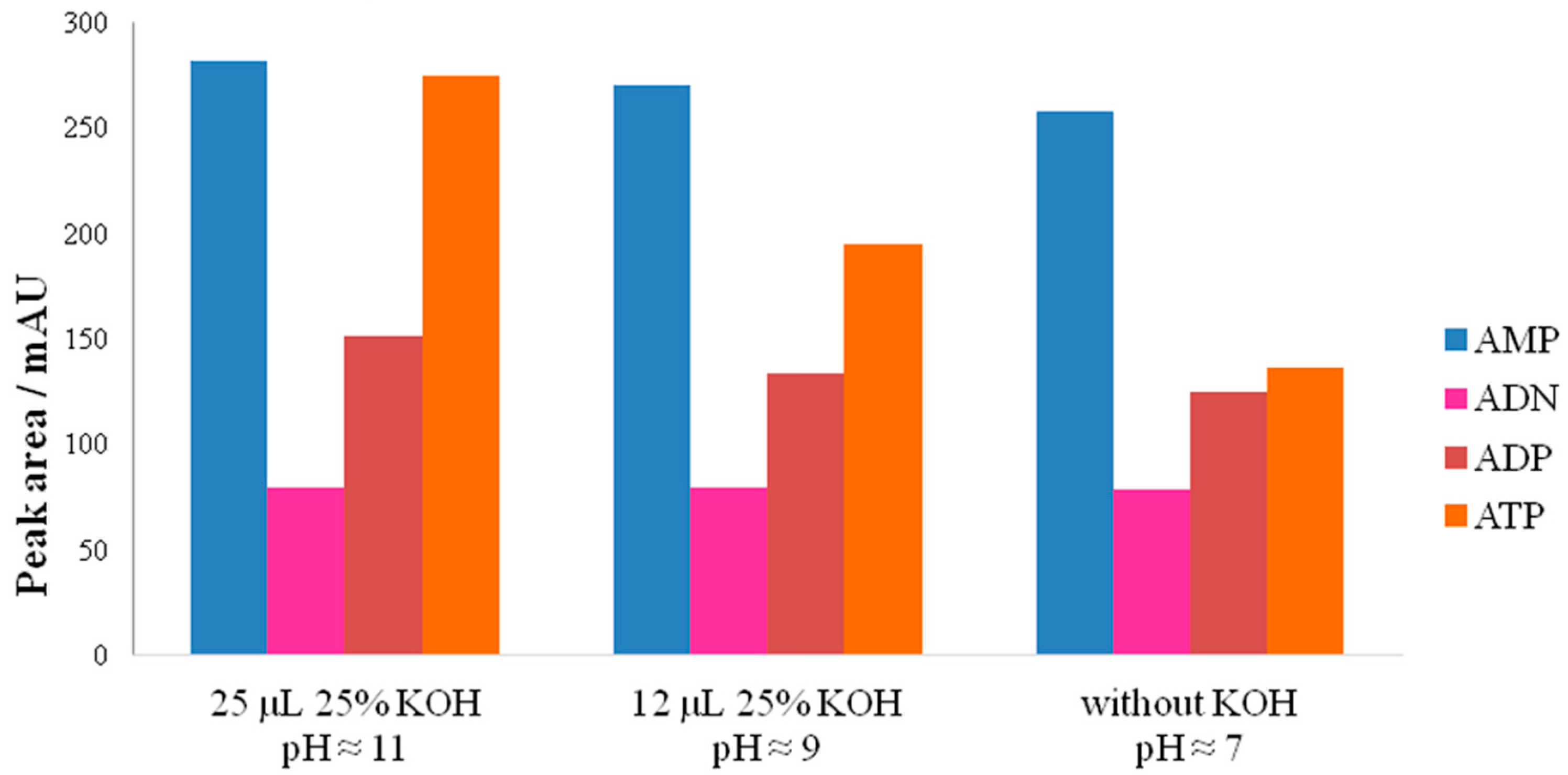
3.2.2. Influence of pH on Adenylate Stability
3.3. Improved Sensitivity with PP µSPE
3.4. Application of AEC to Environmental Issues
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AND | adenosine |
| ADP | adenosine 5′-diphosphate |
| AEC | adenylate energy charge |
| AMP | adenosine 5′-monophosphate |
| ATP | adenosine 5′-triphosphate |
| GTP | guanosine 5′-triphosphate |
| IP-RP-HPLC | ion-pair reverse phase high performance liquid chromatography |
| PCA | perchloric acid |
| PP µSPE | positive pressure micro-solid phase extraction |
| UTP | uridine 5′-triphosphate |
| S/N ratio | signal-to-noise ratio |
| TBA | tetrabutylammonium hydroxide |
References
- Friberg, N.; Bonada, N.; Bradley, D.C.; Dunbar, M.J.; Edwards, F.K.; Grey, J.; Hayes, R.B.; Hildrew, A.G.; Lamouroux, N.; Trimmer, M.; et al. Biomonitoring of human impacts in freshwater ecosystems: The good, the bad and the ugly. Adv. Ecol. Res. 2011, 44, 1–68. [Google Scholar] [CrossRef]
- Fierro, P.; Valdovinos, C.; Vargas-Chacoff, L.; Bertrán, C.; Arismendi, I. Macroinvertebrates and fishes as bioindicators of stream water pollution. In Water Quality; Tutu, H., Ed.; IntechOpen: London, UK, 2017; pp. 24–38. [Google Scholar] [CrossRef]
- Altermatt, F.; Alther, R.; Fišer, C.; Jokela, J.; Konec, M.; Küry, D.; Mächler, E.; Stucki, P.; Westram, A.M. Diversity and distribution of freshwater amphipod species in Switzerland (Crustacea: Amphipoda). PLoS ONE 2014, 9, e110328. [Google Scholar] [CrossRef]
- Little, C.J.; Altermatt, F. Species turnover and invasion of dominant freshwater invertebrates alter biodiversity–ecosystem-function relationship. Ecol. Monogr. 2018, 88, 461–480. [Google Scholar] [CrossRef]
- Bundschuh, M.; Zubrod, J.P.; Schulz, R. The functional and physiological status of Gammarus fossarum (Crustacea; Amphipoda) exposed to secondary treated wastewater. Environ. Pollut. 2011, 159, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, D.E. Energy charge of adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry 1968, 7, 4030–4034. [Google Scholar] [CrossRef] [PubMed]
- Prado, S.; Villamarín, A.; Ibarguren, I. Simultaneous determination of adenosine and related purines in tissues and hemolymph of mussel by HPLC. J. Liq. Chromatogr. Relat. Technol. 2013, 36, 470–485. [Google Scholar] [CrossRef]
- Díaz Enrich, M.J.; Villamarín, J.A.; Ramos Martínez, J.I.; Ibarguren, I. Measurement of adenosine 3′,5′-cyclic monophosphate and guanosine 3′, 5′-cyclic monophosphate in mussel (Mytilus galloprovincialis Lmk.) by high-performance liquid chromatography with diode array detection. Anal. Biochem. 2000, 285, 105–112. [Google Scholar] [CrossRef]
