Abstract
This work consists of a study to detect prostate cancer using E-senses devices based on electronic tongue and electronic nose systems. Therefore, two groups of confirmed prostate cancer and control patients were invited to participate through urine and exhaled breath samples, where the control patients group was categorized as Benign Prostatic Hyperplasia, Prostatitis, and Healthy patients. Afterward, the samples were subsequently classified using Pattern Recognition and machine learning methods, where the results were compared through clinical history, obtaining a 92.9% success rate in the PCa and control samples’ classification accuracy by using eTongue and a 100% success rate of classification using eNose.
1. Introduction
Prostate cancer (PCa) is a malignant tumor diagnosed in the male population, ranking second only to lung cancer, as reported by GLOBOCAN (Global Cancer Observatory) in 2020; it is the second most frequent cancer in more than half (112 cases out of 185) of the countries of the world and the fifth cause of death in men [1]. In many cases, stage PCa normally grows very slowly and often produces no symptoms or problems for years; however, the symptoms produced by this tumor arise when the disease is locally advanced or metastatic, which makes early diagnosis of the disease a public health challenge since diagnosing this type of cancer in its early stage can improve the mortality rate and quality of life [2].
Currently, the main diagnostic and monitoring tests for PCa are prostate-specific antigen (PSA), a physical examination of the prostate gland, rectal examination (DRE), and transrectal biopsy, which is the standard commonly used to confirm the disease. However, in recent years, there has been controversy as to the effectiveness and efficiency of early diagnosis of PCa using these methods [3]. PSA is recognized as an important tumor marker for the detection and monitoring of PCa, as it is a protein produced by normal cells as well as by malignant cells in the prostate gland [4]. According to the literature, in the past, the serum PSA levels accepted as a normal reference range for all age groups was 4 ng/mL (nanograms per milliliter of blood). However, it has been shown that this test is not always sensitive since studies have indicated that some men with PSA concentrations less than 4.0 ng/mL had PCa and that many men with higher concentrations did not have the disease [5,6]; that is why between 10% and 12% of men who undergo periodic PSA tests will obtain a false positive result. The second important aspect to consider is that the PSA is not specific to PCa since its concentration can be elevated for several reasons, such as age, ethnicity, urinary retention, and benign prostate diseases, such as Prostatitis and Benign Prostatic Hyperplasia (BPH). As reported, in patients with Prostatitis, PSA increases exceeding 30 ng/mL can be detected; this value normalizes after six to eight weeks. In patients with BPH, 25% to 50% see PSA levels increase >4 ng/mL, but no higher than 10 ng/mL [7,8]. As mentioned above, the inability of this test to distinguish on its own between benign prostate diseases and localized tumors can be demonstrated, leading to unnecessary false-positive prostate biopsies [9], overdiagnosis [10], and overtreatment against PCa, causing considerable side effects [11], including erectile dysfunction and urinary incontinence [12].
With regard to the digital rectal examination (DRE), it is the oldest and cheapest screening tool. However, it is a technique with low reproducibility due to the variability between examiners who can interpret the results differently [13,14], since the presence of rectal touch abnormalities can include prostate enlargement, the appearance of suspicious nodules, lobular asymmetry, and hard consistency, among others; the above are not necessarily related to PCa, so they may be related to some other prostate disease. Another disadvantage presented by the DRE test is that the PCa in its early stage may not have the necessary size and rigidity to be palpable [15]. This test additionally causes considerable discomfort, is uncomfortable and embarrassing for patients, and is less accurate than the PSA test [16]. That is why, when there are abnormalities found in the DRE or PSA, it is advisable to perform a prostate biopsy; this is an invasive test that involves the rectal insertion of an ultrasound probe to diagnose and confirm PCa, to evaluate the histological architecture using the Gleason grade [17].
On the other hand, transrectal ultrasound (TRUS)-guided systematic biopsy (SB) is the most reliable and currently used method to ensure the accurate sampling of prostate tissue in men considered at high risk of having PCa [18,19]. About 25% of the male population undergoing prostate biopsy due to elevated PSA or abnormal DRE have prostate cancer, indicating that the other 75% of the study population is undergoing this test following a false positive generated by PSA or DRE [20]. This subjects patients to uncomfortable procedures and causes complications such as rectal bleeding, urinary retention, erectile dysfunction, infections, pain, stress, anxiety, costs, and long waiting times [21,22].
Because of the problems mentioned, there is a need to find and apply new tools for the early diagnosis of the disease. These tests should be effective, non-invasive, easily affordable, reliable, and reproducible, with a high accuracy, sensitivity, specificity, and precision, where applicable and feasible at a low cost to patients who are of a risk low level.
In recent years, interest has increased in studying metabolic alterations that are a distinctive characteristics of cancer cells, since they imply significant changes in cellular metabolism (increased glycolysis, lipid metabolism, altered amino acids, oxidative stress, and Reactive Oxygen Species (ROS)) compared with normal cells. These metabolic alterations not only support cancer growth but also contribute to the generation of chemical compounds that can be detected in different biological fluids, and their presence or altered levels could serve as biomarkers for cancer detection, monitoring disease progression, predicting disease recurrence, and therapeutic treatment effectiveness, among others [23,24,25,26]. The analysis and identification of biomarkers is conducted through analytical platforms such as Nuclear Magnetic Resonance Spectroscopy (NMR) [27,28,29], Gas Chromatography coupled with Mass Spectrometry (GC-MS) [29,30,31,32], Liquid Chromatography coupled with Mass Spectrometry (LC-MS) [33,34,35], High-Performance Liquid Chromatography (HPLC) [36,37], and computer and statistical tools. All these analytical techniques can detect and quantify different compounds with low concentrations. However, they have several drawbacks since they are complex, take a long time to diagnose, are expensive, require trained personnel, and are not portable [38].
Sensory perception systems such as the electronic nose and electronic tongue combined with chemometric and pattern recognition tools [39,40,41,42,43] have evolved over the years to stand out as promising tools for the non-invasive, rapid, and potentially inexpensive diagnosis of health conditions, such as cancer. In addition, they have portability characteristics, facilitating their flexibility [44,45,46,47].
Today, the electronic nose (eNose) represents a potential tool capable of mimicking the olfactory system of dogs, who were the pioneers in demonstrating the detection capacity of PCa-specific Volatile Organic Compounds (VOCs) in healthy patients, with a sensitivity and specificity above 90%; even so, according to the promising results obtained, the researchers recognized certain disadvantages in the test, due to the time it took to train the canine (approximately 12 months) and the high cost of the procedure [48,49,50,51,52,53]. The eNose system is composed of gas sensors with partially overlapping sensitivities that generate electrical signals which are acquired by means of basic and programmable electronics to generate a set of data characteristic of the compound or odor to be analyzed by means of a pattern recognition unit based on machine learning algorithms and artificial intelligence [54]. In the field of PCa diagnosis, Bernabei et al. investigated the operation of an electronic nose based on quartz crystal microbalance (QCM) gas sensors, coated with different metalloporphyrins, to assess urine headspace for the detection of patients with PCa, bladder cancer, and controls. The data were processed by principal component analysis (PCA) and discriminant analysis using partial least squares (PLS-DA), achieving good discrimination between the groups [55]. In another study reported by D’Amico et al., urine samples from PCa patients and controls were analyzed using an eNose with eight non-selective gas sensors, coated with metalloporphyrins. Subsequently, the data were processed by PLS-DA and a qualitative graph was presented to evaluate the discrimination achieved [56]. Roine et al. applied a commercial ChemPro 100 Nose to discriminate patients with PCa and patients with BPH using urine samples, achieving a sensitivity and specificity of 78% and 67%, respectively [57]. Filianoti, et al. in 2022 conducted a study to determine the efficiency of the urinary volatiloma profile to distinguish patients with PCa from healthy controls, using a commercial nose called Cyranose C320, in which a sensitivity of 82.7% was obtained, and a specificity of 88.5% for the technique [58]. Likewise, other studies have been reported in different sources [59,60,61,62].
Another widely studied sample type is breath, which is widely used to diagnose cancer [63,64,65]. In a recent study, Waltman et al. conducted a study that histologically confirmed PCa patients and control patients with negative biopsies and diseases related to prostate enlargement, for which exhaled breath samples were collected from 85 patients: 32 with PCa and 53 controls. An analysis of exhaled breath samples was obtained, which could be detected with an electronic nose called Aeonose and was able to differentiate between patients with PCa and healthy patients. However, the authors need to conduct further tests to confirm the findings [66].
Regarding the electronic tongue (eTongue), it is an analytical device inspired by the biological systems of taste. These devices are defined as a set of non-selective chemical electrodes with partial sensitivity to different components, which are capable of performing quantitative and qualitative analyses of complex solutions. eTongue systems, like eNose, are technologies that, in recent years, have been applied to develop various research in the health sector to analyze biological samples [67]. However, the use of these systems has been little explored in the detection of prostate cancer. Pascual et al. evaluated the potential of an eTongue using cyclic voltammetry with metal electrodes for the detection of PCa from urine samples. In total, they analyzed 71 urine samples from patients with PCa before surgery, 26 urine samples from patients with PCa after surgery, and 17 urine samples from patients with BPH, in which they achieved a sensitivity of 91% and a specificity of 73%, respectively, to distinguish the urine of cancer and non-cancer patients [68].
In another study, a simple multi-sensor potentiometric system was developed to distinguish urine samples from patients with diagnosed PCa, and a group of samples from healthy patients. Therein, 28 potentiometric electrodes with different types of sensor membranes, including PVC-plasticized, chalcogenide glass, and polycrystalline membranes were used due to the variety of cationic and anionic species in the urine, as well as the presence of Redox pairs. A total of 89 urine samples were studied (43 from cancer patients confirmed using prostate biopsy and 46 from the healthy control group), and different classification techniques were applied for data analysis. The best results were obtained with the logistic regression model, which reached a sensitivity of 100% and a specificity of 93% in the set of independent test samples [69].
In the present study, two different sensory perception systems, or E-senses, were used, one based on an electronic nose developed especially for this study called “VOCaP” and an electronic tongue for detecting prostate cancer from exhaled breath and urine samples. In this new study, it is important to highlight, and compare with the previously mentioned works, that the detection of prostate cancer was carried out using both devices applied to patients in a non-invasive way, based on the detection of volatile organic compounds and chemical changes in both breath and urine samples.
2. Materials and Methods
Figure 1 illustrates a schematic diagram of the methodology used in prostate cancer detection, which consisted of 4 differentiated stages: (1) selection of patients, (2) conditioning of biological samples for measurement with E-senses devices, (3) data acquisition, and (4) data processing.
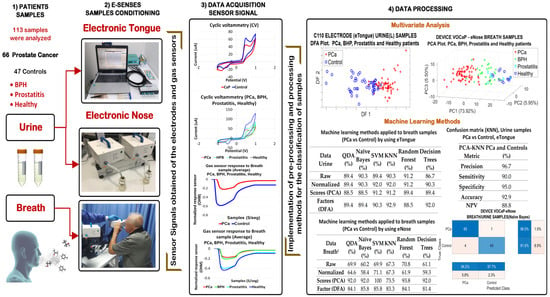
Figure 1.
Scheme of the methodology to detect PCa and control patients through E-senses.
2.1. Measurement Protocol
The methodology used for the selection of the volunteer patients in the study is described in detail below.
2.1.1. Patient Selection
Breath and urine samples from patients were collected at the Uronorte S.A clinic in Cúcuta, Norte de Santander (Colombia). The study population was a total of 113 cases, which were divided into two groups: (i) PCa group (66 cases) and (ii) control group (47 cases), where the inclusion criteria for the PCa group were patients proven histopathologically through prostate biopsies, and aged between 50 and 89 years. Concerning the control group, people with prostate-related diseases (BPH and Prostatitis) and patients with a negative history of urinary symptoms and without evidence of any neoplastic disease (Healthy) were included. The general characteristics of all volunteer patients confirmed through the medical history of both PCa and controls are described in Table 1.

