Abstract
Zero inflation and overfitting can reduce the accuracy rate of using machine learning models for characterizing binary data sets. A zero-inflated Bernoulli (ZIBer) model can be the right model to characterize zero-inflated binary data sets. When the ZIBer model is used to characterize zero-inflated binary data sets, overcoming the overfitting problem is still an open question. To improve the overfitting problem for using the ZIBer model, the minus log-likelihood function of the ZIBer model with the elastic net regularization rule for an overfitting penalty is proposed as the loss function. An estimation procedure to minimize the loss function is developed in this study using the gradient descent method (GDM) with the momentum term as the learning rate. The proposed estimation method has two advantages. First, the proposed estimation method can be a general method that simultaneously uses - and -norm terms for penalty and includes the ridge and least absolute shrinkage and selection operator methods as special cases. Second, the momentum learning rate can accelerate the convergence of the GDM and enhance the computation efficiency of the proposed estimation procedure. The parameter selection strategy is studied, and the performance of the proposed method is evaluated using Monte Carlo simulations. A diabetes example is used as an illustration.
Keywords:
expectation-maximization algorithm; gradient descent method; learning rate; maximum likelihood estimation; zero-inflated model MSC:
62-08; 62-11
1. Introduction
Zero-inflated models help characterize binary data sets with excess zero counts. Structural and sampling zeros can be found in a zero-inflated data set. When the response variable cannot take positive values due to inherent constraints or conditions, the zero is called structural zero. In a medicate setting, Diop et al. [] investigated the infection response related to some diseases where indicates the individual was infected; otherwise, . Assume that an immunity agency controls whether an individual is infected. The individual is immune and labeled by if the individual cannot undergo the interesting outcome. In both examples, is not a random zero and can be a structural zero. Ridout et al. [] has also commented on the distinction between structural zero and random zero. In contrast to the structural zero, the zeros raised from random chance are the sampling zero. Using a mixture mechanism, the zero-inflated model can account for structural and sampling zero sources and performs better than the traditional logistic, Poisson, and non-negative binomial regression models.
Occasionally, the highly disproportionate distribution of categories also needs to be improved for data analysis. Such data sets are also referred to as imbalanced data. In binary classifications, the size of one category significantly dominates the other. Those binary data sets could be inherently imbalanced in real-world scenarios for various reasons.
The initial zero-inflated model was the zero-inflated Poisson (ZIP), proposed by Lambert [], where a zero-inflated Poisson (ZIP) regression model was investigated via maximum likelihood estimation method to make statistical inferences. Moreover, a data set about the soldering defect on the printed wiring board was used to illustrate the proposed ZIP regression model. To overcome the drawback of over-dispersion from the Poisson model, the zero-inflated negative Binomial (ZINB) model was another competitive model. Reducing the impacts of random effects on the zero-inflated data sets, Hall [] conducted case studies for the ZIP and zero-inflated binomial (ZIB) regression models. Hall [] successfully showed that random effects help account for the correlation between observations within clusters. Cheung [] explored the theoretical foundations of zero-inflated models, illustrated the practical applications with examples, and analyzed the implications of using these models for precise inference in the context of growth and development studies. Gelfand and Citron-Pousty [] demonstrated the utilization of zero-inflated models for spatial count data in environmental and ecological statistics. Additionally, they investigated how these models address both spatial correlation and the frequent occurrence of excess zero counts in environmental and ecological applications. Rodrigues [] and Ghosh et al. [] studied the applications of using Bayesian techniques to estimate the parameters of zero-inflated models.
Harri and Zhao [] introduced a zero-inflated-ordered probit model to process data sets with ordered response variables and excess zeros. They also utilized a tobacco consumption data set for modeling illustration. Loeys et al. [] considered ZIP, ZINB, Poisson logit hurdle model (PLH), and Binomial logit hurdle model (NBLH). Diop et al. [] investigated the maximum likelihood estimation method for the logistic regression model with a cure fraction. Staub and Winkelmann [] conducted inferences to study the consistency of the parameter estimation of the zero-inflated count model. He et al. [] investigated the relevance of the structural zeroes to a zero-inflated model. Diop et al. [] conducted simulation-based inference for the zero-inflated binary regression model and proposed the maximum likelihood estimation procedure. Zuur and Ieno [] gave comprehensive guidance for users to use R codes to apply zero-inflated models.
