Abstract
Progressive first-failure censoring is a flexible and cost-efficient strategy that captures real-world testing scenarios where only the first failure is observed at each stage while randomly removing remaining units, making it ideal for biomedical and reliability studies. By applying the -power transformation to the exponential baseline, the proposed model introduces an additional flexibility parameter that enriches the family of lifetime distributions, enabling it to better capture varying failure rates and diverse hazard rate behaviors commonly observed in biomedical data, thus extending the classical exponential model. This study develops a novel computational framework for analyzing an -powered exponential model under beta-binomial random removals within the proposed censoring test. To address the inherent complexity of the likelihood function arising from simultaneous random removals and progressive censoring, we derive closed-form expressions for the likelihood, survival, and hazard functions and propose efficient estimation strategies based on both maximum likelihood and Bayesian inference. For the Bayesian approach, gamma and beta priors are adopted, and a tailored Metropolis–Hastings algorithm is implemented to approximate posterior distributions under symmetric and asymmetric loss functions. To evaluate the empirical performance of the proposed estimators, extensive Monte Carlo simulations are conducted, examining bias, mean squared error, and credible interval coverage under varying censoring levels and removal probabilities. Furthermore, the practical utility of the model is illustrated through three oncological datasets, including multiple myeloma, lung cancer, and breast cancer patients, demonstrating superior goodness of fit and predictive reliability compared to traditional models. The results show that the proposed lifespan model, under the beta-binomial probability law and within the examined censoring mechanism, offers a flexible and computationally tractable framework for reliability and biomedical survival analysis, providing new insights into censored data structures with random withdrawals.
Keywords:
α-powered exponential; cancer; reliability; censoring; β-binomial; symmetry bayesian; Markov chain; Monte Carlo test MSC:
62F10; 62F15; 62N01; 62N02; 62N05
1. Introduction
Progressive first-failure censoring (P-FFC), introduced by Wu and Kuş [1], was developed to address practical challenges in life-testing and reliability studies, where recording every failure time can be prohibitively expensive, time-consuming, or logistically impractical. This scheme integrates two well-known censoring mechanisms: the first-failure censoring design of Balasooriya [2] and the Type-II progressive censoring (TIIPC) framework described by Balakrishnan and Cramer [3]. Unlike conventional methods that observe all failure times, P-FFC records only the earliest failure within each group. This greatly reduces data collection effort while retaining sufficient information for reliable inference. Conceptually, it can be viewed as an adaptation of TIIPC to situations where only the minimum failure time from each group is observed. This design is particularly attractive for applied settings—such as biomedical or reliability experiments—where tracking every failure is infeasible, but recording first failures remains manageable. By focusing on early failures, the scheme achieves a balance between experimental efficiency and statistical reliability.
Wu and Kuş [1] illustrated its practicality using the Weibull lifetime model as an example. Under the P-FFC setup, n independent groups are formed, each containing m identical test units, yielding a total of units. At each observed failure time, only the first failure within the active group is recorded. After this event, a specified number of groups, , are randomly removed from the experiment, in addition to the group where the failure occurred. This process continues until a predetermined number of failures, k, is observed. For example, when the first failure occurs, the affected group plus additional groups are removed, leaving groups under observation. Similarly, when the second failure is observed, its corresponding group and additional groups are removed, reducing the number of groups to . This continues until the k-th failure , after which the remaining groups and the group containing the k-th failure are removed. Although this scheme improves experimental efficiency, it assumes that the number of groups removed at each step under the TIIPC design is fixed and known in advance—an assumption that may not hold in real-world experiments. For example, in a clinical trial, after the first patient death, some participants may voluntarily withdraw due to fear or loss of confidence in the treatment. Subsequent deaths may trigger further withdrawals, causing deviations from the planned censoring schedule. While the study may still terminate after observing k failures, the actual number of removals at each stage becomes random and unpredictable. To address this limitation, several extensions of P-FFC have been proposed to incorporate stochastic removal mechanisms. Examples include discrete uniform removals (Huang and Wu [4]), binomial removals (Ashour et al. [5]), and beta-binomial (BB) removals (Elshahhat et al. [6]). These random-removal schemes better capture the inherent uncertainty in withdrawals or unit eliminations encountered in practical testing scenarios. Section 2 provides a detailed explanation of the sampling process under P-FFC with BB-based removals (denoted P-FFC-BB).
The alpha-power transformation technique, proposed by Mahdavi and Kundu [7], offers a systematic and flexible mechanism for constructing new families of probability distributions from a given baseline model. By introducing an additional shape parameter , this transformation enhances skewness, improves tail behavior, and increases modeling flexibility while preserving mathematical tractability. One member of this transformation is the -power exponential (APE) distribution, which generalizes the classical exponential model while maintaining closed-form expressions for both of its characteristic functions. Let Y denote the lifetime of a test unit that follows the two-parameter APE distribution, where . Subsequently, the probability density function (PDF) , cumulative distribution function (CDF) , reliability function (RF) , and hazard rate function (HRF) , evaluated at a mission time , are expressed as follows:
respectively, where (scale) and (shape).