Table 1.
Overall characteristics of volunteer patients.
This study was endorsed by the ethics committee of the urology center of Uronorte SA, where each volunteer was aware of the scope of the research and, therefore, an informed consent document was signed. Then, each of the patients underwent a survey in order to obtain the basic information, and a technical sheet was also filled out to learn about additional aspects (e.g., family history with PCa, comorbidities, medications, if they were smokers, among others).
2.1.2. Collection and Storage of Samples
To minimize some factors that could affect the results of the study through the E-senses systems, the patients were provided with some recommendations that they should take into account before collecting the breath and urine samples, for example, fasting 10 h before the test, avoiding using tobacco, alcohol, or food the previous night that could alter the breath and urine tests, not using perfumes, not brushing their teeth, and not applying aerosol deodorants.
For the collection of urine samples, each patient collected the first urine in the morning in two sterile containers of 50 mL each, and subsequently they were stored at a temperature of −20 °C, until their corresponding analysis.
On the other hand, the collection of exhaled breath samples was carried out in a room located on the clinic premises, at an ambient temperature of 24 °C, with relative humidity between 47% and 50% RH, at 07:00 and 10:00 a.m. The above was to improve the accuracy and efficiency of the analysis of breath samples. It should be noted that before the breath samples collection, the volunteers remained at rest for 10 min to maintain the same conditions a priori to performing the tests.
2.2. eNose-VOCaP
For this study, the prototype VOCaP was used, as mentioned above, developed by the research group of Multisensory Systems and Pattern Recognition (GISM), Electronic Engineering program of the Universidad de Pamplona, and mainly applied for the detection of prostate cancer (see Figure 2).
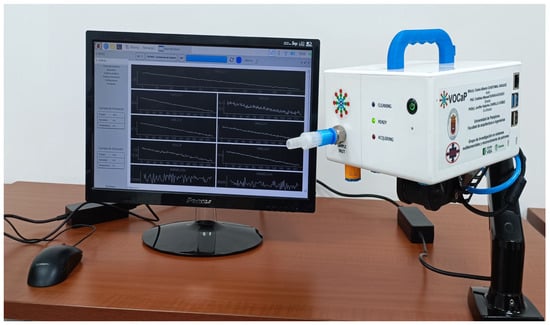
Figure 2.
Electronic nose (VOCaP) for breath and urine samples acquisition.
The VOCaP system is a multi-sensory device composed mainly of a gas sample conditioning chamber, and a digital and analog gas sensor chamber.
The data acquisition process begins when the sample gaseously enters the device through a disposable nozzle or system inlet. To carry out the above, a sample preparation stage was carried out, where different parameters such as temperature, humidity, pressure, and flow were controlled, the above being to obtain the same operating conditions for the acquisition of the measurements. In addition, the device contains an electric pump that draws the sample into the sensor chamber to maintain the operating conditions, and thus ensure sensitivity and repeatability during each of the measurements made by the multisensory device.
The measurement chamber is composed of two arrays of sensors (digital and analog) of MOX (metal oxide) class, based on Microhotplates, and MEMS (manufactured with micro-electro-mechanical system)-type technology, which has a sensitive layer of metal oxides that can detect VOCs emitted by biological samples, for which it uses a detection mechanism, composed of three phases: (1) diffusion of the target gas molecules on the surface of the metal oxide, (2) adsorption of the gas molecules on the metal oxide, and (3) reaction between the gas and the metal oxide [70].
The digital sensor array is composed of 4 different references of MOX-MEMS sensors, such as (a) 2 × CCS811 with a sensitive layer (manufactured by Sciosense, Premstaetten, Austria), (b) 2 × BME-680 with a sensitive layer for gases and temperature, humidity, and pressure sensors (manufactured by BOSH, Reutlingen, Germany), and (c) 2 × SPG30 with two independent sensitive layers (manufactured by Sensirion, Staefa, Switzerland). On the other hand, the analog sensor array is also composed of 4 MOX-MEMS sensor references, whose references and quantities are as follows: 1 × MICS 4514 with 2 independent sensory layers (manufactured by SGX sensortech, Corcelles, Switzerland), 1 × MICS 6814 with 3 independent sensory layers (manufactured by SGX sensortech, Corcelles, Switzerland), 1 × GM-502B with a sensory layer (manufactured by Winsen Electronics, Zhengzhou, China), and 1 × CCS801 with a sensory layer (manufactured by Sciosense, Premstaetten, Austria). Therefore, a total of 15 independent sensory layers were available (among all the sensors), which have the capacity to record 15 signals from a single gas sample, and in this way cover a wide spectrum of VOCs, with the aim of identifying a sample in a more significant and differentiated way with respect to others, to detect the PCa disease.
MEMS technology, being compact, has a low energy consumption and, being built as a Surface Mounting Device (SMD), the structure of the sensors is given in small dimensions which allows the increase in the density or quantity of these components in a small space. This allows the sensor chamber to have an approximate volume of 10 mL, ensuring that the concentration of the sample is homogeneous due to its cylindrical shape; therefore, the exposure of each sensor to the sample is the same. In addition, some of the sensors, such as the BME-680, have other sensors to measure temperature, humidity, and pressure, which makes it possible not only to measure VOCs but also to record these variables to make corrections and compensations due to drifts generated in the sensors [70].
For the operation of the eNose, a graphical interface was developed through the free Python software (exclusive Version 3.9.2), with the necessary acquisition functionalities for the registration of the category and the type of sample, such as the case of exhaled breath and the urine headspace. Also, the software, apart from performing the acquisition of the samples, automatically performs and controls each of the devices of the multisensory system, due to the configuration capacity through the flexible interface and with “on-line” monitoring with an Internet of Things (IoT) connection service.
2.2.1. Measurement Protocol with eNose for Urine Samples
A total of 113 urine samples were stored at −20 °C, and subsequently thawed at 4 °C; they were then transferred in aliquots to 20 mL falcon tubes to be centrifuged at 2500 rpm for 5 min, to concentrate the sediments and eliminate them.
As the aim of the eNose is to analyze gaseous samples, it was necessary to generate the headspace of each of the urine samples. To do this, 20 mL of the centrifuged urine was added into a glass container with septum, and then heated to a constant temperature of 60 °C using a hotplate over a period of 30 min. The compounds generated by the urine headspace were taken to the sampling chamber, stabilizing its interior at an approximate temperature of 40 °C, to reduce the humidity inside the device and prevent condensation in the measurement chamber to improve the release of the sample VOCs towards the gas sensors.
For the sample detection, a constant flow of 0.9 L/min, controlled by an electric pump, was used to drag the compounds towards the sensor chamber, maintaining a temperature between 39 °C and 41 °C and a humidity of 30% RH; at this stage the responses of the sensors due to the passage of the sample in the measurement chamber were recorded. The recording of the different stages of the acquisition of a sample was performed at different acquisition times of the initial baseline, for a period of 2 min, and entering ambient air or pressurized oxygen was used to stabilize the gas sensors. Then, upon entering the sample, it was recorded for 3 min; additionally, the flow and pressures in the sampling and measurement chambers were monitored, so that the sensors detected the sample due to the VOCs, and not due to the change in flow. At the end, ambient air was supplied for a period of 2 min, to clean the gas sensors and eliminate the possible memory effect on the internal pipe and the measuring chamber.
It is important to note that the cleaning of the sampling and measurement chambers was carried out through the intake of ambient air, where an Ultraviolet-Light Emitting Diodes (UV-LED) array was subsequently activated, which was located in the sampling chamber with an exposure time of 4 min, to effectively clean the system due to the measurement previously carried out. In addition, the gas residues were evacuated to the atmosphere through an activated carbon outlet filter and a high efficiency particulate air (HEPA) filter as well. The reason for using UV-LED was to avoid contamination of patients and medical personnel due to the SARS-CoV-2 virus (COVID-19).
2.2.2. Measurement Protocol for Breath Samples Using the eNose
For breath samples, each patient was asked to exhale directly into the VOCaP device, through a disposable mouthpiece, where 2 exhalations per patient were performed, acquiring a total of 226 samples and with a cleaning period of 5 min between each exhalation of the patient. Each patient was asked to exhale continuously, in order to obtain the alveolar air sample, which is the last portion of the exhaled breath that contains high concentrations of endogenous, and for being the essential part of the exhalation that is detected by the multisensory device.
On the other hand, a pressure sensor was located in the system to confirm that the breath sample was considered correct; therefore, in the patient, at the time of exhaling, the sensor detected a threshold value and, in turn, a flow sensor determined if air was entering the measurement chamber. It should be clarified that throughout the measurement process the patient was given instructions and was being monitored during the breath measurement, which took 15 min in total.
2.3. eTongue
For the electrochemical measurements of urine samples, a portable multi-channel potentiostat (see Figure 3) model μStat 8000 manufactured by the company DropSens was used, composed of 8 channels that operate independently, so it was possible to use up to 8 electrochemical sensors at the same time.