2. Motivation and Organization
Zero inflation and overfitting can reduce the accuracy rate of using machine learning models for characterizing binary data sets. A zero-inflated Bernoulli (ZIBer) model can be the right model to characterize zero-inflated binary data sets. To our best knowledge, few works have studied the ZIBer model for binary classification, and overcoming the overfitting problem of the ZIBer model is an open question. Recent works about the applications of the ZIBer model can be summarized as follows: When covariates missing at random is found in the ZIBer regression model, Lee et al. [] proposed a validation likelihood method to estimate the model parameters. Pho [] proposed a goodness-of-fit test for the ZIBer model besides revisiting the goodness-of-fit test of the logit model. Li and Lu [] proposed an implementing procedure and consistent estimation method to obtain the variances of the regression parameter estimators. Then, Monte Carlo simulation methods are used to evaluate their proposed method. Pho [] proposed a new zero-inflated probit Bernoulli model and constructed the parameter estimation process. Lu et al. [] proposed a penalized estimation method for partially linear additive models to ZIBer outcome data. The B-spline method is employed to approximate unknown nonparametric components. Moreover, they proposed a two-stage iterative EM algorithm to obtain the penalized spline estimates. When the response variables in a zero-inflated data set follow a Bernoulli distribution, Chiang et al. [] suggested an expectation-maximum (EM) maximum likelihood estimation process for the ZIBer model to obtain reliable estimates of the model parameters. We denote the EM maximum likelihood estimation method proposed by Chiang et al. [] as the EM-ZIBer method.
The EM-ZIBer method of Chiang et al. [] can be an easy method to obtain more reliable maximum likelihood estimates (MLEs) of the ZIBer model than the typical maximum likelihood estimation method. However, the EM-ZIBer method loses feature selection efficiency when the zero-inflated data set size is vast and has an imbalanced structure. This drawback reduces the feasibility of applying the EM-ZIBer method to an extensive zero-inflated data set. In this study, the gradient descent method (GDM) with momentum term as learning rate is used to minimize the loss function, which combines the minus log-likelihood function and using elastic net regularization rule for an overfitting penalty. The proposed estimation procedure has two advantages: First, the proposed estimation method simultaneously uses - and -norm terms for penalty and includes the ridge and least absolute shrinkage and selection operator (lasso) methods as special cases. Hence, the proposed estimation procedure is a general method. Second, the momentum learning rate can accelerate the convergence of the GDM and enhance the computation efficiency of the proposed estimation procedure. For simplicity, we named the new parameter estimation method ENR-ZIBer method. Because the lasso regulation uses -norm for a penalty. The target function for optimization is not a convex function. Hence, the GDM replaces the Newton method for the loss function minimization in this study.
Steps to implement the proposed ENR-ZIBer method will be analytically studied in Section 3.3. Second, to accelerate the convergence of the proposed ENR-ZIBer method, we suggest using the momentum term to the learning rate parameter to implement the gradient descent method. The implementation of using the momentum term in the ENR-ZIBer method will be analytically studied in Section 3.4.
The rest of this article is organized as follows. The ZIBer model is addressed in Section 3. The maximum likelihood estimation is discussed in Section 3.2. Moreover, the proposed elastic net penalty regularization that includes the - and -norm rules to the maximum likelihood estimation for the ZIBer model, are studied in Section 3.3. To accelerate the convergence, the GDM with the momentum term as the learning rate is used to the GDM for the elastic net regularization rule is examined in Section 3.4. In Section 4, the quality of the proposed ENR-ZIB method is evaluated using Monte Carlo simulations. One example is used in Section 5 for illustration. Finally, some conclusions are given in Section 6.
3. The Statistical Model and Optimization
The ZIBer model will be established and the inference procedure of model parameters will be addressed in this section.