The APE distribution extends the classical exponential model by introducing a shape parameter that captures diverse hazard rate patterns, including increasing, decreasing, and bathtub shapes (see Figure 1a). It enhances skewness and tail flexibility while retaining closed-form density and distribution functions, ensuring mathematical simplicity (see Figure 1b). Consequently, it has gained significant attention in reliability analysis, lifetime modeling, and survival studies due to its ability to represent a broad spectrum of hazard rate shapes; see, for example, Salah [8] based on TIIPC; Salah et al. [9] based on Type-II hybrid; Alotaibi et al. [10] based on adaptive TIIPC (ATIIPC); Elbatal et al. [11] based on improved ATIIPC (IATIIPC); and Elsherpieny and Abdel- Hakim [12] based on unified hybrid, among others.
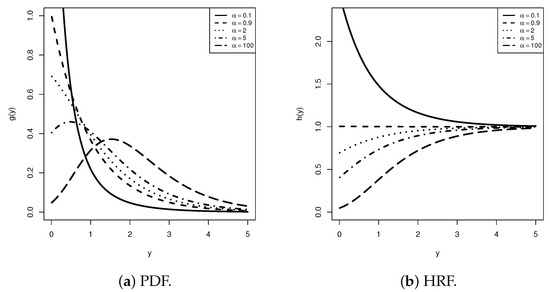
Figure 1.
Several shapes for the APE density and failure rate (when ).
Existing alpha-power distributions offer flexibility but are mostly limited to fixed censoring schemes and have not been developed for complex random removal mechanisms such as the beta-binomial structure. The proposed APE–P-FFC-BB strategy provides closed-form likelihoods and a Bayesian inference framework, improving computational tractability under heavy censoring. This extension fills a gap in biomedical applications by enabling practical analysis of oncology data collected through the P-FFC framework.
Recall that the P-FFC plan assumes fixed or deterministic removal schemes, which may not capture the inherent uncertainty in real clinical environments. To overcome this disadvantage, P-FFC-BB was introduced to offer an efficient compromise by recording only the earliest failures from each group while randomly removing unobserved groups to reduce experimental burden. Moreover, the exponential distribution—although widely used due to its simplicity—often lacks the flexibility required to model heterogeneous survival patterns. To address these limitations, this study integrates the alpha-powering transformation of the exponential model, which enhances its shape flexibility, with a BB random removal mechanism under a P-FFC plan. This censoring strategy provides a more realistic and computationally tractable model for analyzing survival data from heterogeneous cancer populations, offering deeper insights into early-failure behavior under complex removal dynamics. Despite the APE model’s flexibility and practical utility, which have been discussed and applied in several life-testing scenarios in the literature, further investigation is warranted in the presence of samples gathered by the proposed censoring mechanism. To address this gap, we summarize the study objectives sixfold as follows:
- Applying the alpha-powering transformation of the exponential distribution within the proposed censoring scheme, adding flexibility to model various hazard rate shapes for biomedical survival data.
- The joint likelihood under such a censoring setup is derived, and both maximum likelihood and Bayesian estimation procedures are developed for the model parameters and key reliability metrics of the APE distribution, as well as of the BB parameters.
- Bayesian estimation of the same unknown parameters is performed under the assumption of independent gamma and beta priors. The analysis is carried out using both symmetric (squared error) and asymmetric (generalized entropy) loss functions, with the Markov Chain Monte Carlo (MCMC) method employed to approximate the posterior distributions.
- In the interval estimation setup, two types of asymptotic confidence intervals and two Bayesian credible intervals are constructed for all model parameters.
- Extensive Monte Carlo simulations are conducted to assess the performance of the estimators under varying test conditions using several precision metrics.
- Three survival datasets—myeloma, lung, and breast cancer—are analyzed to demonstrate the suitability of the proposed model and its applicability to real-world scenarios.
This work primarily develops maximum likelihood and Bayesian estimation under the P-FFC-BB framework, and alternative inferential approaches such as penalized likelihood, EM-type algorithms, or nonparametric Bayesian methods were not explored but may further enhance robustness. Future research could investigate these estimation strategies, particularly for small or highly censored samples, to complement the inference methods presented here.
The remaining sections of this work are arranged as follows: Section 2 depicts the P-FFC-BB design. Section 3 and Section 4 introduce the ML estimators (MLEs) and Bayes estimators, respectively. Section 5 reports the simulation outcomes. Section 6 analyzes the survival times of leukemia and breast cancer patients. Section 7 ultimately concludes the study.
2. The P-FFC-BB Plan
Let denote the set of observed independent order statistics failure times together with their associated removals collected under the P-FFC-BB removal plan. Assume that the individual lifetimes in follow a continuous distribution with PDF () and CDF (); then, the likelihood function (LF) of the observed data, where , for simplicity, can be expressed as
where and .
Suppose that, at the ith observed failure (), the number of groups removed, denoted by , follows a binomial distribution with parameters and success probability p. Then, the corresponding probability mass function (PMF) of is
where , , and for .
Now, assume that the removal probability p is not constant during the experiment but is, instead, treated as a random variable following a distribution, where . The corresponding beta density function of p is then given by
where is the beta function; see Singh et al. [13] for additional details. Thus, from (6) and (7), the unconditional distribution of becomes
After simplifying, we get
The PMF in (8) corresponds to the BB distribution, denoted by , where represents the total number of trials. Accordingly, the joint probability distribution of the BB removals can be written as
We further assume that the random removals are independent of the corresponding recorded failure times for all . Under this independence assumption, and by combining (5) and (10), the complete LF, denoted by , can be expressed as
where represents the joint LF that depends only on the parameter vector , and is the joint likelihood independent of that depends solely on . This factorization shows that the parameters and can be estimated separately. In practice, one may maximize for independently to obtain the corresponding MLE(s). By setting in (11), the TIIPC BB-based plan (introduced by Singh et al. [13]) emerges as a special case.