- Blanco, S.L.; Suárez, M.P.; San Juan, F. Seasonal changes of nucleotides in mussel (Mytilus galloprovincialis) mantle tissue. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2006, 143, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xue, C.; Li, Z.; Fu, X.; Xu, J.; Xue, Y. Changes in the contents of ATP and its related breakdown compounds in various tissues of oyster during frozen storage. J. Ocean. Univ. China 2007, 6, 407–412. [Google Scholar] [CrossRef]
- Woll, A.K.; Bakke, S. Stress and mortality in the supply chain of live scallops Pecten maximus L., from scuba diver to market. Aquac. Res. 2017, 48, 594–607. [Google Scholar] [CrossRef]
- Anacleto, P.; Maulvault, A.L.; Barrento, S.; Mendes, R.; Nunes, M.L.; Rosa, R.; Marques, A. Physiological responses to depuration and transport of native and exotic clams at different temperatures. Aquaculture 2013, 408–409, 136–146. [Google Scholar] [CrossRef]
- Schäfer, S.; Abele, D.; Weihe, E.; Köhler, A. Sex-specific biochemical and histological differences in gonads of sea urchins (Psammechinus miliaris) and their response to phenanthrene exposure. Mar. Environ. Res. 2011, 71, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Guida, L.; Walker, T.I.; Reina, R.D. The adenylate energy charge as a new and useful indicator of capture stress in chondrichthyans. J. Comp. Physiol. B 2016, 186, 193–204. [Google Scholar] [CrossRef]
- Napolitano, M.J.; Shain, D.H. Quantitating adenylate nucleotides in diverse organisms. J. Biochem. Biophys. Methods 2005, 63, 69–77. [Google Scholar] [CrossRef]
- Czarnecka, J.; Cieślak, M.; Michał, K. Application of solid phase extraction and high-performance liquid chromatography to qualitative and quantitative analysis of nucleotides and nucleosides in human cerebrospinal fluid. J. Chromatogr. B 2005, 822, 85–90. [Google Scholar] [CrossRef]
- Zur Nedden, S.; Eason, R.; Doney, A.S.; Frenguelli, B.G. An ion-pair reversed-phase HPLC method for determination of fresh tissue adenine nucleotides avoiding freeze-thaw degradation of ATP. Anal. Biochem. 2009, 388, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Schill, R.O.; Köhler, H.R. Does the environment or the source of the population define stress status and energy supply in the freshwater amphipod, Gammarus fossarum? Ecotoxicology 2004, 13, 683–695. [Google Scholar] [CrossRef]
- Moal, J.; Le Coz, J.R.; Samain, J.F.; Daniel, J.Y. Nucleotides in bivalves: Extraction and analysis by high-performance liquid chromatography (HPLC). Comp. Biochem. Physiol. B 1989, 93, 307–316. [Google Scholar] [CrossRef]