3.1. The Statistical Model
Regarding the binary classification modeling, the response variable Y can be labeled as 0 or 1, where indicates an individual infected by a disease; otherwise, . Let the probability of depend upon the covariate with ; that is, , where p is a function of . Therefore, , where denotes the Bernoulli distribution with the conditional success probability . To construct the ZIBer model, the logit (or named logistic regression) function is used as the link and defined by
where . It can be shown that
In a zero-inflated model, the zero inflation occurs in the analysis of counts if the data-generating process results in too many zeros. Failure to account for extra zeros could produce unreasonable estimates and inferences. To remove the drawback, two possible random resources of are considered here. The structural zero occurs with probability , and the non-structural zero happens with the probability . Hence, the unconditional probability of can be obtained by
It is easy to obtain the probability
Assume that depends on the latent covariate , where . Using the logit model again by
where , it can be shown that
Given the covariate information , and let denote the probability of . We can represent
The odd can be presented by
Therefore, the ZIBer model can be defined by the following: the response variable , where is defined by Equation (8).
3.2. Maximum Likelihood Estimation
Let . Given independent responses, , of Y with the corresponding observed and latent covariates, , where and for . The likelihood function can be presented by
The log-likelihood function of the ZIBer model can be presented by
The first derivative of with respect to and are
and
respectively. The typical maximum likelihood estimates of and can be obtained by
where is the solution space of .
3.3. Optimization with Regularization
The -norm regularization uses lasso rule to penalize the model overfitting. The norm is defined by . Assume that we want to minimize with the -norm penalty, the lasso target function can be modified as
where is a constant, which is used to control the regularization strength.
The -norm regularization uses ridge regression to prevent the model overfitting. The norm is defined by . Without loss of generality, we use the term to develop the ridge regularization for the zero-inflated binary model. Assume that we want to minimize with the -norm penalty, the ridge target function can be modified as
where is a constant for controlling the regularization strength, and the constant is used to reduce the computation loading.
Lasso regularization using -norm can result in a parsimonious model. Large can help to result in a sparse model by setting some coefficients to zero, reducing the number of effective parameters. However, lasso regularization rule is sensitive to outliers because of taking the absolute value for the penalty.
The -norm penalty in ridge regularization discourages large coefficients. The -norm penalty pushes the model’s coefficients toward zero but never exactly reaches zero. Compared with the -norm regularization rule, the ridge regularization rule is less sensitive to outliers because it takes a square value for the penalty.
It is a good idea to use a regularization method that covers the strength of the lasso and ridge regularization rules. Elastic net regularization uses the penalties from the lasso and ridge techniques to regularize the target model. Taking advantage of the - and -norms, the elastic net regularization rule combines the lasso and ridge regularization rules and learns from their shortcomings to improve the regularization of statistical models. The loss function based on the log-likelihood function and elastic net regulation can be defined by
where and are constants to control the regularization strength. It is obvious that reduces to if , and reduces to if . When , the elastic net regularization rule reduces to the lasso regularization rule. When , the elastic net regularization rule reduces to the ridge regularization rule. Therefore, the elastic net regularization rule covers the lasso and ridge regularization rules as a special case. The elastic regularization rule is a generalized rule for practical applications.
The first derivative of with respect to can be obtained by
where . If , then ; otherwise, . The first derivative of with respect to can be obtained by
The gradient descent method is used to obtain the estimates of and . Let and denote the solutions of and at the tth iteration. Let
and
The values of and at the th iteration can be updated by
and
respectively, for ; and are learning rates. The update process stops if the condition
is satisfied, where is a small positive number. Then, and are the required solution of and , respectively.
3.4. Convergence Acceleration for the Gradient Descent Method
Constant learning rates have been commonly used to implement the gradient descent method. However, implementing a gradient descent method with a constant learning rate for optimization could take more computation time to reach convergence. Improving the learning rate based on the one-step ahead estimation results to the next step can be a good idea to accelerate the convergence of the gradient descent method. In this study, the momentum learning rate method is adapted to accelerate the convergence speed of the gradient descent method. More comprehensive discussions for using the momentum term to the learning rates can be found in Yu and Chen [], Attoh-Okine [], Qian [], Wang et al. [], Liu et al. [], Friedman et al. [], and Simon et al. [].
At the initial step of iteration , let
and
where is the initial solution vector of . and are the initial solutions of and , respectively. For iteration , the values of and are updated by
and
where m is the momentum term that has is a constant in [0,1). Various existing studies have verified the appropriate value of the momentum term can be ; see Liu et al. []. Then and can be updated at the iteration t by
and
respectively.