Recently, several studies on progressive censoring schemes with BB removals have been reported in the literature. For example, Kaushik et al. [14] and Sangal and Sinha [15] investigated progressive Type-I interval and progressive hybrid Type-I plans, respectively. Using TIIPC with BB-based removals, Singh et al. [13], Usta and Gezer [16], and Vishwakarma et al. [17] examined the generalized Lindley, Weibull, and inverse Weibull distributions, respectively. More recently, Elshahhat et al. [6] analyzed the Weibull lifespan model using a P-FFC-BB strategy. The growing interest in BB-based schemes can be explained by several factors: the inherently complex formulation of the joint likelihood function, the larger number of unknown parameters that need to be estimated, and the additional computational challenges these models introduce.
3. Classical Inference
This section addresses the classical point estimation and asymptotic interval estimation of the APE and BB parameters based on data generated under the P-FFC-BB strategy. The same frequentist methods are also applied to estimate the RF and HRF .
3.1. Point Estimation
Let denote a P-FFC-BB dataset of size k, collected from the APE distribution with the PDF and CDF given in (1) and (2), respectively. By substituting (1) and (2) into (5), the LF (5) can then be expressed, up to proportional, as
where its associated natural logarithm (say, ) is
and . To derive the MLEs of and , denoted by and , respectively, we differentiate the log-likelihood function in (12) with respect to each parameter. Setting these first-order partial derivatives equal to zero yields a system of nonlinear equations, given by
and
where and .
From (14) and (15), it is clear that closed-form solutions for and are not attainable. Therefore, a numerical approach, such as the Newton–Raphson (NR) algorithm, is recommended to iteratively solve these equations. After evaluating the MLEs and , we apply their invariance property to derive the corresponding estimates of the RF and HRF. Specifically, by substituting and into the respective functional forms of and , the MLEs of and at any specified mission time , denoted by and , are expressed as
On the other hand, by applying the beta function where denotes the gamma function, the natural logarithm of in (9) can be written as
As a result, from (16), the MLEs can be derived by simultaneously solving the following two nonlinear normal expressions:
and
where is the digamma function.
It is essential to investigate the existence and uniqueness of the MLEs for the APE () and BB () model parameters. However, due to the intricate form of the log-likelihood functions (), given in (12) and (16), establishing these properties analytically is intractable. To address this limitation, we simulate a P-FFC-BB sample from APE (2, 1) and BB (1, 1) under the setup . From the simulated data, the estimated MLEs for the APE parameters are obtained as (2.6456, 0.6637), while the BB parameters are estimated as (0.0194, 0.1266). Figure 2 presents the likelihood contours of and , which clearly demonstrate that the proposed MLEs, and , both exist and are unique.
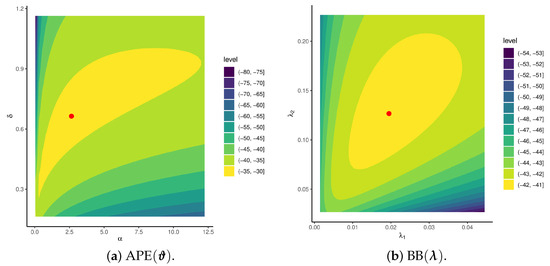
Figure 2.
Contours of APE and BB model parameters from P-FFC-BB data.
3.2. Interval Estimation
To acquire the ACI estimators for the APE parameters , , , and , as well as for the BB parameters , we utilize the asymptotic properties of their MLEs based on large-sample theory. Under standard regularity conditions, the joint asymptotic distribution of is bivariate normal with mean vector and variance–covariance (VC) matrix , where denotes the Fisher information (FI) matrix. However, due to the complex structure of the second-order partial derivatives of the log-likelihood function , obtaining a closed-form expression for the FI matrix is generally infeasible. Consequently, the observed information matrix evaluated at is used to approximate as
with the items, , expressed and reported in Appendix A.
Constructing the ACIs for and requires estimates of the variances of and . A commonly employed approach for this purpose is the delta method, which provides a convenient way to approximate the variance of functions of estimated parameters. For an in-depth discussion of the delta method, refer to Greene [18]. Using this technique, the approximate variances of the estimators for and are obtained as
We first obtain and , where
and
On the other hand, by replacing with their , the approximated VC matrix of the BB parameters (symbolized by ) is given by
with the items, , expressed and reported in Appendix B.
It is evident from the VC matrices in (19) and (21) that the likelihood equations for APE and BB cannot be expressed in closed form. Therefore, again, we recommend using the NR iterative procedure to compute . Hence, at a significance percentage , the two-sided ACI limits of , , , , or (say, ℧ for brevity) are
where represent the approximated variances of , , , , and , given by (19) and (20), and is the upper th percentile point of the standard Gaussian distribution.
4. Bayesian Inference
The Bayesian framework allows prior beliefs or expert knowledge to be incorporated into the inference process for unknown parameters. In this setting, the parameters and of the APE lifetime distribution are treated as random variables, with their prior distributions representing any available prior information. A particularly convenient and flexible choice is the gamma conjugate prior, as highlighted by Kundu [19] and Dey et al. [20]. The gamma distribution provides substantial modeling versatility, making it suitable for encoding a broad range of subjective or empirical prior beliefs. Independent gamma priors are assigned to and to ensure positive support and conjugacy, offering flexibility in specifying both weakly and moderately informative beliefs. Here, we assume that and follow independent gamma priors, specified as and , where for . These hyperparameters are selected to reflect prior information relevant to the distribution parameters. Under this independence assumption, the joint prior PDF of and , denoted by , takes the form
where and are assumed to be known. By substituting (12) and (23) into the Bayes’ theorem, the joint posterior PDF of APE, denoted by , can be expressed as
where its normalized term, say , is given by
Similarly, for the BB parameters, we assume that they are mutually independent and that each follows a gamma prior distribution, denoted by and , respectively. The hyperparameters and for are assumed to be known. Under this independence assumption, the joint gamma PDF of , denoted by , is given by
The squared error loss (SEL) is a commonly used symmetric loss function for Bayesian estimation. Under this criterion, the Bayes estimator, denoted by , corresponds to the posterior mean based on the observed data. Formally, the SEL function (written as ) and the resulting Bayes estimator can be defined as
and
respectively; see Martz and Waller [21].