Figure 3.
μStat 8000 potentiostat used as eTongue.
The potentiostat was controlled by the “DropView 8400” software, version 3.6, that allows processing the electrochemical measurements and analyzing the acquired data, so in this study, screen-printed type C110 electrodes with different electrode materials (working electrode (4 mm diameter) and Auxiliary = Carbon, and reference electrode = Silver) were used. The carbon electrode is suitable for working with microvolumes or by dipping them in solution, which is ideal to be used in platforms for decentralized assays or when developing bio-sensors.
Figure 4 shows the electrode dimensions; it should be clarified that in this study tests were carried out with other types of sensors, from which good results were not obtained.
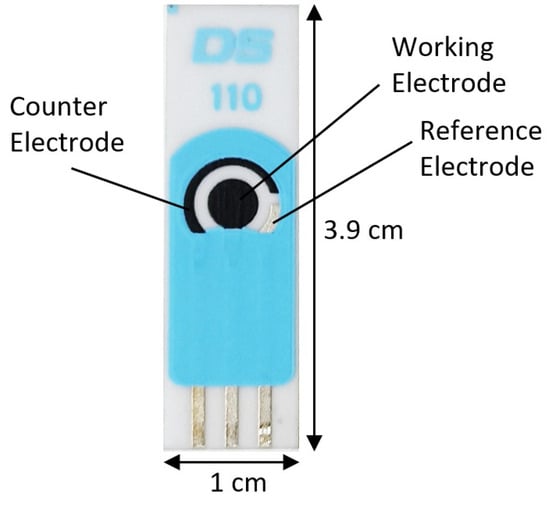
Figure 4.
C110 Screen-printed Carbon Electrode.
To obtain a reproducible response of the eTongue system to the urine sample, for the analyses, a quantity of 50 µL of sample was applied with a micropipette, where the technique used was cyclic voltammetry by setting the following measurement parameters: Ebegin: −1 VDC; scan start potential, Evtx1: −1 VDC; scan investment potential, Evtx2: +1 VDC; voltage with scan stop and the number of scans = 1.
2.4. Data Processing Methods
As mentioned above, for the use of the eNose, 226 breath samples were acquired where they were subsequently averaged in order to match the amount of data with respect to the amount of urine samples acquired with the eTongue (133 samples). In addition, when obtaining the average of the two measurements, some variations were corrected at the time of data acquisition, due to flow changes during each exhalation performed by the patients and other experimental errors.
Once the samples were acquired by the E-senses systems, the characteristic extraction of the signals from both devices was performed through the following parameters: , , , where Rmax = maximum resistance value of the gas sensor responses, Rmin = minimum resistance value of the gas sensor responses, = maximum current value of the electrode response, = minimum current value of the electrode response, Cfinal= final current value of the electrode response, and = initial current value of the electrode response. It is important to note that in the eTongue, y were applied, while in the eNose, only was used; this is because the best results were obtained by selecting these parameters for each of the two sensory devices.
Subsequently, the data were normalized from the mean-centering function, providing a scale value to the data in a similar range. It consists of subtracting the mean-value of the data from each observation, so that the new mean is equal to zero. On the other hand, in the data acquired by eNose, the analysis was carried out by applying the Orthogonal Signal Correction (OSC) algorithm to make the drift correction for the breath and urine dataset. Once the drift correction was performed, two classic multivariate analysis techniques, Principal Component Analysis (PCA) and Discriminant Function Analysis (DFA), were applied.
On the other hand, different machine learning algorithms such as Quadratic Discrimination Analysis (QDA), Naïve Bayes, Support Vector Machine (SVM), K-Nearest Neighbor (K-NN), Random Forests, and Decision Trees were used with the corresponding kernel previously selected.
Once the data were loaded with each learning model, a cross-validation (CV) algorithm was applied with k-fold = 5 to avoid overfitting in the machine learning algorithm. K-fold cross-validation is a technique used in machine learning to evaluate the performance of a model more accurately and robustly; in addition, it was helpful in this study due to the limited data and was used efficiently and without excessive consumption of resources to train and evaluate the model. In this study, 113 measurements were split into groups, such as 50% for training, 25% for validation, and 25% for testing; therefore, with CV, the first 75% were used for training/validation, and every sample of 75% was used for training and validation randomly. This means that 25% of remaining samples were used for test data.
At the end of the processing, the device’s responses were analyzed through the metrics calculated by the confusion matrix to obtain the accuracy, precision, sensitivity, specificity, and negative predictive value of the predictions made by the classification model.
3. Results
The analysis was carried out in two stages, the first by dividing the database into the PCa category (66 samples) and the control category (47 samples). On the other hand, the second analysis was made to split the control category into different subgroups (BPH, Prostatitis, and Healthy), as described in Table 1.
3.1. Results with eNose (VOCaP)
The measurement process made with the eNose for each type of sample (exhaled breath and urine headspace) was carried out according to the aforementioned protocols, which allowed the standardization of each sample acquisition and thus avoided significant variations during the sampling time (samples/second), which could have negatively affected the results. In this way, the responses of the sensor array to a sample were acquired, as can be seen in Figure 5a.
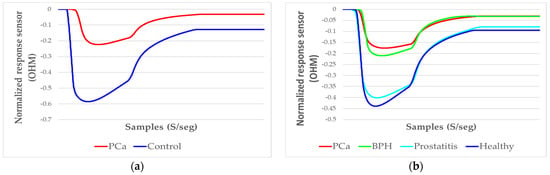
Figure 5.
Signals representative of the average resulting from the sensor array responses, normalized to an exhaled breath sample: (a) sensor responses to a PCa and a control sample; (b) sensor responses to the PCa, BPH, Prostatitis, and Healthy samples.
Therefore, a clear difference was observed between the response (amplitude) of a PCa sample and a control sample. On the other hand, Figure 5b illustrates the analysis of PCa and the control subgroups: BPH, Prostatitis, and Healthy, applying a normalization of data scaling to observe the variation in amplitude of each category.
3.1.1. Analysis of Breath Samples with eNose (PCa vs. controls)
From the feature extraction of the sensor signals by ∆R and applying the mean-centering normalization, the correction of the drifts was conducted through OSC, where the PCA analysis was subsequently applied to the dataset, obtaining a 79.89% variation for the PCa and control groups (see Figure 6a).
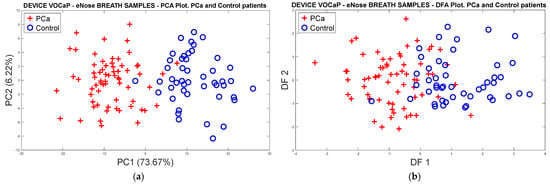
Figure 6.
Analysis of the PCa and control groups with exhaled breath samples acquired with the eNose device: (a) PCA analysis, PCa vs. control patients, (b) DFA analysis, PCa vs. control patients.
On the other hand, Figure 6b shows the response of the DFA classifier where there is a slight overlap of the measurements; however, it was possible to obtain a good separation trend in both categories.
Applying different classification methods through the cross-validation technique with k-fold = 5 to the data acquired from the PCa and control samples, the best results were obtained with SVM and PCA scores, reaching 100% accuracy in the classification of the samples, as shown in Table 2

Table 2.
Machine learning methods applied to breath samples (PCa vs. control) by using eNose.
Taking the best classification model (PCA and SVM) as a reference, Figure 7 shows the confusion matrix results, in which the model’s performance in classifying exhaled breath samples using the eNose is represented. Likewise, Table 3 lists the values of each of the metrics.
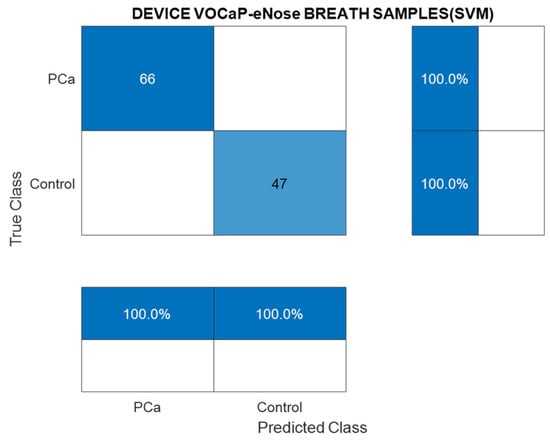
Figure 7.
Confusion matrix of the SVM classification model through breath samples (PCa vs. control) by using eNose.

Table 3.
Metrics obtained from the confusion matrix (SVM), breath samples (PCa vs. control).
3.1.2. PCa vs. BPH, Prostatitis, and Healthy Patients
In the following analysis, different subgroups (BPH, Prostatitis, and Healthy) of the control category were created, where an 80% variance in the discrimination of the samples was reached through the PCA analysis with two PCs (see Figure 8a). Likewise, the selectivity in the discrimination of each category is observed, as some overlaps were generated due to cases of patients with different conditions that make it difficult to separate the categories. However, it was possible to identify each of the control subgroups and the PCa observations through the projection of the measures using three first main components, and obtaining a better discrimination of data with a variance of 85.4%, as seen in Figure 8b.
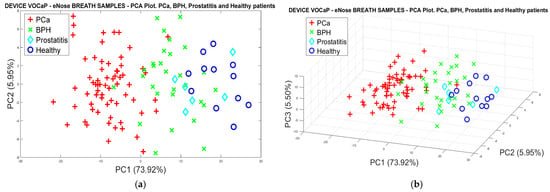
Figure 8.
(a) PCA analysis, breath samples (PCa vs. BPH, Prostatitis, Healthy) by using eNose, (b) PCA analysis using 3-PCs with breath samples for Pca vs. (BPH, Prostatitis, Healthy) discrimination.
Applying the same procedure of the PCA analysis, Figure 9a represents the classification results with the DFA model using two factors, where a better trend in separating the categories is observed despite the dispersion presented in some observations, such as the Prostatitis group.
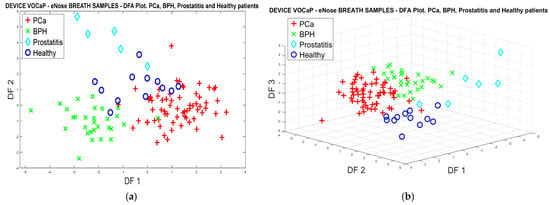
Figure 9.
(a) DFA analysis through breath samples, for PCa vs. (BPH, Prostatitis, Healthy) classification using eNose, (b) DFA analysis using 3-PCs through breath samples for Pca vs. (BPH, Prostatitis, Healthy) classification.
On the other hand, a better projection of the Pca categories and control subgroups was achieved through three PCs, which are shown in Figure 9b.
It is important to note that PCA analysis provides a progressive result in relation to the patient cases with Pca (horizontal axis at point 0, to the left), up to the cases of healthy patients (without prostate involvement) (horizontal axis at point 0 to the right), and, among them, the BPH and Prostatitis classes, obtaining a 70% success rate of classification.
Table 4 lists the classification results through the machine learning methods using PCA scores and DFA factors, where the DFA classification method reached 95.6% success rate in classifying the categories and subsequently using the KNN model.

Table 4.
Breath samples classification of Pca vs. (BPH, Prostatitis, Healthy) samples.
Figure 10 shows the results of the two most relevant metrics extracted from the confusion matrix of the DFA method and KNN model, where it is established that the Prostatitis class has 66.7% sensitivity, 100% accuracy, and 100% specificity.
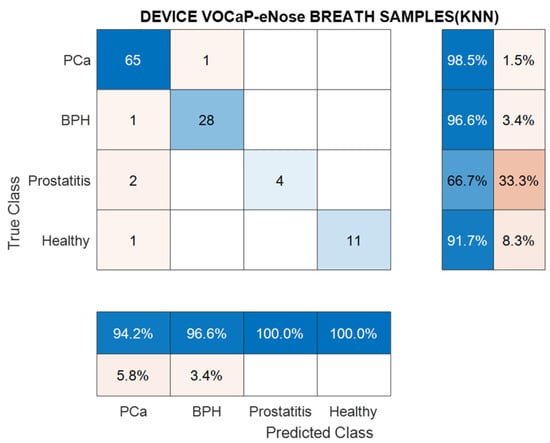
Figure 10.
Confusion matrix generated from KNN classification model through breath samples for Pca vs. BPH, Prostatitis, and Healthy categories by using eNose.
Regarding the results of Prostatitis, it should be clarified that the low representative number of samples of this category was a predominant factor in the low classification rate of the samples.
By means of the DFA method, it was possible to achieve a high classification of the measurements through the KNN model, where an average accuracy of 95.5% was achieved, in each of the categories. The metrics for breath samples in each category are listed in Table 5.