The optimization to minimize depends on the value of and . We use the same setting as the R package glmnet. Let and ; see [,]. We can see that reduces if and reduces to if . In this study, we suggest to set . Then, search the value of over the interval [0,1] for the optimal values of and . We will study the performance of the ENR-ZIB method in Section 4 using the Monte Carlo simulation method.
4. Monte Carlo Simulations
To assess the quality of the proposed ENR-ZIBer estimation method, we generated zero-inflated data sets from the ZIBer model. Let for , and . The logit model based on and is defined by
where , , , , and , , , , for , and denotes the normal distribution with . Let and The logit model based on and is defined by
where , , and , for .
The values of and are used for the simulations. Before generating from , and are standardized for each and , respectively. Moreover, the coefficient vectors of and are also normalized by and , respectively. The learning rates and are used for the gradient descent method. Moreover, the momentum term is used to update the values of and in Equations (27) and (28), respectively, at each iteration run to accelerate the convergence speed of the numerical computation.
To make the proposed ENR-ZIBer method a general model that can cover the results of the lasso and ridge regularization penalty model, k values of are first generated, in which values of are used for (), (), and and (), respectively. Hence, the lasso, ridge, and elastic net regularization penalty models are included in the pool to compete with the best model. The cross-validation rule is used in this study to make the competition fair. Seventy percent of the sample was used as the training sample for establishing the target model. Then, the other third percent of the sample was used as the testing sample for the error rate evaluation. The best model with the lowest error rate can be screened out based on the testing sample among the model competition.
First, we determine how many repetitions are required to obtain a reliable error rate for the convergence of the mean error rate (MER) based on the simulation study with 5000 repetitions. The obtained MERs are reported in Figure A1. From Figure A1, we find that the MER converges at 1000 or more repetitions for the sample size 300 and 500. Hence, 1000 repetitions should be enough for the performance comparison of competitive models.
The ZIBer model is a weak classifier because it uses an analytical nonlinear function for classification. It is expected to enhance the accuracy rate of the ZIBer model by using regularization rules to penalize the model overfitting. Because the GDM works stable for large samples. Training a weak learner does not need a huge sample. However, the cross-validation rule asks to cut the original sample into training and testing samples. To achieve these two goals, at least 150 cases and up to 500 cases are considered for implementing the proposed method in simulations.
Second, the performance of the proposed ENR-ZIBer method is compared with the EM-ZIBer method and logit model for the sample sizes , 300, and 500. 70% observations in the sample were used as the training sample to establish the models, and the other 30% observations were used as the testing sample for evaluation. The mean error rates (MERs) of the EM-ZIBer method, the logit model, and the proposed ENR-ZIBer method were evaluated based on the testing sample using the models established with the training sample. Then, the model performance is evaluated based on 1000 repetitions. All simulation results are reported in Table 1.

Table 1.
The MER of the ENR-ZIBer, EM-ZIBer, and logistic regression methods for and in 1000 repetitions.
Table 1 shows that the MER is stable under each sample size for and 150. The best ENR-ZIBer model is selected from 30 (for ) and 50 (for ) lasso, ridge, and elastic net penalty regularization models, respectively. Because the cross-evaluation rule is used. The model is established based on the training sample, which contains 70% observations. The error rate is evaluated based on the testing sample, which is composed of the other 30% observation. The sample size cannot be small. A small testing sample will make the MER estimation unstable. We also find that the proposed ENR-ZIBer method works to generate a stable MER and smaller standard deviation if . This finding will be used for the data analysis in Section 5.
The logit model in Table 1 has the largest MER compared with its competitors of the ENR-ZIBer and EM-ZIBer methods. The proposed ENR-ZIBer method with can well characterize zero-inflated data and outperform the EM-ZIBer method and logit model regarding the MER and its standard deviation in most cells based on 1000 repetitions. Hence, is recommended for the implementation of the ENR-ZIBer method. The ENR-ZIBer method can include 50 lasso ( or ), 50 ridge ( or ), and 50 elastic net ( and ; or ) penalty regulations sets with 50 random generated values of , respectively, in which and , to compete the best model.