- Khlyntseva, S.V.; Bazel’, Y.R.; Vishnikin, A.B.; Andruch, V. Methods for the determination of adenosine triphosphate and other adenine nucleotides. J. Anal. Chem. 2009, 64, 657–673. [Google Scholar] [CrossRef]
- Ribeiro, C.; Esteves da Silva, J.C. Kinetics of inhibition of firefly luciferase by oxyluciferin and dehydroluciferyl-adenylate. Photochem. Photobiol. Sci. 2008, 7, 1085–1090. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Guo, R.; Li, S.; Chang, A.; Zhu, Z.; Tu, P. Optimized bioluminescence analysis of adenosine triphosphate (ATP) released by platelets and its application in the high throughput screening of platelet inhibitors. PLoS ONE 2019, 14, e0223096. [Google Scholar] [CrossRef]
- Aragon-Martinez, O.H.; Galicia, O.; Isiordia-Espinoza, M.A.; Martinez-Morales, F. A novel method for measuring the ATP-related compounds in human erythrocytes. Tohoku J. Exp. Med. 2014, 233, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Logotheti, M.; Theochari, K.; Kostakis, M.; Pasias, I.N.; Thomaidis, S. Development and validation of a HILIC-UV method for the determination of nucleotides in fish samples. Food Chem. 2018, 248, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Dudley, E.; Bond, A.E. Proteomics in Biomedicine and Pharmacology. In Advances in Protein Chemistry and Structural Biology; Donev, R., Ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 95, pp. 25–69. [Google Scholar]
- Mora, L.; Hernández-Cázares, A.S.; Aristoy, M.C.; Toldrá, F. Hydrophilic interaction chromatographic determination of adenosine triphosphate and its metabolites. Food Chem. 2010, 123, 1282–1288. [Google Scholar] [CrossRef]
- Furusawa, N. Organic solvent-free and simple method for determining cyromazine and its metabolite, melamine, in cow’s milk. J. Anal. Sci. Meth. Instrum. 2012, 2, 68–73. [Google Scholar] [CrossRef]
- Monasterio, R.P.; Londonio, J.A.; Farias, S.S.; Smichowski, P.; Wuilloud, R.G. Organic solvent-free reversed-phase ion-pairing liquid chromatography coupled to atomic fluorescence spectrometry for organoarsenic species determination in several matrices. J. Agric. Food Chem. 2011, 59, 3566–3574. [Google Scholar] [CrossRef]
- Robles-Romo, A.; Arjona, O.; Racotta, I.S. Influence of sampling, storage, processing and optimal experimental conditions on adenylate energy charge in penaeid shrimp. Arch. Biol. Sci. 2014, 66, 651–666. [Google Scholar] [CrossRef]
- Wei, H.; Tian, Y.; Lin, Y.; Maeda, H.; Yamashita, T.; Yu, K.; Takaki, K.; Yuan, C. Condition-dependent adenosine monophosphate decomposition pathways in striated adductor muscle from Japanese scallop (Patinopecten yessoensis). J. Food Sci. 2020, 85, 1462–1469. [Google Scholar] [CrossRef]