In contrast to the symmetric SEL, the generalized entropy loss (GEL) introduces an asymmetric estimation criterion. This loss function is particularly suitable when the consequences of overestimation and underestimation are not equally severe. The GEL, denoted by , is defined as
where denotes the derived Bayes estimator from GEL.
As seen from (28), the generalized entropy loss is minimized when . Notably, by setting in (28), the asymmetric Bayes estimator reduces to the symmetric Bayes estimator . When , the GE loss penalizes overestimation more heavily than underestimation, whereas for , the opposite effect occurs. Based on (28), the Bayes estimator of is given by
For more details, see Dey et al. [22]. Due to the nonlinear form of the posterior distribution, , presented in (24), closed-form expressions for the Bayes estimators of , , or against both SEL and GEL functions are analytically intractable.
It is worth mentioning that the SEL function is a natural default choice due to its symmetry and simplicity, yielding posterior means as Bayes estimators. In contrast, the GEL function introduces asymmetry to accommodate scenarios where overestimation and underestimation incur unequal costs.
Consequently, we utilize the MCMC technique to approximate the Bayes estimators and and to construct the associated BCI estimators. To generate such samples, it is first necessary to derive the full-conditional posterior PDFs of and , as follows:
and
respectively.
From (29) and (30), it is clear that the full conditional PDFs of and do not belong to any standard family of statistical distributions. Therefore, direct sampling from the conditional densities and using conventional sampling methods is not possible. To handle this difficulty, as shown in Figure 3, we employ the Metropolis–Hastings (MH) algorithm with normally distributed proposal densities to generate posterior samples. This allows us to obtain Bayesian point estimates and credible intervals for the APE model parameters , , , and ; see Algorithm 1.
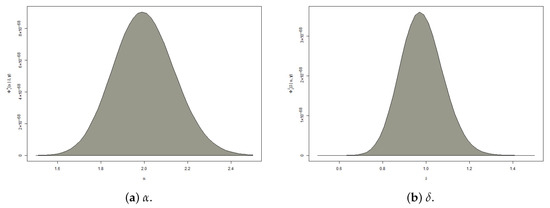
Figure 3.
The posterior PDFs of and from APE (2, 1) and BB (1, 1).
On the other hand, from (26), the full-conditional PDFs of the BB parameters , can be formulated as
and
respectively.
As expected from (31) and (32), the conditional PDFs of do not correspond to any standard statistical distribution. Consequently, direct sampling from these posteriors using conventional methods is not feasible. To overcome this, again, we employ the MH algorithm, following the same procedure described in Algorithm 1, to generate MCMC samples for .
| Algorithm 1 The MCMC (MH-based) Generation Steps |
|
5. Monte Carlo Comparisons
This section presents a detailed evaluation of the proposed theoretical results, focusing on point and interval estimation for the APE parameters. It also considers related reliability measures, including the RF and FRF .
5.1. Simulation Setups
A Monte Carlo simulation is conducted based on samples generated from two populations of APE, namely Pop-A:(0.8, 0.5) and Pop-B:(2, 1). To achieve this, the proposed P-FFC-BB strategy is replicated 2000 times. To generate data from the APE lifetime model under the proposed censoring plan, we follow the steps outlined in Algorithm 2. At , the true values of and are approximately 0.94588 and 0.55494 from Pop-A and approximately 0.93181 and 0.71897 from Pop-B, respectively, serving as reference values for assessing estimation accuracy. The performance of the estimators for , , , and is investigated under varying experimental settings by changing (BB coefficients), (group size), (total number of test units), and different choices of k (number of failed units). According to the mechanism of the proposed censoring, the life test ends when the observed number of failures k is reached for each given n.
Following the MCMC strategy described in Section 4, the simulation uses iterations, discarding the first as burn-in. Posterior estimates, along with their 95% BCI/HPD interval estimates, are obtained for , , , and . For further analysis, after collecting 2000 P-FFC-BB samples, we employ two packages within the R software version 4.2.2 environment:
- A ‘maxLik’ (by Henningsen and Toomet [23]) for maximum likelihood calculations;
- A ‘coda’ (by Plummer et al. [24]) for simulating and analyzing posterior samples.
It should be noted here that the MLEs and (obtained by solving the score Equations (14) and (15)) and the MLEs (obtained by solving the score Equations (18) and (17)) are computed using the NR iterative algorithm using ‘maxLik’. Computations are performed in R, and convergence is declared when the absolute change in the log-likelihood between successive iterations is less than .
To further evaluate the sensitivity of the estimation procedures with respect to the BB removal mechanism, we examine two parameter configurations of the BB distribution, namely BB-1:(0.5, 0.5) and BB-2:(1.5, 1.5). One of the key challenges in Bayesian inference is the appropriate specification of hyperparameters. In this study, we follow the prior elicitation approach outlined by Kundu [19] to determine the values of the hyperparameters and in the joint gamma prior distribution. Accordingly, two prior settings are considered:
- For Pop-A:(0.8, 0.5):
- –
- Prior-A:[PA] with for ;
- –
- Prior-B:[PB] with for .
- For Pop-B:(2, 1):
- –
- Prior-A:[PA] with for ;
- –
- Prior-B:[PB] with for .
The corresponding hyperparameters for each unknown parameter are determined using the method of moments, based on simulated censoring proportions. Two prior settings are considered—PA (weakly informative) and PB (more informative)—to evaluate sensitivity and robustness. This strategy ensures that posterior inference remains primarily data-driven while offering additional stability in scenarios involving small sample sizes or heavy censoring.
| Algorithm 2 P-FFC-BB generation procedure |
|
We now evaluate the introduced estimators obtained for the parameters , , , and (say, for short) using the following performance criteria:
- Root Mean Squared Error (RMSE):
- Average Relative Absolute Bias (ARAB):
- Average Interval Length (AIL):where and denote the (lower and upper) 95% interval bounds.
- Coverage Percentage (CP):where is the indicator function, taking the value 1 if lies within the interval ; otherwise, it is 0.
5.2. Simulation Results and Interpretation
From Table 1, Table 2, Table 3, Table 4, Table 5, Table 6, Table 7 and Table 8, for BB-1, we focus on estimation methods yielding the lowest RMSE, ARAB, and AIL values while maintaining the highest CP values. For brevity, the remaining simulation results based on BB-2 are provided in the Supplementary File. The main findings can be summarized as follows:

Table 1.
The RMSE (1st Col.) and ARAB (2nd Col.) results of from BB-1.

Table 2.
The RMSE (1st Col.) and ARAB (2nd Col.) results of from BB-1.

Table 3.
The RMSE (1st Col.) and ARAB (2nd Col.) results of from BB-1.

Table 4.
The RMSE (1st Col.) and ARAB (2nd Col.) results of from BB-1.

Table 5.
The AIL (1st Col.) and CP (2nd Col.) results of from BB-1.

Table 6.
The AIL (1st Col.) and CP (2nd Col.) results of from BB-1.

Table 7.
The AIL (1st Col.) and CP (2nd Col.) results of from BB-1.

Table 8.
The AIL (1st Col.) and CP (2nd Col.) results of from BB-1.
- The point and interval estimates for , , , and all exhibit satisfactory performance across the simulated settings.
- As (or equivalently n) increases, the accuracy of all estimates improves, indicating greater statistical efficiency with higher failure proportions.
- As m increases, the following patterns are noted:
- –
- The RMSE, ARAB, and AIL findings of , , and tend to decrease, whereas those of increase;
- –
- The CP findings of decreased, whereas those of , , and increased.
- Bayesian estimators based on gamma conjugate priors consistently outperform their frequentist counterparts due to the use of prior information in terms of lowest RMSE, ARAB, and AIL values and highest CP values.
- Furthermore, the asymmetric Bayes results under the GEL-based method yield better results than the symmetric Bayes results obtained using the SEL-based method.
- Additionally, due to the informative gamma priors, both credible interval estimates (including BCI and HPD) consistently yield narrower AILs and higher CPs compared to both proposed ACI estimates.
- For and , asymmetric Bayes estimates under GEL outperform those obtained under GEL. Conversely, for and , asymmetric Bayes estimates with GEL perform better than those with GEL. This observation holds for Pop-i for .
- For Pop-i for , the following is noted:
- –
- All Bayesian point results obtained under PB outperform those based on PA;
- –
- All BCI/HPD results against PB achieve better performance than those under PA;
- –
- This fact is attributed to PB being more informative, with a smaller prior variance compared to PA.
- As the levels of in the APE model increased, the following was observed:
- –
- The RMSE and ARAB findings of increased, whereas those for and decreased;
- –
- The RMSE findings of increased, whereas the associated ARAB findings decreased;
- –
- The AIL findings of , , and increased, whereas those for decreased;
- –
- The CP findings of , , and decreased, whereas those for increased.
- As the levels of in the BB model increased, for Pop-i for , the following was observed:
- –
- The RMSE and ARAB findings of , , , and increased;
- –
- The AIL findings of and increased, whereas those for and decreased;
- –
- The CP findings of and decreased, whereas those for and increased.
- Across most simulation scenarios, the estimated CPs for all parameters remain close to the nominal 95% level, confirming good interval estimation properties.
- Overall, the Bayesian approach with informative gamma priors is recommended for efficiently estimating APE life parameters when a dataset is gathered under the protocol of progressive first-failure censoring with beta-binomial random removal.
6. Cancer Data Applications
To illustrate the practical applicability and relevance of the proposed methodologies, we analyzed three real cancer datasets. The first dataset consists of the survival times of patients in a study on multiple myeloma, the second corresponds to remission times of leukemia patients, and the third involves recurrence-free survival times of breast cancer patients. These case studies demonstrate how the developed approaches can effectively address real-world challenges in medical research and survival analysis. These applications can be represented as follows:
- Application 1: Multiple myeloma is a type of blood cancer that arises from malignant plasma cells in the bone marrow, leading to excessive production of abnormal antibodies and damage to bones, kidneys, and the immune system. This application investigates the survival experience of 48 patients diagnosed with multiple myeloma and treated with alkylating agents at the West Virginia University Medical Center. The patients, aged between 50 and 80 years, were followed prospectively to record their survival times in months, with censoring applied to those still alive at the end of the observation period; see Collett [25].
- Application 2: Lung cancer is a malignant disease in which abnormal cells in the lungs grow uncontrollably, forming tumors that can interfere with normal lung function and spread to other parts of the body. This application examines the survival data of 44 patients with advanced lung cancer who were randomly assigned to receive the standard chemotherapy regimen; see Lawless [26]. The “standard” treatment represented the control arm in a randomized clinical trial designed to assess the effectiveness of alternative chemotherapeutic protocols.
- Application 3: Breast cancer is a malignant condition marked by abnormal, uncontrolled proliferation of cells within breast tissue, most commonly originating in the milk ducts or lobular structures. It predominantly affects women and may present clinically as palpable masses and structural or morphological changes in the breast. This application analyzes recurrence-free survival times (measured in days) for 42 breast cancer patients with tumors exceeding 50 mm in size, all of whom received hormonal therapy; see Royston and Altman [27].
In Table 9, the survival times associated with myeloma, lung, and breast cancer patients are reported. Firstly, before analyzing the myeloma dataset, we ignore the survival times that are still censored (denoted by ‘+’). Additionally, for computational purposes, each time point in the breast cancer data is transformed by dividing survival times by 100. Table 10 provides a detailed summary of descriptive statistics for the datasets listed in Table 9. The summary includes minimum, maximum, mean, mode, three quartiles (), standard deviation (Std.Dv.), and skewness, offering a comprehensive overview of the data distribution.

Table 9.
Survival times of myeloma, lung, and breast cancer data.

Table 10.
Statistical summary of myeloma, lung, and breast cancer data.
Before evaluating the theoretical estimators of , , , and , it is worth demonstrating the superiority of the proposed APE lifespan model compared to one of its most common competitors. To achieve this goal, the suitability of the APE distribution is first assessed alongside the Weibull () distribution (see Bourguignon et al. [28]) using the three cancer datasets summarized in Table 9. To verify model adequacy, this fitting analysis employed the Kolmogorov–Smirnov () test, along with its corresponding p-value, as well as several model selection criteria: negative log-likelihood (NLL), Akaike criterion (AC), consistent Akaike criterion (CAC), Bayesian criterion (BC), and Hannan–Quinn criterion (HQC). In addition, the MLEs of and , together with their standard errors (Std.Ers), for both the APE and Weibull (W) models are computed and reported in Table 11. The results in Table 11 also demonstrate that the APE model provides the lowest estimated values for all the information criteria considered compared to the Weibull model based on the myeloma dataset, whereas the opposite conclusion is reached for the other two datasets. Table 11 also shows that the APE model produces the highest p-value and the smallest statistic compared to the Weibull model for all considered datasets, indicating that the APE distribution provides an excellent fit to the myeloma, lung, and breast cancer datasets.