5. An Example
In this section, the proposed ENR-ZIBer method is illustrated by a diabetes data set. A total of 768 cases with eight covariate variables are included in this data set. The purpose is to predict whether each case will have a positive or negative response to diabetes. Hence, the response variable is Diabetes, labeled by y, and can be either a positive () or negative () response. The covariate variables are
- (1)
- Pregnant (): pregnancy numbers;
- (2)
- Glucose(): plasma glucose concentration based on the 2-h oral glucose tolerance test;
- (3)
- Pressure(): diastolic blood pressure (mm Hg);
- (4)
- Triceps(): Triceps skin fold thickness (mm);
- (5)
- Insulin(): Two-hour serum insulin ( U/mL);
- (6)
- Mass(): Body mass index;
- (7)
- Pedigree(): Diabetes pedigree function;
- (8)
- Age(): The age in years.
This data set can be obtained from the R package “mlbench”. The R codes for this sample analysis are given in Appendix B.
First, we cleaned the data set to remove 376 cases that contained incomplete covariate information. After data cleaning, 392 cases are kept as the final data set for modeling. In summary for the response variable in the cleaned sample, the diabetes rate is 0.332. Following the suggestion of Chiang et al. [], the “pressure”, “mass”, and “Age” are selected as the covariate variables of the second logit model, labeled by , and . The and are a m-dimensional vector with entry 1. Using the EM algorithm proposed by Chiang et al. [], the EM-ZIBer model was established and reported in Table 2.

Table 2.
The estimates of the model coefficients for the diabetes data set.
Chiang et al. [] suggest the best cutting probability of the ZIBer is for , which results in an accuracy of 80.6% for this data set. The logit model was also established for the diabetes data set and reported at the third column of Table 2. The best cutting probability of the typical logit model , , which results in the accuracy is 79.8%.
The ENR-ZIBer method with was established to compete with the logit model and the EM-ZIBer methods for the best model. The model coefficients of the ENR-ZIBer model are reported at the forth column of Table 2. As discussed in Section 4, the ENR-ZIBer method with includes 50 lasso, 50 ridge, and 50 elastic net penalty regularization models with and for the model selection to compete with the best model. The MLEs of the EM-ZIBer method were used as the initial solutions of the model parameters to implement the ENR-ZIBer method. Finally, we obtained the best model with the lowest error rate, which has . Moreover, the best cutting probability based on the ENR-ZIBer method is 0.45, which results in an error rate of 0.188 or accuracy 1 − 0.188 = 81.1%. The accuracy of the ENR-ZIBer model beats the logit model and the EM-ZIBer method. We also found that the Triceps() contributes less to the binary classification using the ENR-ZIB model than other covariates of .
6. Concluding Remarks
In this study, we established the loss function that is composed of the log-likelihood function of the ZIBer model and the - and -norm terms for penalty. The computation procedure to obtain reliable estimates of the proposed ENR-ZIBer method was also developed using the GDM to overcome the computation difficulties caused by the complicated target function for minimization. To accelerate the convergence of the GDM for zero-inflated data, the momentum term is used as the learning rate for the numerical computation to implement the proposed ENR-ZIBer method. We also find the ENR-ZIBer method with works well. This model can include the lasso and ridge regularization rules for a penalty in the pool to compete with the best model.
Monte Carlo simulations were conducted to evaluate the performance of the proposed method. Moreover, the performance of the proposed ENR-ZIBer method is compared with the EM-ZIBer method and the logit model. Simulation results show that the proposed ENR-ZIBer method with outperforms its competitors with the lowest MRE. We recommend using the ENR-ZIBer with for characterizing zero-inflated data with a feature selection consideration.
The proposed method fills the gap of the EM-ZIBer method proposed by Chiang et al. [] to reach a compromise of high accuracy and feature selection for the zero-inflated data set. Other feasible learning rates could have similar performance as the momentum term. Moreover, using the elastic net regulation rule to other zero-inflated models can be another important topic. These two topics will be studied in the near future.