- Fang, M.; Ivanisevic, J.; Benton, H.P.; Johnson, C.H.; Patti, G.J.; Hoang, L.T.; Uritboonthai, W.; Kurczy, M.E.; Siuzdak, G. Thermal degradation of small molecules: A global metabolomic investigation. Anal. Chem. 2015, 87, 10935–10941. [Google Scholar] [CrossRef]
- Alberty, R.A. Thermodynamics of the hydrolysis of adenosine triphosphate as a function of temperature, pH, pMg, and ionic strength. J. Phys. Chem. B 2003, 107, 12324–12330. [Google Scholar] [CrossRef]
- Simão, A.M.; Bolean, M.; Hoylaerts, M.F.; Millán, J.L.; Ciancaglini, P. Effects of pH on the production of phosphate and pyrophosphate by matrix vesicles’ biomimetics. Calcif. Tissue Int. 2013, 93, 222–232. [Google Scholar] [CrossRef]
- Liu, X.C.; Scouten, W.H. Boronate affinity chromatography. Methods Mol. Biol. 2000, 147, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Le Moullac, G.; Quéau, I.; Le Souchu, P.; Pouvreau, S.; Moal, J.; Le Coz, J.R.; Samain, J.F. Metabolic adjustments in the oyster Crassostrea gigas according to oxygen level and temperature. Mar. Biol. Res. 2007, 3, 357–366. [Google Scholar] [CrossRef]
- Louis, F.; Rocher, B.; Barjhoux, I.; Bultelle, F.; Dedourge-Geffard, O.; Gaillet, V.; Bonnard, I.; Delahaut, L.; Pain-Devin, S.; Geffard, A.; et al. Seasonal monitoring of cellular energy metabolism in a sentinel species, Dreissena polymorpha (bivalve): Effect of global change? Sci. Total. Environ. 2020, 725, 138450. [Google Scholar] [CrossRef] [PubMed]
- Delaporte, M.; Soudant, P.; Lambert, C.; Moal, J.; Pouvreau, S.; Samain, J.F. Impact of food availability on energy storage and defense related hemocyte parameters of the Pacific oyster Crassostrea gigas during an experimental reproductive cycle. Aquaculture 2006, 254, 571–582. [Google Scholar] [CrossRef]
| Nucleotide | LR a (µM) | Slope (α) (µM−1) | y-Intercept | Sa b | Sb c | Sx/y d | r2 | RSS e |
|---|---|---|---|---|---|---|---|---|
| AMP | 4.41–300 | 2.0506 | −1.3661 | 0.4755 | 0.0033 | 0.9036 | 1.000 | 4.0823 |
| ADP | 3.04–300 | 1.9620 | −0.7240 | 0.3142 | 0.0022 | 0.5972 | 1.000 | 1.7835 |
| ATP | 2.67–300 | 2.3340 | −1.8763 | 0.3274 | 0.0023 | 0.6222 | 1.000 | 1.9359 |
| ADN | 2.74–300 | 2.5320 | −0.9110 | 0.3646 | 0.0025 | 0.6930 | 1.000 | 2.4015 |
| Nucleotide | Theoretical Conc. (µM) | Intra-Day | Inter-Day | ||
|---|---|---|---|---|---|
| Observed Conc. (µM) | RSD (%) | Observed Conc. (µM) | RSD (%) | ||
| AMP | 10 | 10.43 ± 0.01 | 0.10 | 10.46 ± 0.04 | 0.38 |
| 12 | 12.34 ± 0.08 | 0.65 | 12.46 ± 0.08 | 0.64 | |
| 25 | 25.04 ± 0.04 | 0.16 | 25.12 ± 0.05 | 0.20 | |
| 50 | 49.48 ± 0.04 | 0.08 | 49.46 ± 0.02 | 0.04 | |
| 100 | 99.41 ± 0.15 | 0.15 | 99.07 ± 0.32 | 0.32 | |
| 200 | 200.15 ± 0.06 | 0.03 | 200.21 ± 0.12 | 0.06 | |