Table 5.
Metrics obtained from DFA-KNN through breath samples for Pca vs. (BPH, Prostatitis, Healthy).
3.1.3. Analysis of Urine Samples with the eNose Device
Regarding the protocol established for the measurement of urine samples previously mentioned, the different analyses were carried out from the intercalated sampling of the Pca and control samples.
PCa vs. Control
In this analysis, the OSC and PCA methods were applied, as depicted in Figure 11, where in the Pca and control groups, 98.7% of data variation was reached by PC1 and PC2. In addition, it is illustrated that in the data distribution, an overlap was generated in the two categories, where 5.3% was achieved, since the projection generated an overlap greater than 50% in variation, where the discriminant trend in the categories could be observed, in the same way as that achieved with the breath samples. In this analysis, better discrimination was achieved by the PCA, since in the classification with DFA, there were several Pca measures and misclassified controls.
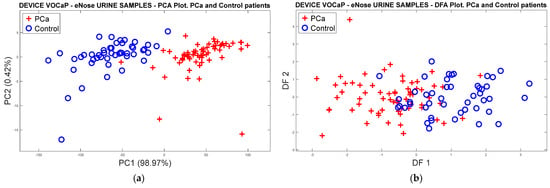
Figure 11.
Plot analysis of PCa and control categories of urine samples by using eNose. (a) PCA analysis of urine samples through Pca vs. control patients, (b) DFA analysis of urine samples, Pca vs. control patients.
The classification based on machine learning applied to raw data obtained an average of 55% classification and with the averaged normalized data, a 54.7% accuracy was accomplished. On the other hand, with the average of PCA scores calculated for each model, a classification accuracy of 92.2% was achieved. Likewise, an 80.1% success rate was reached based on the DFA factors. It should be further noted that the cross-validation method with k-fold = 5 was applied to each learning model. Therefore, the Pca and control urine samples were preconditioned to be used with the classification methods (see Table 6).

Table 6.
Machine learning applied to urine samples using eNose, Pca vs. control classification.
Figure 12 represents the result of the confusion matrix through the SVM model and the DFA factors, where a high accuracy was accomplished, and the metrics were obtained from the confusion matrix.
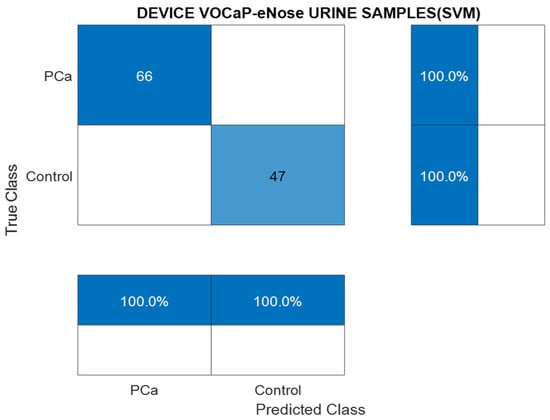
Figure 12.
Confusion matrix generated through SVM model with urine samples, Pca vs. control.
Likewise, with the data from the urine samples, a good performance of the binary classification model was achieved.
The metrics calculated with the PCA analysis and SVM model for urine samples in each category are listed in Table 7.

Table 7.
Confusion matrix obtained of urine samples through SVM, Pca vs. control using eNose.
Pca vs. (BPH, Prostatitis, Healthy)
In the following analysis, urine data were labeled in Pca and subcategories (BPH, Prostatitis, and Healthy) of the control category. Therefore, dimensionality reduction was performed through the PCA model, where Figure 13a illustrates 98% of the data variation through the first two PCs. The location and dispersion of each category in the plot can also be determined in a similar way to that generated with the PCA model of the exhaled breath samples, maintaining the separation of the Pca samples and the subgroups of the control samples. Like breath samples, a separation trend and projection of categories is indicated in the 3D plot (see Figure 13b).
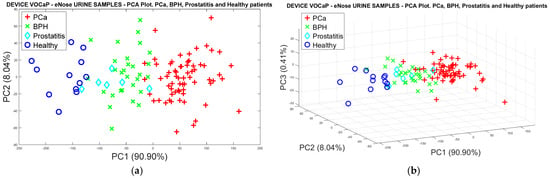
Figure 13.
(a) PCA analysis with urine samples through the eNose device for the discrimination of PCa vs. BPH, Prostatitis, Healthy categories, (b) Discrimination of the samples based on the selection of three main components.
As performed with the PCA analysis, the data were analyzed through the DFA classifier for the four categories of urine samples. Figure 14a shows the projection and, again, the trend in discrimination of the Pca, BPH, and Healthy categories, where the Prostatis category has an uneven behavior since the samples have a slight dispersion that allows them to be identified more clearly. However, there is an increase in poorly classified samples from the other Healthy categories for Pca and BPH, where it is also possible to compare the results obtained with breath samples. Consequently, the 3D plot of the DFA method was developed (Figure 14b) where the separation of healthy patients is better illustrated, resulting in a better projection of the measurements with three PCs.
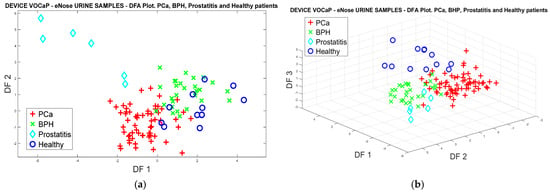
Figure 14.
(a) DFA analysis of urine samples, for PCa vs. (BPH, Prostatitis, Healthy) classification by using eNose, (b) DFA analysis using 3-PCs of urine samples for Pca vs. (BPH, Prostatitis, Healthy) classification.
In general, the results with the classification models with the raw data and the normalized data reached 53.8% and 70.35% accuracy, using the k-fold = 5, with the first two PCs of PCA. Likewise, for the factors based on the DFA analysis, an average of 88.2% accuracy was obtained with the DFA model applying the Decision Trees and Random Forest classification methods with the highest accuracy, reaching a 91.2% success rate for the urine headspace samples (see Table 8).

Table 8.
Machine learning applied to urine samples using eNose for Pca vs. (BPH, Prostatitis, Healthy) classification.
Regarding the confusion matrix developed from the DFA classification technique with the Decision Trees model, a smaller number of samples was classified due to the overlapping with the Pca category, which corresponds to 10% of the data with respect to the control category (see Figure 15). Therefore, the BPH category had a higher number of errors with respect to Pca and the other categories.
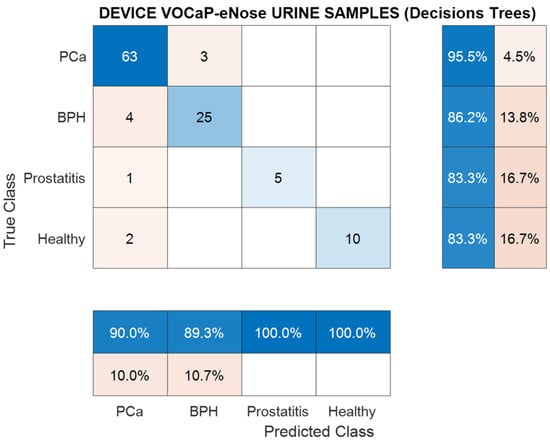
Figure 15.
Confusion matrix, classification using Decision Trees, urine samples for Pca vs. (BPH, Prostatitis, Healthy) classification.
From the metrics calculated through the confusion matrix (see Table 9), it can be concluded that the Random Forest model performed well in the dataset by having an accuracy rate of 91.2% and a sensitivity rate of 88.3%.

Table 9.
Metrics from confusion matrix of urine samples for Pca vs. (BPH, Prostatitis, Healthy) categories classification using DFA and Random Forest model.
3.1.4. Data Fusion Analysis of Breath and Urine Samples Acquired with the eNose Device
Once the models with exhaled breath samples and urine headspace acquired with the eNose system were analyzed, the two datasets were combined, in order to perform a more complete analysis of their performance, using the information extracted from each type of sample; for this, the fusion was initiated from the scores extracted with PCA and the DFA factors with greater accuracy, in order to be merged and introduced as input parameters for the new analysis through machine learning methods.
PCa vs. Control
The PCA scores and the DFA factors of each category corresponding to Pca and controls were merged, and the PCA fusion projections were made. Figure 16 and Figure 17 depict the two categories of data projected, where a variance of 96.74% was obtained through two first PCs, where a slight increase in the classification of the two categories through DFA is also observed, decreasing the number of misclassified samples to 5.26% with DFA, and 2.25% with PCA, respectively, of the variation in the data. In both cases, a good selectivity of the categories was obtained, with PCA resulting in the best selectivity achieved. Therefore, it is concluded that the discrimination results of the measurements improved successfully, including information from urine and breath samples.
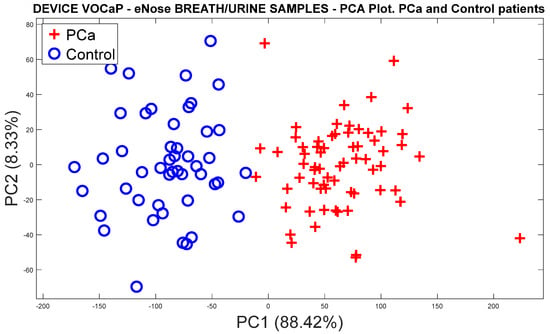
Figure 16.
PCA analysis, breath, and urine data merged for Pca vs. control discrimination by using eNose.
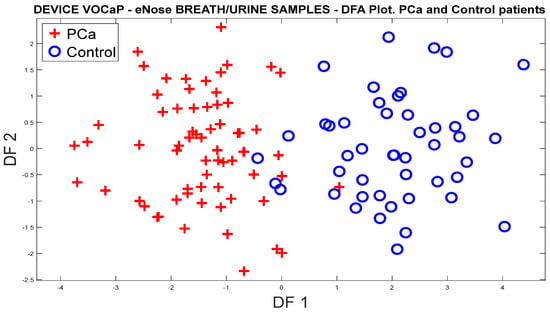
Figure 17.
DFA analysis, breath, and urine data merged for DFA vs. control classification by using eNose.
With the classification methods, an average of 94.5% of success was achieved, with k-fold = 5 for the two PCA and DFA models, from which the Decision Trees and KNN classification methods were selected and applied, with which the best successes were achieved in the classification of Pca and control measures (see Table 10).

Table 10.
Machine learning applied to breath and urine samples merged, Pca vs. control.
According to the confusion matrices of Figure 18a, it is observed how the samples were appropriately classified through the Decision Tree’s classification model applied with PCA; on the other hand, the Naïve Bayes model with the DFA incorrectly classified 8.5% of the control category concerning the Pca category. In the end, a sample was classified as incorrect in the Pca category, which represented a 1.5% accuracy, as illustrated in Figure 18b.
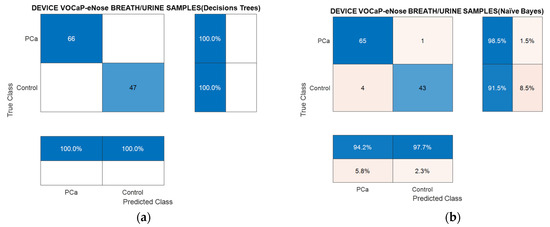
Figure 18.
Confusion matrix using Decision Trees (a) and Naïve Bayes (b) through breath and urine data merged for PCa vs. controls classification using eNose.
The calculated metrics from the confusion matrix in Table 11 denote the classification models for PCA and DFA resulting from the fusion of the breath and urine data, where it is confirmed that, by combining the information of each type of biological sample, the results improved successfully, because in the individual analyses, a similar trend was observed in the representation of the data of each category. Therefore, with more information in the dataset, it is possible to increase the probability that it will be achieved in detecting the Pca and control. In this case, the PCA analysis provided better results in relation to DFA, which confirms the projection of the measurements in Figure 15.

