Error in Figures/Table
In the original publication [1], there were mistakes in Figures 8 and 9 and Table 2 as published. The mistake involves the use of “Non-structural protein 3 (NSP3)” instead of “novel SH2-containing protein 3 (NSP3)”. The corrected Figure 8 and Figure 9 and Table 2 appears below.
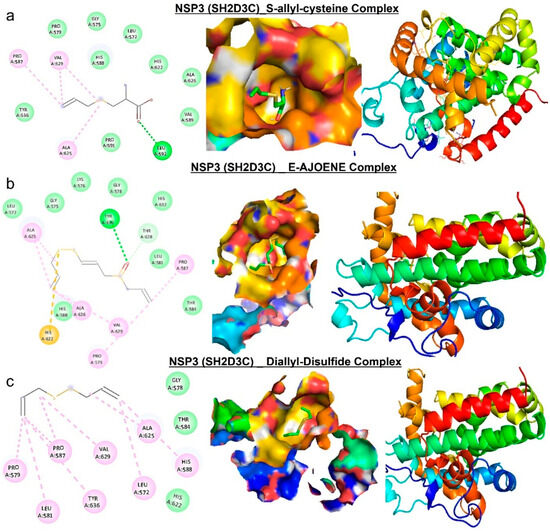
Figure 8.
Molecular docking profile on NSP3 with the organosulfur small molecule from Allium sativum. Two-dimensional (2D) structure and binding surface flip of the ligand−receptor interactions between NSP3 (SH2D3C) and (a) S-allyl-cysteine, (b) E-ajoene, and (c) diallyl sulfide.
Figure 8.
Molecular docking profile on NSP3 with the organosulfur small molecule from Allium sativum. Two-dimensional (2D) structure and binding surface flip of the ligand−receptor interactions between NSP3 (SH2D3C) and (a) S-allyl-cysteine, (b) E-ajoene, and (c) diallyl sulfide.
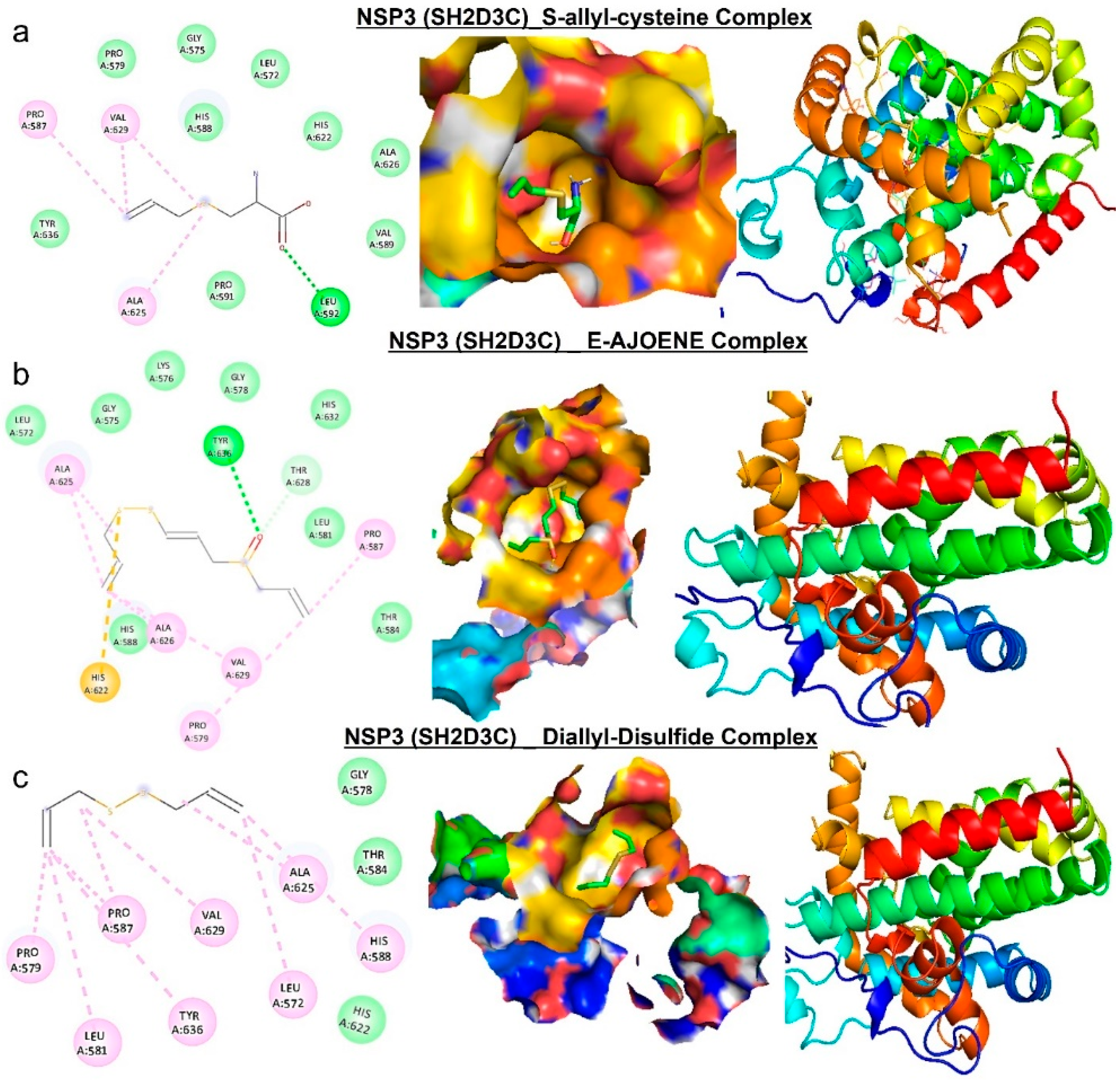
Figure 9.
Molecular docking profile on novel SH2-containing protein 3 (NSP3) with the organosulfur small molecule from Allium sativum. Three- (3D) and two-dimensional (2D) structure of the ligand−receptor interactions between NSP3 (SH2D3C) and alliin, allicin, and 2-vinyl-4H-1,3-dithiin.
Figure 9.
Molecular docking profile on novel SH2-containing protein 3 (NSP3) with the organosulfur small molecule from Allium sativum. Three- (3D) and two-dimensional (2D) structure of the ligand−receptor interactions between NSP3 (SH2D3C) and alliin, allicin, and 2-vinyl-4H-1,3-dithiin.

Table 2.
Docking profile of NSP3 (SH2D3C) with the organosulfur small molecule from Allium sativum.
Table 2.