Table 11.
Fitting the APE and W models from myeloma, lung, and breast cancer data.
To further substantiate the model assessment, Figure 4 provides a comprehensive visual evaluation for both the APE and W lifespan models using four diagnostic tools: (i) fitted density curves overlaid on data histograms, (ii) the fitted versus empirical probability–probability (PP) plot, (iii) the fitted versus empirical quantile–quantile (QQ) plot, and (iv) the fitted versus empirical reliability function (RF). Figure 4a–d corroborate the numerical results, demonstrating that the APE lifetime model provides the best fit to all three cancer datasets and highlighting that the APE model is a strong competitor to the commonly used W lifespan model.
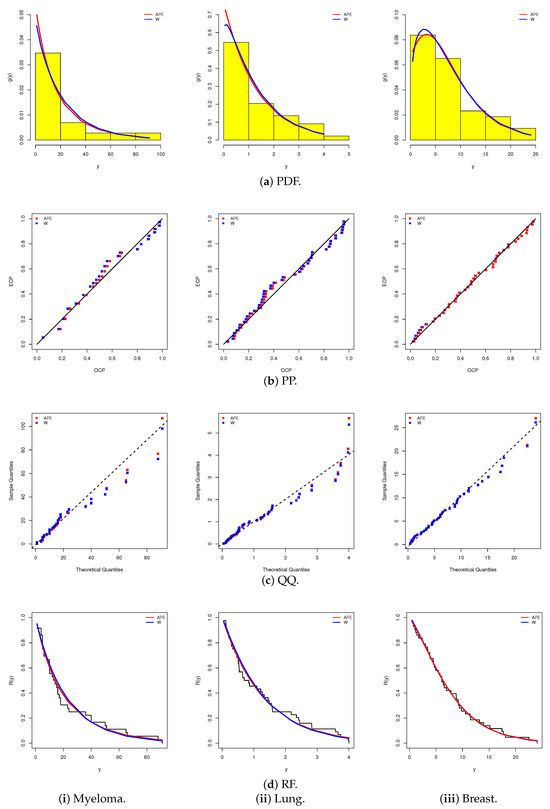
Figure 4.
Fitting diagrams for the APE and W models from myeloma, lung, and breast cancer data.
Figure 5 displays the log-likelihood contour plot and the violin boxplot of the APE model. Moreover, the contour plot in Figure 5a confirms the existence and uniqueness of the fitted MLEs of and , thereby reinforcing the model’s stability and identifiability. The estimates and are recommended as robust initial values for subsequent analytical procedures involving this dataset. Figure 5b reveals that the (i) myeloma data is right-skewed, with a wider spread and several high-value outliers, indicating greater variability in survival times; the (ii) lung data is nearly symmetric, with a tighter interquartile range and fewer mild outliers, showing lower variability; and the (iii) breast data is mildly right-skewed, with a moderate spread and fewer outliers, reflecting more consistent survival times.
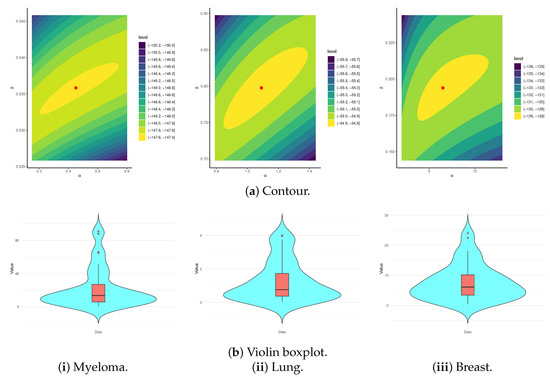
Figure 5.
The contour and violin boxplot diagrams for the APE model from myeloma, lung, and breast cancer data.
To assess the theoretical estimation performance of the APE model parameters (, , , and ) together with the BB distribution parameters (), the complete myeloma, lung, and breast cancer datasets were used in a simultaneous, life-testing experiment. These datasets were randomly partitioned into groups, respectively, each containing units, as shown in Table 12, where starred items denote the first failure in each group. By fixing and considering different options of k, two artificial P-FFC-BB samples were generated from the myeloma, lung, and breast cancer datasets (see Table 13).

Table 12.
Random grouping of myeloma, lung, and breast cancer data.

Table 13.
Artificial P-FFC-BB samples from myeloma, lung, and breast cancer data.
In progressive censoring studies, the independence assumption between random removals and observed failure times is common because withdrawals are typically driven by administrative or external factors (e.g., patient drop-out or loss to follow-up) rather than the exact time-to-failure of other units.
Using the P-FFC-BB samples (with ) generated from the myeloma, lung, and breast cancer datasets reported in Table 13, the corresponding Spearman rank correlation statistics (with their associated p-values) between the observed failure times and removal counts are estimated as −0.714 (0.020), −0.678 (0.030), and −0.679 (0.031), respectively. These results indicate a statistically significant negative association: groups with earlier failures tended to have higher numbers of removals. This finding suggests that the assumption of independence between removals and failure times may not hold strictly for these datasets.
In the absence of prior information on the APE parameters and the BB parameters, a vague joint gamma prior with hyperparameters was adopted. Following Algorithm 1, a total of iterations were run, discarding the first iterations as burn-in. Posterior summaries were then obtained for , , , , and , including MCMC estimates under both SEL and GEL loss functions with , as well as their 95% BCI/HPD interval estimates. For and 1 in the myeloma, lung, and breast cancer datasets, respectively, all estimates of and were computed. The resulting point estimates (with their Std.Ers) and the 95% interval estimates (with their ILs) are summarized in Table 14 and Table 15. In comparison to classical likelihood-based estimates of , , , , and , the point and interval estimation findings developed from the Bayesian approach showed clear superiority, producing smaller Std.Ers and shorter ILs for all parameters.

Table 14.
The point estimates of , , , , and from myeloma, lung, and breast cancer datasets.