Table 11.
Metrics of confusion matrix obtained from PCA–Decision Trees and DFA–Naïve Bayes of breath and urine data merged for Pca vs. controls classification using eNose.
PCa vs. BPH, Prostatitis, and Healthy Patients
Through the PCA scores calculated from the original data of the breath samples, and the data of the urine samples, the data fusion was performed, obtaining an approximate variance of 98% employing the first three PCs. Therefore, by comparing the responses of Figure 8 and Figure 13, it is possible to observe a better separation of the categories from the data fusion of the two types of samples (See Figure 19a,b). This indicates the difference that exists in the samples’ discrimination with Pca and healthy patients, which determines the high selectivity of the eNose system in the detection of the disease.
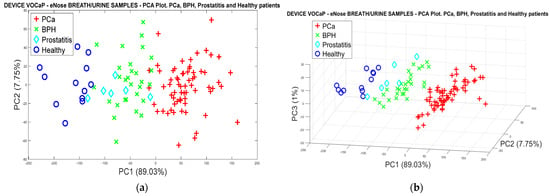
Figure 19.
(a) PCA analysis, breath and urine data merged for PCa vs. (BPH, Prostatitis, Healthy), (b) PCA analysis, breath and urine data merged for Pca vs. (BPH, Prostatitis, Healthy).
By means of the factors extracted with DFA, in Figure 20a,b, the results of the projection of each of the breath and urine samples are depicted, where the data fusion was performed, achieving a better classification of the categories compared with the results projected in Figure 8 and Figure 13. These results confirm the relationship that exists in the two types of biological samples, since by merging the information of the characteristics of the data of both breath and urine, it is possible to improve the response of the eNose in the Pca detection and other related diseases.
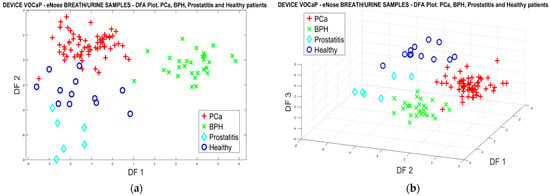
Figure 20.
(a) DFA analysis, breath and urine data merged for PCa vs. (BPH, Prostatitis, Healthy) classification, (b) DFA analysis 3D, breath and urine data merged for Pca vs. (BPH, Prostatitis, Healthy).
With the classification models evaluated with k-fold = 5, the success rate in the classification was increased, as can be seen in Table 12, where the average accuracy achieved for the PCA analysis was 84.4% and for DFA, it achieved a 96.6% success rate.

Table 12.
Metrics of confusion matrix, breath, and urine data merged, Pca vs. (BPH, Prostatitis, Healthy), using eNose.
Therefore, it is established that the data fusion applied to two biological samples of both breath and urine through the eNose provides more characteristics of the data, which allow us to find greater similarities between samples of the same category, thus improving the prediction of them.
The results are then represented through two confusion matrices with the highest-performing ranking models. Figure 21 illustrates that the SVM model presented difficulty in predicting the Prostatitis category, misclassifying 50% of the samples in this category, having fewer representative samples. Unlike SVM, the KNN method obtained only one BPH misclassified sample.
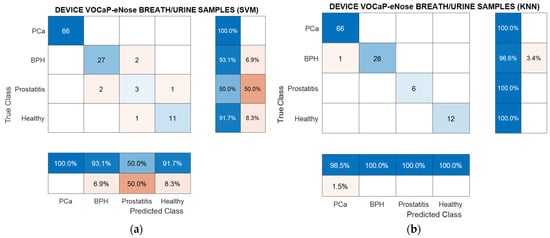
Figure 21.
(a) Confusion matrix, breath and urine samples by using eNose through PCA– SVM, PCa vs. (BPH, Prostatitis, Healthy) classification, (b) Confusion matrix obtained from DFA-KNN model.
As shown in Table 13, the values of the metrics were similar with success rates of 95%, with the exception of the Prostatitis category.

Table 13.
Metrics of confusion matrix obtained from PCA–SVM, breath and urine samples merged, PCa vs. (BPH, Prostatitis, Healthy) classification.
As mentioned, DFA and KNN reached an outstanding result as most of the metrics gained above 95% success rates of classification (see Table 14).

Table 14.
Metrics of confusion matrix obtained from DFA–KNN, breath and urine samples merged, PCa vs. (BPH, Prostatitis, Healthy).
3.2. Analysis of Urine Samples through eTongue
Next, the results of the eTongue based on the C110 electrode response are described below through pattern recognition methods and classification algorithms. Initially, the analysis was performed with the identified prostate cancer categories and control. Then, another analysis was performed, where three types of control patients (i.e., BPH, Prostatitis, and Healthy), and the PCa category were formed. It should be clarified that the groupings were made based on the results achieved with the eNose.
Figure 22 shows the variations in the amplitudes and shapes of the voltammogram signals acquired by the eTongue from patients with PCa and one of the control groups (Healthy), which reveal that there are differences in the Oxidation–Reduction (REDOX) reaction for the urine samples. For example, minerals such as Zinc in the urine could be related and contribute to the discrimination of the data since some studies have proposed it as a possible compound or biomarker of PCa. In addition, the healthy prostate contains high concentrations of Zinc and these levels drastically decreased during the development of PCa, including when it is in the early stage [71,72,73,74].
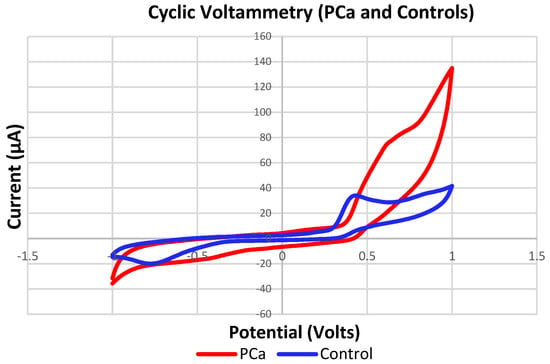
Figure 22.
Signals of measurements acquired with the eTongue based on the C110 electrode in detecting PCa samples (Red) and controls (Blue).
3.2.1. PCa vs. Controls
Figure 23 illustrates the PCA plot performed with the data previously acquired by the potentiostat, where the categories of PCa and controls were discriminated, although with some overlaps between some measures of the two categories. In addition, in the graph, the measurements are projected with almost 100% of the variance captured, where the totality occurs in PC1.
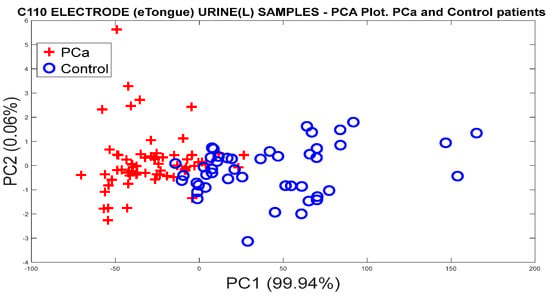
Figure 23.
PCA analysis for the discrimination of PCa and controls measurements from the C110 carbon electrode responses.
It is important to note that two main characteristics were extracted once the signals were acquired, taking the current signal (µA) from the measurements. Therefore, the maximum, minimum, initial, and final values of each of the signals were extracted, similar to what was achieved with the eNose, where the variations of the signals were subsequently calculated to obtain the static parameters. The method calculates the difference between the maximum signal value and the baseline, and the difference between the final and the initial value. On the other hand, the data were initially normalized by means of “mean-centering”, and subsequently, the processing methods were applied to obtain the relevant information of the data from the main components or factors of PCA and DFA.
Figure 24 represents the discrimination of the categories of PCa and control, where a better separation of the clusters with respect to the PCA response is noted, and overlaps are seen between some samples.
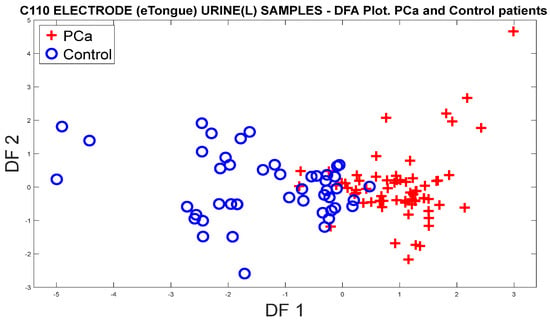
Figure 24.
DFA plot for the discrimination of PCa measurements and controls from the responses of the C110 electrode.
3.2.2. Analysis of the PCa Categories and Split of the Group of Control Patients into BPH, Prostatitis, and Healthy
To discriminate and classify the control groups into different categories, the measures of patients with BPH, Prostatitis, and Healthy were grouped to see the eTongue’s ability in detecting PCa and the other control groups. Figure 25 demonstrates the discrimination response from the PCA plot, where the same normalization was previously applied.
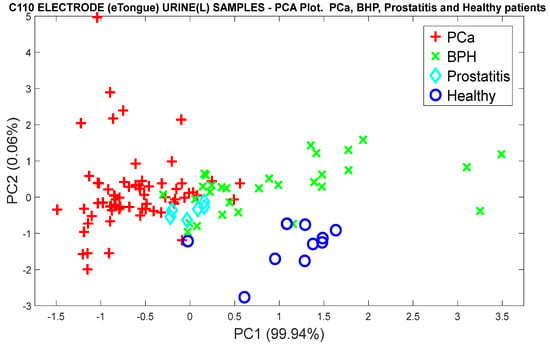
Figure 25.
PCA analysis for the discrimination of PCa measurements and the control groups (BPH, Prostatitis, and Healthy), using the responses of the C110 electrode.
Regarding the classification of the three categories of controls, the DFA algorithm was used, where the high selectivity of the control groups is clearly observed. It should be noted in Figure 26 that there are similar volatile organic compounds in some BPH and Prostatitis samples with the PCa samples, since in the projection of the DFA factors at the point of origin, most of the overlaps of the unclassified measurements appear. However, healthy patients differ from patients with PCa.
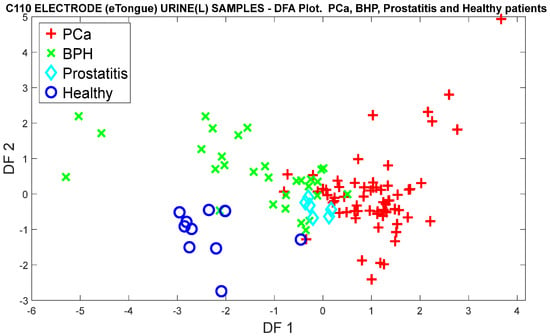
Figure 26.
DFA analysis for the classification of PCa measurements and control categories (BPH, Prostatitis, and Healthy), based on the responses of the C110 electrode.
3.2.3. Data Processing with Classification Methods
This section shows the results of the eTongue with the different classification methods, where the analyses were carried out from the original, normalized data, using the PCA scores and the DFA factors. Table 15 lists the percentages of accuracy calculated with each of the methods for the classification of PCa and control categories.