Docking profile of NSP3 (SH2D3C) with the organosulfur small molecule from Allium sativum.
| S-allyl-cysteine | E-AJOENE | Alliin | Diallyl-Disulfide | Allicin | 2-Vinyl-4H-1,3-dithiin | |
|---|---|---|---|---|---|---|
| ΔG (Kcal/mol) | −4.40 | −4.40 | −6.70 −4.70 | −3.50 | −3.90 | −4.00 |
| Hydrophobic contact | PRO587 (3.85 Ӑ) HIS622A (3.78 Ӑ) ILE131 (3.62 Ӑ) PHE132 (3.61 Ӑ) PHE156 (3.70 Ӑ) | PRO579 (3.50), LEU581 (3.52), THR584 (3.56), PRO587 (3.79), ALA625 (3.62) | Pro579 (3.99), leu581 (3.89), thr584 (3.78), pro587 (3.94) | Leu572 (3.72), pro579 (3.90), leu581 (3.79), thr584 (3.67), pro587 (3.79), his588 (3.75) | Leu572 (3.87), pro579 (3.92), leu581 (3.77), thr584 (3.89), pro587 (3.51), his588 (3.74) | His588 (3.75), his622 (3.76), ala625 (3.53) |
| Conventional H-bond | LEU592 (2.21) | Thr636 (2.61) Thr628 (3.64) | His622, gly575, leu572, his588 | His588 | ||
| Pi-sulfur | his622 | His588, his622 | ||||
| alkyl interaction | VAL626, PRO587, ALA625 | Ala625, ala626, val629, pro579, pro587 | Tyr636, leu581, pro587, pro579 | Pro579, pro587, val629, ala625, leu581, tyr636, leu572, his588 | Pro587, pro579, leu581 | Leu592, ala625 |
| Van der waal forces | Pro579, Gly575, His588, Leu572, His622, Ala626, Val589, Pro591, Tyr636 | Leu572, Gly585, Lys576, Gly578, His632, Leu581, Thr584, His 588 | Val629, Thr584, Ala625, Gly578 | His622, Gly578 | Thr584, Tyr636, Val629, Ala626, Gly578, Gly575,Leu572, His622 | Pro591, Val589, Val629, Pro579, Lys576, Gly578, Leu572, Gly575 |
Text Correction
There were errors in the original publication. The mistake involves the use of “Non-structural protein 3 (NSP3)” instead of “novel SH2-containing protein 3 (NSP3)”.