Table 15.
The 95% interval estimates of , , , , and from myeloma, lung, and breast cancer datasets.
To establish the existence and uniqueness of the MLEs for the parameters of APE and BB , based on the P-FFC-BB datasets summarized in Table 13, Figure 6 depicts the log-likelihood contours of , , and (). These plots demonstrate that the fitted MLEs (reported in Table 14) for both APE and BB exist and are unique. Using these estimates as initial values, the proposed Markov chains were implemented. To assess the convergence behavior of the simulated 40,000 Markov chain variates for , , , , and (), we focused on the P-FFC-BB dataset at (as a representative case) from Table 13. Figure 7 presents the corresponding trace plots and posterior density plots for each parameter. In each plot, the solid line denotes the Bayes’ MCMC estimates calculated by the SEL, and the dashed lines represent the 95% BCI bounds. The trace plots indicate that the simulated chains for all parameters exhibit stable and symmetric behavior, suggesting satisfactory convergence. To further validate this, we summarized key descriptive statistics based on the remaining 40,000 post-burn-in samples for each parameter (see Table 16). These numerical summaries reinforce the visual evidence from Figure 7, providing additional support for convergence and distributional stability across all monitored parameters.
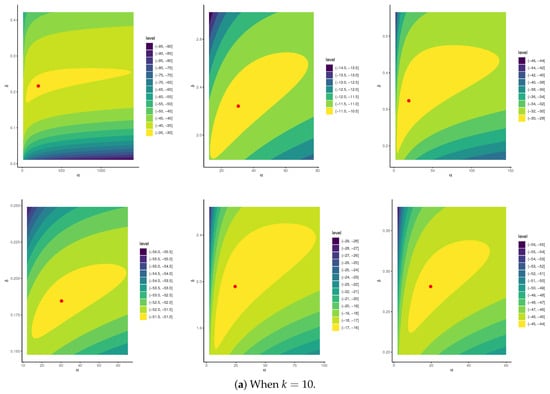
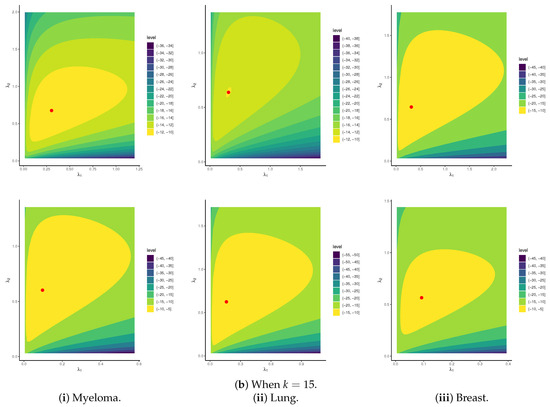
Figure 6.
Contours of APE(,) and BB(,) distributions from myeloma, lung, and breast cancer datasets.
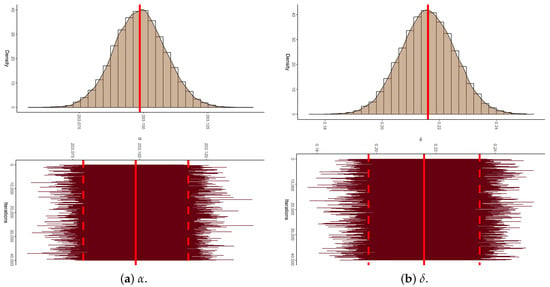
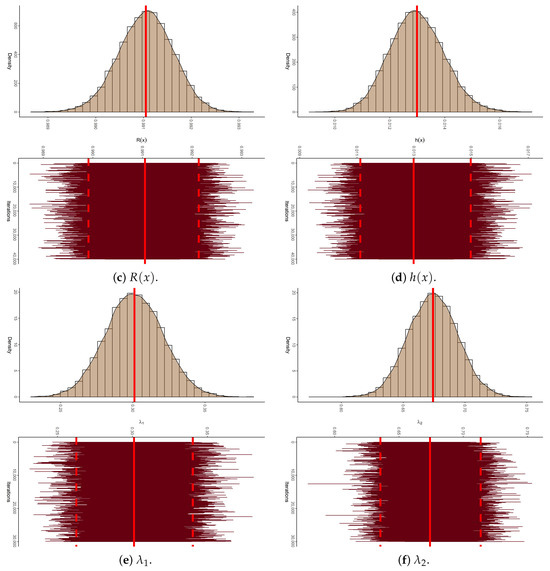
Figure 7.
The density (with its Gaussian curve) and trace diagrams of , , , , and from myeloma data.