Table 15.
Metrics of confusion matrix urine samples, PCa vs. controls classification using eTongue.
Through multivariate analysis methods with machine learning methods, the classification of PCa vs. BPH, Prostatitis, and Healthy categories are represented in Table 16.

Table 16.
Metrics of confusion matrix obtained from urine samples. PCa vs. (BPH, Prostatitis, Healthy) categories using eTongue.
Additionally, the metrics extracted from the confusion matrix are related through the KNN result, when applying the PCA scores (see Table 17). It can be concluded that the accuracy value reached a 92.9% success rate, which indicates the good performance of the eTongue in the classification of cancer patients.

Table 17.
Metrics (KNN) of confusion matrix obtained from urine samples. PCa vs. control categories.
4. Discussion
In this study, the potential of E-senses for the detection of PCa and controls was evaluated. According to the results, these systems stood out for obtaining high precision, sensitivity, and specificity when it comes to discriminating and classifying patients with PCa from the different controls, which represents a promising and supportive tool for the early, non-invasive, rapid, and portable diagnosis of PCa and thus be able to address the problems generated by current diagnostic methods, which lead to overdiagnosis, overtreatment, and complications associated with invasive procedures, among others.
On the other hand, in this study, some endogenous and exogenous factors (temperature, humidity, flow, pressure, sample volume, time of measurements, fasting by patients, rest period before taking the sample, and ambient air, among others) were considered when collecting and analyzing biological samples (breath and urine), in order to standardize and minimize the influence of these confounding factors that could influence the results of the E-senses. However, in the present study, in the PCA and DFA figures, in the case of eNose and eTongue, the measures that were not correctly discriminated or classified may be due to other factors such as shared comorbidities, since some patients diagnosed with PCa also had BPH or Prostatitis, which made it difficult for the data to be properly classified in each of the groups. Likewise, some measures of the controls overlapped in the group of patients with PCa and, as reported in the medical history, these patients presented symptoms, elevated PSA values, and abnormal rectal examination, which, according to the literature, would make them candidates for having PCa; however, after a biopsy, it was shown that they had prostate-related diseases.
Additionally, and as evidenced in Table 1, patients diagnosed with PCa and controls shared other diseases; the most relevant and those repeated in the majority of volunteers were diabetes and hypertension, which could also affect the composition of VOCs and non-volatile chemical compounds. However, to substantiate the aforementioned, a more in-depth study would have to be performed to see the influence of other comorbidities. Some studies have shown that men with diabetes have a low risk of being diagnosed with PCa, but only several years after the diagnosis of diabetes [75,76,77,78].
In addition, other factors such as age, race, diet, and medications may also influence the analysis of VOCs and chemical compounds. In the present study, patients with PCa and controls with BPH and Prostatitis shared some drugs in their treatment, among the most prominent of which was Tamsulosin, which belongs to the class of drugs called alpha blockers. For example, Tamsulosin is used to treat and reduce symptoms of an enlarged prostate (BPH), including difficulty and pain with urination, and is commonly used to treat advanced prostate cancer. So far, some studies reported in the literature focused on detecting PCa using alternative techniques such as nose or electronic tongue and biofluids such as urine or breath.
It should be noted that this proposal is the first study where both sensory systems have been evaluated to analyze urine and breath samples to detect Prostate Cancer [54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69]. In this study, a dataset was generated from E-senses devices to create ML models using 113 measurements; however, despite this limited number of cases, this representative dataset can provide us with an excellent pathway to generate models to detect possible PCa cases in the future.
Thus, this limitation can be addressed by making a campaign arranged by the local health sector (i.e., Uronorte S.A, hospitals, clinical centers, etc.), which allows us to conduct further studies and acquire many measurements with the devices to enhance the reliability and feasibility of these alternative and innovative methods for PCa detection in Colombian patients.
5. Conclusions
In this study, two E-senses systems were implemented, composed of an electronic nose and electronic tongue for the detection of previously acquired samples from patients with prostate cancer and other related diseases such as Prostatitis, BPH, and Healthy patients.
The results of the classification of breath and urine samples independently through the eNose-VOCaP in the PCa and control samples reached the maximum success rate (100%), where the Prostatitis, BPH, and Healthy samples achieved 95.5% (DFA-KNN) and 91.2% (DFA-Random Forest) in the classification of the breath and urine samples, respectively.
The eNose system obtained a good performance in the classification of the samples of both breath and urine, where the data were merged to improve both the discrimination and classification of the PCa measures and controls, reaching success rates in the classification of measures of 100% through the PCA–SVM model, improving the separation and projection of the samples based on the PCA, DFA, and machine learning methods graphs.
In addition, in the data fusion of the eNose with breath and urine samples, by dividing the control samples into sub-categories such as Prostatitis, BPH, and Healthy, it was possible to obtain a 95.5% accuracy, maintaining the same eNose result with the analysis of exhaled breath samples through the DFA–KNN model.
On the other hand, the eTongue results for the classification of PCa samples and controls obtained a 92.9% accuracy, which was achieved with the DFA–KNN model, confirming the performance of the model in the classification of the two types of biological samples in the detection of prostate cancer. Likewise, the classification of the samples of the PCa vs. (Prostatitis, BPH, and Healthy) categories obtained an 89.4% success rate of classification in urine measurements.
Both digital and analog gas sensors gave good sensitivity and selectivity in the detection of VOCs since the measurements were discriminated and classified despite having variations in the samples.
The “C110” carbon electrode performed well in patients confirmed with prostate cancer and controls (Healthy), which was observed in multivariate analyses with PCA and DFA. Although the results were good with the use of the C110 electrode, it is important to make tests using other screen-printed electrodes with different materials, such as gold, silver, platinum, and others, to obtain more information about the targets or molecules of the measurements, for the detection of PCa.
As mentioned above, the eNose–VOCaP performed very well, which could be a low-cost and reliable tool to be used in the health sector. However, to become a standard tool, it is necessary to continue evaluating the classification capacity of the eNose system through a study where a high number of samples are acquired as a social campaign, in order to plan the early diagnosis of PCa.
In future work, to validate the results, the capacity, performance, and reliability of the E-senses systems in the detection of CaP, it will be necessary to use classic analytical techniques such as GC-MS and LC-MS, which can identify and quantify several individual compounds in a sample that are statistically significant between PCa and control patients. The above can be used as potential biomarkers, which can help the health sector to pursue solutions in the prevention and diagnosis of PCa. The amount and type of biomarkers identified in a given study depend on the analytical technique chosen and the biological matrix (urine and breath).
Therefore, urine and breath samples contain many VOCs, which can be released as metabolic by-products or due to physiological processes within the body. These compounds can potentially serve as biomarkers for different diseases or health conditions. However, there are some variations in the concentration types of volatile compounds found in urine and breath, and among the factors that can influence are the following: (1) metabolism, since there are different metabolic pathways in the body that can lead to the production of different volatile compounds, and (2) excretions due to urine being a waste material that contains metabolic decomposition products of a wide range of foods, beverages, medications, environmental pollutants, endogenous waste metabolites, and bacteria.
On the other hand, the breath generates volatile compounds to be transported from the bloodstream to the lungs (by a diffusion mechanism through the pulmonary alveolar membrane) for exhalation. Another factor is (3) sampling techniques and technologies, because they can influence the identification and quantification of volatile compounds. For example, some volatile compounds associated with the PCa and control groups have been reported in the literature. For example, in urine compounds such as formaldehyde, 2,6-dimethyl-7-octen-2-ol, 3-octanone, 2-octanone, pentanal, furan, xylene, hexanal, 2,5-dimethylbenzaldehyde, 4-methylhexan-3-one, dihydroedulan IA, methylglyoxal, 3-phenylpropionaldehyde, and furan-3-methanol, among others, have been reported in the research [26,30,79,80,81]. In addition, there are a few studies of VOCs in patients with PCa and controls that use breath, where some compounds used are Toluene, 2-amino-5-isopropyl-8-methyl-1-azulenecarbonitrile, p-xylene, and 2,2-dimethyldecane [65].
Author Contributions