A correction has been made to Abstract:
(1) Original: The multi-domain non-structural protein 3 (NSP3) is an oncogenic molecule that has been concomitantly implicated in the progression of coronavirus infection.
Revised: The novel SH2-containing protein 3 (NSP3) is an oncogenic molecule that has been concomitantly associated with T cell trafficking.
(2) Original: −4.3~−6.70 Ă
Revised: −3.5~−6.70 Ă
(3) Removed “However, S-allyl-cysteine interaction with NSP3 (SH2D3C) is unfavorable and hence less susceptible to NSP3 ligandability.”
A correction has been made to Result, Section 3.9:
Removed: However, S-allyl-cysteine interaction with NSP3 (SH2D3C) is unfavorable (Figure 8A) and hence the least ΔG (−4.30 Kcal/mol) and RF (−4.31 pKd) values, respectively.
A correction has been made to Discussion, the first Paragraph:
Removed: Notwithstanding, NSP3 was proposed to be a promising therapeutic target in both cancer and COVID-19.
A correction has been made to Introduction, the third Paragraph:
Original: Non-structural protein 3 (NSP3) is a multi-domain, multifunctional protein that is an essential component of the replication/transcription complex (RTC), responsible for the synthesis and processing of RNA, and interference with the innate immune system of host cells [20,21]. NSP3 has been implicated in cancer progression and metastasis [22,23]. It is a key component in coronavirus replication and has thus played a pivotal role in the coronavirus disease 2019 (COVID-19) pandemic. It thus could be an attractive target for the development of therapeutic strategies for treating cancer and coronavirus infections [24].
Revised: The novel SH2-containing protein 3 (NSP3) is an oncogenic molecule that regulates T cell receptor signaling [20]. It acts as an adapter protein that mediates cell signaling pathways involved in cellular functions such as cell adhesion and migration, tissue organization, and the regulation of the immune response [20–22]. It thus could be an attractive target for the development of therapeutic strategies for treating cancer.
References
Remove References [20–23] from the original manuscript [1] and use References [2,3,4] as [20–22]. With this correction, the order of some references has been adjusted accordingly.
The authors state that the scientific conclusions are unaffected. This correction was approved by the Academic Editor. The original publication has also been updated.
References
- Yeh, Y.-C.; Lawal, B.; Hsiao, M.; Huang, T.-H.; Huang, C.-Y.F. Identification of NSP3 (SH2D3C) as a Prognostic Biomarker of Tumor Progression and Immune Evasion for Lung Cancer and Evaluation of Organosulfur Compounds from Allium sativum L. as Therapeutic Candidates. Biomedicines 2021, 9, 1582. [Google Scholar] [CrossRef] [PubMed]
- Dodelet, V.C.; Pazzagli, C.; Zisch, A.H.; Hauser, C.A.; Pasquale, E.B. A novel signaling intermediate, SHEP1, directly couples Eph receptors to R-Ras and Rap1A. J. Biol. Chem. 1999, 274, 31941–31946. [Google Scholar] [CrossRef]
- Sakakibara, A.; Ohba, Y.; Kurokawa, K.; Matsuda, M.; Hattori, S. Novel function of Chat in controlling cell adhesion via Cas-Crk-C3G-pathway-mediated Rap1 activation. J. Cell Sci. 2002, 115, 4915–4924. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Vervoort, V.; Wallez, Y.; Coré, N.; Cremer, H.; Pasquale, E.B. The SRC homology 2 domain protein Shep1 plays an important role in the penetration of olfactory sensory axons into the forebrain. J. Neurosci. 2010, 30, 13201–13210. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