Table 16.
Statistical summary for MCMC iterations of , , , , and from myeloma, lung, and breast cancer data.
7. Conclusions
In this work, we examined a novel alpha-powering model based on the exponential distribution within the framework of the P-FFC-BB removal plan. This contribution provides a flexible and realistic mechanism for capturing uncertainty in unit withdrawals, a prevalent challenge in reliability and biomedical studies. Comprehensive inference procedures were developed, including both maximum likelihood and Bayesian estimation methods. To address the intractability of the posterior distributions under squared-error and generalized entropy loss functions, a tailored Metropolis–Hastings algorithm was implemented, enabling accurate approximation of posterior summaries and credible intervals. To evaluate the performance of the proposed estimators, we conducted an extensive Monte Carlo simulation study, systematically assessing the effects of varying censoring levels, prior specifications, and group sizes. The simulation results revealed that Bayesian methods—particularly those based on asymmetric loss functions with informative priors—outperform the classical approach, yielding more accurate and stable estimates under complex censoring scenarios. Furthermore, asymptotic confidence intervals and Bayesian credible intervals have been derived and demonstrated satisfactory coverage probabilities across diverse settings. The practical relevance of the proposed model has been validated through applications to three real-world survival datasets from myeloma, lung, and breast cancer patients, where progressive censoring and drop-out variability are inherently present. In all cases, the proposed model exhibited superior goodness of fit, predictive reliability, and improved hazard rate estimation, underscoring its robustness in modeling complex survival mechanisms. While classical methods, such as the Kaplan–Meier estimator, Cox proportional hazards model, and Weibull-based parametric approaches, are well-established for right-censored data, they do not naturally accommodate the progressive first-failure censoring scheme with the stochastic beta–binomial removals considered here. The proposed framework specifically addresses this complexity by deriving tractable likelihoods and flexible hazard structures under such censoring, providing insights beyond the reach of conventional methods. Future work may include empirical comparisons with these classical approaches to highlight further the contexts where our method offers distinct advantages. Future research directions include extending the proposed framework to other baseline distributions with non-monotonic hazard rates, incorporating covariate effects within a regression structure, and exploring accelerated life-testing designs under similar censoring schemes. Future research could extend the proposed censoring framework to other flexible lifetime models—such as log-logistic, log-normal, generalized gamma, or Burr families—to better capture non-monotonic and heavy-tailed hazard structures.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/math13183028/s1. Table S1: The RMSE (1st Col.) and ARAB (2nd Col.) results of from BB-2; Table S2: The RMSE (1st Col.) and ARAB (2nd Col.) results of from BB-2; Table S3: The RMSE (1st Col.) and ARAB (2nd Col.) results of from BB-2; Table S4: The RMSE (1st Col.) and ARAB (2nd Col.) results of from BB-2; Table S5: The AIL (1st Col.) and CP (2nd Col.) results of from BB-2; Table S6: The AIL (1st Col.) and CP (2nd Col.) results of from BB-2; Table S7: The AIL (1st Col.) and CP (2nd Col.) results of from BB-2; Table S8: The AIL (1st Col.) and CP (2nd Col.) results of from BB-2.
Author Contributions
Methodology, A.E., O.E.A.-K. and H.S.M.; Funding acquisition, H.S.M.; Software, A.E.; Supervision H.S.M.; Writing—original draft, A.E. and O.E.A.-K.; Writing—review & editing H.S.M. and O.E.A.-K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project, number (PNURSP2025R175), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. The FI Components of α and δ
References
- Wu, J.W.; Hung, W.L.; Chen, C.Y. Approximat MLE of the scale parameter of the truncated Rayleigh distribution under the first failure-censored data. J. Inf. Optim. Sci. 2004, 25, 221–235. [Google Scholar]
- Balasooriya, U. Failure–censore reliability sampling plans for the exponential distribution. J. Stat. Comput. Simul. 1995, 52, 337–349. [Google Scholar] [CrossRef]
- Balakrishnan, N.; Cramer, E. The Art of Progressive Censoring. Statistics for Industry and Technology; Springer: Birkhäuser, Germany; New York, NY, USA, 2014. [Google Scholar]
- Huang, S.R.; Wu, S.J. Estimation of Pareto distribution under progressive first-failure censoring with random removals. J. Chin. Stat. Assoc. 2011, 49, 82–97. [Google Scholar]
- Ashour, S.K.; El-Sheikh, A.A.; Elshahhat, A. Inference for Weibull parameters under progressively first-failure censored data with binomial random removals. Stat. Optim. Inf. Comput. 2021, 9, 47–60. [Google Scholar] [CrossRef]
- Elshahhat, A.; Sharma, V.K.; Mohammed, H.S. Statistical analysis of progressively first-failure censored data via β-binomial removals. AIMS Math. 2023, 8, 22419–22446. [Google Scholar] [CrossRef]
- Mahdavi, A.; Kundu, D. A new method for generating distributions with an application to exponential distribution. Commun. Stat.-Theory Methods 2017, 46, 6543–6557. [Google Scholar] [CrossRef]
- Salah, M.M. On Progressive Type-II Censored Samples from Alpha Power Exponential Distribution. J. Math. 2020, 2020, 2584184. [Google Scholar] [CrossRef]
- Salah, M.M.; Ahmed, E.A.; Alhussain, Z.A.; Ahmed, H.H.; El-Morshedy, M.; Eliwa, M.S. Statistica inferences for type-II hybrid censoring data from the alpha power exponential distribution. PLoS ONE 2021, 16, e0244316. [Google Scholar] [CrossRef]
- Alotaibi, R.; Elshahhat, A.; Rezk, H.; Nassar, M. Inference for alpha power exponential distribution using adaptive progressively type-II hybrid censored data with applications. Symmetry 2022, 14, 651. [Google Scholar] [CrossRef]
- Elbatal, I.; Nassar, M.; Ben Ghorbal, A.; Diab, L.S.G.; Elshahhat, A. Reliabilit analysis and its applications for a newly improved type-II adaptive progressive alpha power exponential censored sample. Symmetry 2023, 15, 2137. [Google Scholar] [CrossRef]
- Elsherpieny, E.A.; Abdel-Hakim, A. Statistica analysis of Alpha-Power exponential distribution using unified Hybrid censored data and its applications. Comput. J. Math. Stat. Sci. 2025, 4, 283–315. [Google Scholar]
- Singh, S.K.; Singh, U.; Sharma, V.K. Expecte total test time and Bayesian estimation for generalized Lindley distribution under progressively Type-II censored sample where removals follow the beta-binomial probability law. Appl. Math. Comput. 2013, 222, 402–419. [Google Scholar]
- Kaushik, A.; Singh, U.; Singh, S.K. Bayesia inference for the parameters of Weibull distribution under progressive Type-I interval censored data with beta-binomial removals. Commun. Stat.-Simul. Comput. 2017, 46, 3140–3158. [Google Scholar] [CrossRef]
- Sangal, P.K.; Sinha, A. Classical estimation in exponential power distribution under Type-I progressive hybrid censoring with beta-binomial removals. Int. J. Agric. Stat. Sci. 2021, 17, 1973–1988. [Google Scholar]
- Usta, I.; Gezer, H. Paramete estimation in Weibull distribution on progressively Type-II censored sample with beta-binomial removals. Econ. Bus. J. 2016, 10, 505–515. [Google Scholar]
- Vishwakarma, P.K.; Kaushik, A.; Pandey, A.; Singh, U.; Singh, S.K. Bayesia estimation for inverse Weibull distribution under progressive Type-II censored data with beta-binomial removals. Austrian J. Stat. 2018, 47, 77–94. [Google Scholar] [CrossRef]
- Greene, W.H. Econometric Analysis, 4th ed.; Prentice-Hall: New York, NY, USA, 2000. [Google Scholar]
- Kundu, D. Bayesia inference and life testing plan for the Weibull distribution in presence of progressive censoring. Technometrics 2008, 50, 144–154. [Google Scholar] [CrossRef]
- Dey, S.; Elshahhat, A.; Nassar, M. Analysi of progressive type-II censored gamma distribution. Comput. Stat. 2023, 38, 481–508. [Google Scholar] [CrossRef]
- Martz, H.F.; Waller, R.A. Bayesian Reliability Analysis; Wiley and Sons: New York, NY, USA, 1982. [Google Scholar]
- Dey, D.K.; Ghosh, M.; Srinivasan, C. Simultaneou estimation of parameters under entropy loss. J. Stat. Plan. Inference 1987, 15, 347–363. [Google Scholar] [CrossRef]
- Henningsen, A.; Toomet, O. maxLik: A package for maximum likelihood estimation in R. Comput. Stat. 2011, 26, 443–458. [Google Scholar] [CrossRef]
- Plummer, M.; Best, N.; Cowles, K.; Vines, K. CODA: Convergence diagnosis and output analysis for MCMC. R News 2006, 6, 7–11. [Google Scholar]
- Collett, D. Modelling Survival Data in Medical Research; Chapman and Hall/CRC: Boca Raton, FL, USA, 2023. [Google Scholar]
- Lawless, J.F. Statistical Models and Methods for Lifetime Data; John Wiley and Sons: New York, NY, USA, 2011. [Google Scholar]
- Royston, P.; Altman, D.G. Externa validation of a Cox prognostic model: Principles and methods. BMC Med. Res. Methodol. 2013, 13, 33. [Google Scholar] [CrossRef]
- Bourguignon, M.; Silva, R.B.; Cordeiro, G.M. The Weibull-G family of probability distributions. J. Data Sci. 2014, 12, 53–68. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).