Conceptualization, C.M.D.A. and J.K.C.G.; methodology, J.K.C.G., C.M.D.A. and C.A.C.V.; validation, C.A.C.V. and J.K.C.G.; investigation, C.M.D.A., J.K.C.G. and J.R.; writing—original draft preparation, C.M.D.A., J.K.C.G., C.A.C.V. and J.R.; writing—review and editing, C.M.D.A., J.K.C.G. and J.R.; visualization, C.A.C.V. and C.M.D.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Minciencias grant number 112184468047 and contract number 929-2019.
Institutional Review Board Statement
URONORTE S.A conducted the ethical review and approval for studies involving humans.
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data may be requested from the authors by mail.
Acknowledgments
The authors also acknowledge Uronorte Clinic (Cucuta-Colombia) for its cooperation and support during the collecting and acquiring of the patient’s samples.
Conflicts of Interest
The authors have no conflict of interests to declare.
References
- Sung, H.; Ferlay, J.; Siegel, R.; Soerjomataram, I.; Torre, L.; Jemal, A. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Manisha, A.; Stephen, W.; Amit, S. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63. [Google Scholar]
- Jain, M.; Leslie, S.; Sapra, A. Prostate Cancer Screening. StatPearls 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK556081/ (accessed on 7 September 2023).
- Kachuri, L.; Hoffmann, T.; Jiang, Y.; Berndt, S.I.; Shelley, J.P.; Schaffer, K.R.; Machiela, M.J.; Freedman, N.D.; Huang, W.-Y.; Li, S.A.; et al. Genetically adjusted PSA levels for prostate cancer screening. Nat. Med. 2023, 29, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- Tourinho, R.R.; Lima, A.C.; Glina, S. Prostate cancer in Brazil and Latin America: Epidemiology and screening. Int. Braz. J. Urol. 2016, 42, 1081. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.D. Reference range of serum prostate-specific antigen levels in Indian men. Indian J. Med. Res. 2014, 140, 480. [Google Scholar] [PubMed]
- Sarwar, S.; Adil, M.A.; Nyamath, P.; Ishaq, M. Biomarkers of Prostatic Cancer: An Attempt to Categorize Patients into Prostatic Carcinoma, Benign Prostatic Hyperplasia, or Prostatitis Based on Serum Prostate Specific Antigen, Prostatic Acid Phosphatase, Calcium, and Phosphorus. Prostate Cancer 2017, 2017, 5687212. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Zhu, L.; Liu, X.; Ma, S.; Liu, Y.; Wang, X. Differential diagnosis of uncommon prostate diseases: Combining mpMRI and clinical information. Insights Imaging 2021, 12, 79. [Google Scholar] [CrossRef]
- Lumbreras, B.; Parker, L.A.; Caballero, J.P.; Gómez, L.; Puig, M.; López, M.; García, N.; Hernández, I. Variables Associated with False-Positive PSA Results: A Cohort Study with Real-World Data. Cancers 2022, 15, 261. [Google Scholar] [CrossRef]
- Sandhu, G.C.; Andriole, G.L. Overdiagnosis of Prostate Cancer. J. Natl. Cancer Inst. Monogr. 2012, 45, 146. [Google Scholar] [CrossRef]
- Palsdottir, T.; Nordstrom, T.; Karlsson, A.; Grönberg, H.; Clements, M.; Eklund, M. The impact of different prostate-specific antigen (PSA) testing intervals on Gleason score at diagnosis and the risk of experiencing false-positive biopsy recommendations: A population-based cohort study. BMJ Open 2019, 9, e027958. [Google Scholar] [CrossRef]
- Kim, H.; Park, S.; Jeong, I.G.; Song, S.H.; Jeong, Y.; Kim, C.S.; Lee, K.H. Noninvasive Precision Screening of Prostate Cancer by Urinary Multimarker Sensor and Artificial Intelligence Analysis. ACS Nano 2021, 15, 4054–4065. [Google Scholar] [CrossRef] [PubMed]
- Okpua, N.C.; Okekpa, S.I.; Njaka, S.; Emeh, A.N. Clinical diagnosis of prostate cancer using digital rectal examination and prostate-specific antigen tests: A systematic review and meta-analysis of sensitivity and specificity. Afr. J. Urol. 2021, 27, 32. [Google Scholar] [CrossRef]
- Gosselaar, C.; Kranse, R.; Roobol, M.J.; Roemeling, S.; Schröder, F.H. The interobserver variability of digital rectal examination in a large randomized trial for the screening of prostate cancer. Prostate 2008, 68, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Irekpita, E.; Achor, G.O.; Alili, U. Assessment of the value of the different variants of abnormal digital rectal examination finding in predicting carcinoma of the prostate: A preliminary report of a two-center study. Afr. J. Urol. 2020, 26, 3. [Google Scholar] [CrossRef]
- Ilic, D.; Neuberger, M.M.; Djulbegovic, M.; Dahm, P. Screening for prostate cancer. Cochrane Database Syst. Rev. 2013, 2013, CD004720. [Google Scholar] [CrossRef] [PubMed]
- Gravestock, P.; Shaw, M.; Veeratterapillay, R.; Heer, R. Prostate Cancer Diagnosis: Biopsy Approaches. Urol. Cancers 2022, 12, 141–168. [Google Scholar]
- Shariat, S.F.; Roehrborn, C.G. Using Biopsy to Detect Prostate Cancer. Rev. Urol. 2008, 10, 262. [Google Scholar]
- Moe, A.; Hayne, D. Transrectal ultrasound biopsy of the prostate: Does it still have a role in prostate cancer diagnosis. Transl. Androl. Urol. 2020, 9, 3018. [Google Scholar] [CrossRef]
- López, E.; Ohori, M.; Wheeler, T.M.; Reuter, V.E.; Scardino, P.T.; Kattan, M.W.; Eastham, J.A. Prostate Cancer Diagnosed After Repeat Biopsies Have a Favorable Pathological Outcome but Similar Recurrence Rate. J. Urol. 2006, 175, 923–928. [Google Scholar] [CrossRef][Green Version]
- Coyle, C.; Morgan, E.; Drummond, F.J.; Sharp, L.; Gavin, A. Do men regret prostate biopsy: Results from the PiCTure study. BMC Urol. 2017, 17, 11. [Google Scholar] [CrossRef]
- Loeb, S.; Vellekoop, A.; Ahmed, H.U.; Catto, J.; Emberton, M.; Nam, R.; Rosario, D.J.; Scattoni, V.; Lotan, Y. Systematic review of complications of prostate biopsy. Eur. Urol. 2013, 64, 876–892. [Google Scholar] [CrossRef] [PubMed]
- Detchokul, S.; Frauman, A.G. Recent developments in prostate cancer biomarker research: Therapeutic implications. Br. J. Clin. Pharmacol. 2011, 71, 157–174. [Google Scholar] [CrossRef] [PubMed]
- Manceau, C.; Fromont, G.; Beauval, J.B.; Barret, E.; Brureau, L.; Créhange, G.; Dariane, C.; Fiard, G.; Gauthé, M.; Mathieu, R.; et al. Biomarker in active surveillance for prostate cancer: A systematic review. Cancers 2021, 13, 4251. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Ni, J.; Beretov, J.; Cozzi, P.; Willcox, M.; Wasinger, V.; Walsh, B.; Graham, P.; Li, Y. Urinary biomarkers in prostate cancer detection and monitoring progression. Crit. Rev. Oncol. Hematol. 2017, 118, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.R.; Pinto, J.; Carvalho, C.; Jerónimo, C.; Henrique, R.; Bastos, M.L.; Carvalho, M.; Guedes de Pinho, P. A Panel of Urinary Volatile Biomarkers for Differential Diagnosis of Prostate Cancer from Other Urological Cancers. Cancers 2020, 12, 2017. [Google Scholar] [CrossRef] [PubMed]
- De Visschere, P.J.; Briganti, A.; Fütterer, J.J.; Ghadjar, P.; Isbarn, H.; Massard, C.; Ost, P.; Sooriakumaran, P.; Surcel, C.I.; Valerio, M.; et al. Role of multiparametric magnetic resonance imaging in early detection of prostate cancer. Insights Imaging 2016, 7, 205–214. [Google Scholar] [CrossRef]
- Nematollahi, H.; Moslehi, M.; Aminolroayaei, F.; Maleki, M.; Shahbazi, D. Diagnostic Performance Evaluation of Multiparametric Magnetic Resonance Imaging in the Detection of Prostate Cancer with Supervised Machine Learning Methods. Diagnostics 2023, 13, 806. [Google Scholar] [CrossRef]
- Hua, X.; Wei, W.; Zhi, Y.; Jun, L.; Sheng, L.; Lin, W.; Qing, J.; Lin, J. Metabolomics studies of prostate cancer using gas chromatography-mass spectrometery. Transl. Cancer Res. 2016, 5, 302–314. [Google Scholar]
- Liu, Q.; Fan, Y.; Zeng, S.; Zhao, Y.; Yu, L.; Zhao, L.; Gao, J.; Zhang, X.; Zhang, Y. Volatile organic compounds for early detection of prostate cancer from urine. Heliyon 2023, 9, e16686. [Google Scholar] [CrossRef]
- Lima, A.R.; Araújo, A.M.; Pinto, J.; Jerónimo, C.; Henrique, R.; Bastos, M.L.; Carvalho, M.; Guedes de Pinho, P. GC-MS-Based Endometabolome Analysis Differentiates Prostate Cancer from Normal Prostate Cells. Metabolites 2018, 8, 23. [Google Scholar] [CrossRef]
- McDunn, J.E.; Li, Z.; Adam, K.P.; Neri, B.P.; Wolfert, R.L.; Milburn, M.V.; Lotan, Y.; Wheeler, T.M. Metabolomic signatures of aggressive prostate cancer. Prostate 2013, 73, 1547. [Google Scholar] [CrossRef] [PubMed]
- Pinto, F.G.; Mahmud, I.; Harmon, T.A.; Rubio, V.Y.; Garrett, T.J. Rapid Prostate Cancer Noninvasive Biomarker Screening Using Segmented Flow Mass Spectrometry-Based Untargeted Metabolomics. J. Proteome Res. 2020, 19, 2080–2091. [Google Scholar] [CrossRef] [PubMed]
- Markushin, Y.; Nilesh, G.; HZhang Paul, K.; Rogan, E.; Cavalieri, E.; Trock, B.; Pavlovich, C.; Ryszard, J. Potential biomarker for early risk assessment of prostate cancer. Prostate 2006, 66, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Watson, D.G.; Wang, L.; Abbas, M.; Murdoch, L.; Bashford, L.; Ahmad, I.; Lam, N.Y.; Ng, A.C.; Leung, H.Y. Application of Holistic Liquid Chromatography-High Resolution Mass Spectrometry Based Urinary Metabolomics for Prostate Cancer Detection and Biomarker Discovery. PLoS ONE 2013, 8, e65880. [Google Scholar] [CrossRef] [PubMed]
- Ankerst, D.P.; Liss, M.; Zapata, D.; Hoefler, J.; Thompson, I.M.; Leach, R.J. A case control study of sarcosine as an early prostate cancer detection biomarker Urological oncology. BMC Urol. 2015, 15, 99. [Google Scholar] [CrossRef] [PubMed]
- Wilton, J.H.; Titus, M.A.; Efstathiou, E.; Fetterly, G.J.; Mohler, J.L. Androgenic biomarker profiling in human matrices and cell culture samples using high throughput, electrospray tandem mass spectrometry. Prostate 2014, 74, 722–731. [Google Scholar] [CrossRef]
- Jornet, N.; Moliner, Y.; Molins, C.; Campíns, P. Trends for the Development of In Situ Analysis Devices. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017. [Google Scholar]
- Zaim, O.; Diou, A.; Bari, N.E.; Lagdali, N.; Benelbarhdadi, I.; Ajana, F.Z.; Liobet, E.; Bouchikhi, B. Comparative analysis of volatile organic compounds of breath and urine for distinguishing patients with liver cirrhosis from healthy controls by using electronic nose and voltammetric electronic tongue. Anal. Chim. Acta 2021, 1184, 339028. [Google Scholar] [CrossRef]
- Hidayat, S.; Triyana, K.; Fauzan, I.; Julian, T.; Lelono, D.; Yusuf, Y.; Ngadiman, N.; Veloso, A.C.; Peres, A.M. The Electronic Nose Coupled with Chemometric Tools for Discriminating the Quality of Black Tea Samples In Situ. Chemosensors 2019, 7, 29. [Google Scholar] [CrossRef]