| 300 | 300.15 ± 0.08 | 0.03 | 300.22 ± 0.04 | 0.01 | |
| ADP | 10 | 10.22 ± 0.12 | 1.17 | 10.13 ± 0.02 | 0.20 |
| 12 | 12.21 ± 0.08 | 0.66 | 12.22 ± 0.12 | 0.98 | |
| 25 | 25.07 ± 0.05 | 0.20 | 25.05 ± 0.11 | 0.44 | |
| 50 | 49.84 ± 0.04 | 0.08 | 49.90 ± 0.02 | 0.04 | |
| 100 | 99.64 ± 0.14 | 0.14 | 99.39 ± 0.43 | 0.43 | |
| 200 | 199.68 ± 0.44 | 0.22 | 200.34 ± 0.19 | 0.09 | |
| 300 | 300.33 ± 0.33 | 0.11 | 299.97 ± 0.19 | 0.06 | |
| ATP | 10 | 9.78 ± 0.02 | 0.20 | 9.80 ± 0.08 | 0.82 |
| 12 | 11.84 ± 0.06 | 0.51 | 11.89 ± 0.20 | 1.68 | |
| 25 | 25.12 ± 0.06 | 0.24 | 25.17 ± 0.03 | 0.12 | |
| 50 | 50.10 ± 0.02 | 0.04 | 50.14 ± 0.13 | 0.26 | |
| 100 | 100.07 ± 0.14 | 0.14 | 99.85 ± 0.14 | 0.14 | |
| 200 | 200.40 ± 0.12 | 0.06 | 200.39 ± 0.19 | 0.09 | |
| 300 | 299.70 ± 0.11 | 0.04 | 299.76 ± 0.11 | 0.04 | |
| ADN | 10 | 10.19 ± 0.05 | 0.49 | 10.27 ± 0.12 | 1.17 |
| 12 | 12.22 ± 0.07 | 0.57 | 12.29 ± 0.11 | 0.90 | |
| 25 | 25.07 ± 0.01 | 0.04 | 25.10 ± 0.08 | 0.32 | |
| 50 | 49.78 ± 0.01 | 0.02 | 49.86 ± 0.07 | 0.14 | |
| 100 | 99.54 ± 0.07 | 0.07 | 98.99 ± 0.48 | 0.48 | |
| 200 | 200.10 ± 0.27 | 0.13 | 200.44 ± 0.06 | 0.03 | |
| 300 | 300.10 ± 0.20 | 0.07 | 300.03 ± 0.10 | 0.03 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Redžović, Z.; Erk, M.; Svetličić, E.; Dončević, L.; Gottstein, S.; Hozić, A.; Cindrić, M. Determination of Adenylate Nucleotides in Amphipod Gammarus fossarum by Ion-Pair Reverse Phase Liquid Chromatography: Possibilities of Positive Pressure Micro-Solid Phase Extraction. Separations 2021, 8, 20. https://doi.org/10.3390/separations8020020
Redžović Z, Erk M, Svetličić E, Dončević L, Gottstein S, Hozić A, Cindrić M. Determination of Adenylate Nucleotides in Amphipod Gammarus fossarum by Ion-Pair Reverse Phase Liquid Chromatography: Possibilities of Positive Pressure Micro-Solid Phase Extraction. Separations. 2021; 8(2):20. https://doi.org/10.3390/separations8020020
Chicago/Turabian StyleRedžović, Zuzana, Marijana Erk, Ema Svetličić, Lucija Dončević, Sanja Gottstein, Amela Hozić, and Mario Cindrić. 2021. "Determination of Adenylate Nucleotides in Amphipod Gammarus fossarum by Ion-Pair Reverse Phase Liquid Chromatography: Possibilities of Positive Pressure Micro-Solid Phase Extraction" Separations 8, no. 2: 20. https://doi.org/10.3390/separations8020020
APA StyleRedžović, Z., Erk, M., Svetličić, E., Dončević, L., Gottstein, S., Hozić, A., & Cindrić, M. (2021). Determination of Adenylate Nucleotides in Amphipod Gammarus fossarum by Ion-Pair Reverse Phase Liquid Chromatography: Possibilities of Positive Pressure Micro-Solid Phase Extraction. Separations, 8(2), 20. https://doi.org/10.3390/separations8020020