- Dong, W.; Zhao, J.; Hu, R.; Dong, Y.; Tan, L. Differentiation of Chinese robusta coffees according to species, using a combined electronic nose and tongue, with the aid of chemometrics. Food Chem. 2017, 229, 743–751. [Google Scholar] [CrossRef]
- Xu, M.; Wang, J.; Zhu, L. The qualitative and quantitative assessment of tea quality based on E-nose, E-tongue and E-eye combined with chemometrics. Food Chem. 2019, 289, 482–489. [Google Scholar] [CrossRef]
- Fitzgerald, J.; Fenniri, H. Cutting Edge Methods for Non-Invasive Disease Diagnosis Using E-Tongue and E-Nose Devices. Biosensors 2017, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Christodoulides, N.; McRae, M.P.; Simmons, G.W.; Modak, S.S.; McDevitt, J.T. Sensors that Learn: The Evolution from Taste Fingerprints to Patterns of Early Disease Detection. Micromachines 2019, 10, 251. [Google Scholar] [CrossRef] [PubMed]
- Farraia, M.V.; Rufo, J.C.; Paciência, I.; Mendes, F.; Delgado, L.; Moreira, A. The electronic nose technology in clinical diagnosis: A systematic review. Porto Biomed. J. 2019, 4, e42. [Google Scholar] [CrossRef] [PubMed]
- Tonacci, A.; Scafile, A.; Billeci, L.; Sansone, F. Electronic Nose and Tongue for Assessing Human Microbiota. Chemosensors 2022, 10, 85. [Google Scholar] [CrossRef]
- Bax, C.; Taverna, G.; Eusebio, L.; Sironi, S.; Grizzi, F.; Guazzoni, G.; Capelli, L. Innovative Diagnostic Methods for Early Prostate Cancer Detection through Urine Analysis: A Review. Cancers 2018, 10, 123. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, D.; Kim, W.J.; Kim, G. Detection of volatile organic compounds from human prostate cancer cell using canine olfaction. J. Vet. Behav. 2022, 49, 80–84. [Google Scholar] [CrossRef]
- Janfaza, S.; Khorsand, B.; Nikkhah, M.; Zahiri, J. Digging deeper into volatile organic compounds associated with cancer. Biol. Methods Protoc. 2019, 4, bpz014. [Google Scholar] [CrossRef]
- Taverna, G.; Tidu, L.; Grizzi, F.; Torri, V.; Mandressi, A.; Sardella, P.; La Torre, G.; Cocciolone, G.; Seveso, M.; Giusti, G.; et al. Olfactory system of highly trained dogs detects prostate cancer in urine samples. J. Urol. 2015, 193, 1382–1387. [Google Scholar] [CrossRef]
- Cornu, J.N.; Cancel-Tassin, G.; Ondet, V.; Girardet, C.; Cussenot, O. Olfactory Detection of Prostate Cancer by Dogs Sniffing Urine: A Step Forward in Early Diagnosis. Eur. Urol. 2011, 59, 197–201. [Google Scholar] [CrossRef]
- Park, G.; Kim, D.; Jeong, S.; Park, D.; Kim, G. Odor detection of cancer cell metabolites by scent-detection dogs. Thai J. Vet. Med. 2022, 52, 537–541. [Google Scholar] [CrossRef]
- Ryman-Tubb, T.; Lothion-Roy, J.H.; Metzler, V.M.; Harris, A.E.; Robinson, B.D.; Rizvanov, A.A.; Jeyapalan, J.N.; James, V.H.; England, G.; Rutland, C.S.; et al. Comparative pathology of dog and human prostate cancer. Vet. Med. Sci. 2022, 8, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Guo, L.; Wang, M.; Su, C.; Wang, D.; Dong, H.; Chen, J.; Wu, W. Review on Algorithm Design in Electronic Noses: Challenges, Status, and Trends. Intell. Comput. 2023, 2, 1–19. [Google Scholar] [CrossRef]
- Bernabei, M.; Pennazza, G.; Santonico, M.; Corsi, C.; Roscioni, C.; Paolesse, R.; Di Natale, N.; D’Amico, A. A preliminary study on the possibility to diagnose urinary tract cancers by an electronic nose. Sens. Actuators B Chem. 2008, 131, 1–4. [Google Scholar] [CrossRef]
- Arnaldo, D.; Marco, S.; Giorgio, P.; Rosamaria, C.; Giuseppe, V.; Dario Del, F.; Roberto, P.; Corrado Di, N.; Eugenio, M.; Enrico, F. A Novel Approach for Prostate Cancer Diagnosis using a Gas Sensor Array. Procedia Eng. 2012, 47, 1113–1116. [Google Scholar]
- Roine, A.; Veskimäe, E.; Tuokko, A.; Kumpulainen, P.; Koskimäki, J.; Keinänen, T.A.; Häkkinen, M.R.; Vepsäläinen, J.; Paavonen, T.; Lekkala, J.; et al. Detection of Prostate Cancer by an Electronic Nose: A Proof of Principle Study. J. Urol. 2014, 192, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Filianoti, A.; Costantini, M.; Bove, A.M.; Anceschi, U.; Brassetti, A.; Ferriero, M.; Mastroianni, R.; Misuraca, L.; Tuderti, G.; Ciliberto, G.; et al. Volatilome Analysis in Prostate Cancer by Electronic Nose: A Pilot Monocentric Study. Cancers 2022, 14, 2927. [Google Scholar] [CrossRef]
- Bax, C.; Prudenza, S.; Gaspari, G.; Capelli, L.; Grizzi, F.; Taverna, G. Drift compensation on electronic nose data for non-invasive diagnosis of prostate cancer by urine analysis. iScience 2022, 25, 103622. [Google Scholar] [CrossRef]
- Bax, C.; Capelli, L.; Grizzi, F.; Prudenza, S.; Taverna, G. A novel approach for the non-invasive diagnosis of prostate cancer based on urine odour analysis. In Proceedings of the International Symposium on Olfaction and Electronic Nose, ISOEN 2022—Proceedings 2022, Aveiro, Portugal, 29 May–1 June 2022; pp. 1–4. [Google Scholar]
- Capelli, L.; Bax, C.; Grizzi, F.; Taverna, G. Optimization of training and measurement protocol for eNose analysis of urine headspace aimed at prostate cancer diagnosis. Sci. Rep. 2021, 11, 20898. [Google Scholar] [CrossRef]
- Taverna, G.; Grizzi, F.; Tidu, L.; Bax, C.; Zanoni, M.; Vota, P.; Lotesoriere, B.J.; Prudenza, S.; Magagnin, L.; Langfelder, G.; et al. Accuracy of a new electronic nose for prostate cancer diagnosis in urine samples. Int. J. Urol. 2022, 29, 890–896. [Google Scholar] [CrossRef]
- Scheepers, M.H.M.C.; Al-Difaie, Z.; Brandts, L.; Peeters, A.; van Grinsven, B.; Bouvy, N.D. Diagnostic Performance of Electronic Noses in Cancer Diagnoses Using Exhaled Breath: A Systematic Review and Meta-analysis. JAMA Netw. Open 2022, 5, E2219372. [Google Scholar] [CrossRef]
- Nakhleh, M.K.; Amal, H.; Jeries, R.; Broza, Y.Y.; Aboud, M.; Gharra, A.; Ivgi, H.; Khatib, S.; Badarneh, S.; Har-Shai, L.; et al. Diagnosis and Classification of 17 Diseases from 1404 Subjects via Pattern Analysis of Exhaled Molecules. ACS Nano 2017, 11, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Hakim, M.; Broza, Y.Y.; Billan, S.; Abdah-Bortnyak, R.; Kuten, A.; Tisch, U.; Haick, H. Detection of lung, breast, colorectal, and prostate cancers from exhaled breath using a single array of nanosensors. Br. J. Cancer 2010, 103, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Waltman, C.G.; Marcelissen, T.A.T.; van Roermund, J.G.H. Exhaled-breath Testing for Prostate Cancer Based on Volatile Organic Compound Profiling Using an Electronic Nose Device (AeonoseTM): A Preliminary Report. Eur. Urol. Focus. 2020, 6, 1220–1225. [Google Scholar] [CrossRef] [PubMed]
- Zniber, M.; Vahdatiyekta, P.; Huynh, T.P. Analysis of urine using electronic tongue towards non-invasive cancer diagnosis. Biosens. Bioelectron. 2023, 219, 114810. [Google Scholar] [CrossRef] [PubMed]
- Pascual, L.; Campos, I.; Vivancos, J.L.; Quintás, G.; Loras, A.; Martínez-Bisbal, M.C.; Martínez-Máñez, R.; Boronat, F.; Ruiz-Cerdà, J.L. Detection of prostate cancer using a voltammetric electronic tongue. Analyst 2016, 141, 4562–4567. [Google Scholar] [CrossRef]
- Svetlana, S.; Mikhail, K.; Vitaly, P.; Evgeny, A.; Liudmila, K.; Elena, B.; Andrey, L.; Ping, W.; Hao, W.; Igor, J.; et al. Potentiometric multisensor system as a possible simple tool for non-invasive prostate cancer diagnostics through urine analysis. Sens. Actuators B Chem. 2019, 289, 42–47. [Google Scholar]
- Liu, H.; Zhang, L.; Li, K.H.H.; Tan, O.K. Microhotplates for Metal Oxide Semiconductor Gas Sensor Applications—Towards the CMOS-MEMS Monolithic Approach. Micromachines 2018, 9, 557. [Google Scholar] [CrossRef]
- Maddalone, M.G.; Oderda, M.; Mengozzi, G.; Gesmundo, I.; Novelli, F.; Giovarelli, M.; Gontero, P.; Occhipinti, S. Urinary Zinc Loss Identifies Prostate Cancer Patients. Cancers 2022, 14, 5316. [Google Scholar] [CrossRef]
- Sugimoto, R.; Lee, L.; Tanaka, Y.; Morita, Y.; Hijioka, M.; Hisano, T.; Furukawa, M. Zinc Deficiency as a General Feature of Cancer: A Review of the Literature. Biol. Trace Elem. Res. 2023. [Google Scholar] [CrossRef]
- Eskra, J.N.; Rabizadeh, D.; Pavlovich, C.P.; Catalona, W.J.; Luo, J. Approaches to urinary detection of prostate cancer. Prostate Cancer Prostatic Dis. 2019, 22, 362. [Google Scholar] [CrossRef]
- Medarova, Z.; Ghosh, S.K.; Vangel, M.; Drake, R.; Moore, A. Risk stratification of prostate cancer patients based on EPS-urine zinc content. Am. J. Cancer Res. 2014, 4, 385. [Google Scholar]
- Pierce, B.L. Why are diabetics at reduced risk for prostate cancer? A review of the epidemiologic evidence. Urol. Oncol. Semin. Orig. Investig. 2012, 30, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Kasper, J.S.; Liu, V.; Giovannucci, E. Diabetes Mellitus and Risk of Prostate Cancer in the Health Professionals Follow-Up Study. Int. J. Cancer. J. Int. Du. Cancer 2009, 124, 1398. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, C.; Patel, A.V.; Mondul, A.M.; Jacobs, E.J.; Thun, M.J.; Calle, E.E. Diabetes and Risk of Prostate Cancer in a Prospective Cohort of US Men. Am. J. Epidemiol. 2005, 161, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Calton, B.A.; Chang, S.C.; Wright, M.E.; Kipnis, V.; Lawson, K.; Thompson, F.E.; Subar, A.F.; Mouw, T.; Campbell, D.S.; Hurwitz, P.; et al. History of diabetes mellitus and subsequent prostate cancer risk in the NIH-AARP Diet and Health Study. Cancer Causes Control 2007, 18, 493–503. [Google Scholar] [CrossRef]
- Jiménez-Pacheco, a.; Salinero-Bachiller, M.; Iribar, M.; López-Luque, A.; Miján-Ortiz, J.; Peinado, J. Furan and p-xylene as candidate biomarkers for prostate cancer. Urol. Oncol. 2018, 36, e21–e243. [Google Scholar] [CrossRef]
- Lima, A.; Pinto, J.; Azevedo, A.I.; Barros-Silva, D.; Jeronimo, C.; Henrique, R.; de Lourdes Bastos, M.; de Pinho, P.G.; Carvalho, M. Identification of a biomarker panel for improvement of prostate cancer diagnosis by vola-tile metabolic profiling of urine. Br. J. Cancer 2019, 121, 857. [Google Scholar] [CrossRef]
- Khalid, T.; Aggio, R.; White, P.; De Lacy Costello, B.; Persad, R.; Alkateb, H.; Jones, P.; Probert, C.S.; Ratcliffe, N. Urinary Volatile Organic Compounds for the Detection of Prostate Cancer. PLoS ONE 2015, 10, e0143231. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).