Author Contributions
Conceptualization, investigation, writing, review and editing, and project administration: T.-R.T.; validation, investigation, and review and editing: Y.L.; methodology: H.X.; investigation: H.-C.C. and H.X.; funding acquisition: T.-R.T. and H.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science and Technology Council, Taiwan, grant number NSTC 112-2221-E-032-038-MY2; and Heilongjiang Provincial Natural Science Foundation, grant number LH2023D024.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The diabetes data set can be found from the R package “mlbench”.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Figures for Mean Error Rates
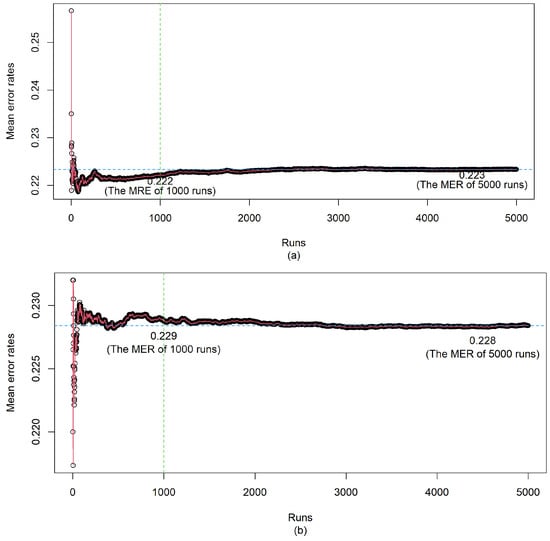
Figure A1.
The MERs from top to bottom for (a) and (b) .
Appendix B. R Codes for the Diabetes Example
rm(list=(ls(all=T)))
library(mlbench)
.
## 'data.frame': 768 obs. of 9 variables:
## $ pregnant: num 6 1 8 1 0 5 ...
## $ glucose : num 148 85 183 89 137 116 ...
## $ pressure: num 72 66 64 66 40 74 ...
## $ triceps : num 35 29 NA 23 35 ...
## $ insulin : num NA NA NA 94 168 NA ...
## $ mass : num 33.6 26.6 23.3 28.1 43.1 25.6 ...
## $ pedigree: num 0.627 0.351 0.672 ...
## $ age : num 50 31 32 21 33 30 4 ...
.
## $ diabetes: Factor w/ 2 levels "neg","pos": 2 1 2 1 2 1 2 1 2 2 ...
.
## ENR-ZIBER functions---------------------------------------
## mx is a n*(p+1) matrix, the first col. is 1's
## mz is a n*(m+1) matrix, the first col. is 1's
## y is a n*1 vector
## be.o: old beta; be.n: updated beta
## the.o: old theta; be.n: updated beta
## be.n: the vector of the gradient of ell w.r.t beta
## the.n: the vector of the gradient of ell w.r.t theta
## Random generate m combinations of lam1 and lam2
## lam1=lam*(1-alp)/2
## lam2=lam*alp
## m: the number of combinations of (lam1, lam2)
.
## Random generate m combinations of lam1 and lam2
## lam1=lam*(1-alp)/2
## lam2=lam*alp
## mm: the number of combinations of (lam1, lam2)
## estimating the parameters using elastic net regularization
## be0, the0: initial solutions of be and the
##
##-----------------------------------------------------------------
## Obtain the estimators of beta and the, and the est. errR ahd y.hat
##-----------------------------------------------------------------
.
## start function--------------------------------------------------
.
# scaling
minmax <- function(x) {
(x - min(x)) / (max(x) - min(x))
} # min-max scaling function
.
## return elastic net regularization target
## -(log-likelihood)+(lam1/2)(norm_2(beta))^2+lam2norm_1(beta)
## log-like= sum(yi*log(pi_i/(1-p_i)) + log(1-pi_i))
## proportional to sum(yi*log(pi_i/(1-p_i)) )
like.f<-function(be, the, lam1, lam2){
n=nrow(x)
log.lik=0
for(i in 1:n){
s.be=sum(be*x[i,])
s.the=sum(the*z[i,])
pi=exp(s.be)/((1+exp(s.the))*(1+exp(s.be)))
log.lik=log.lik+y[i]*log(pi/(1-pi))
}
return(round(-log.lik+(lam1/2)*(sum(be^2))+lam2*sum(abs(be)),5))
}
.
## the first devariates of the elastic net regularization target
dbe_the<-function(be, the, lam1, lam2){
## The 4 terms in the derivatives of ell
## input the ith rows of x, z; be.o, the.o
term.f<-function(xi, zi, be, the){
## exp. terms in the derivative of ell w.r.t. beta
s.be=sum(be*xi); s.the=sum(the*zi)
term1=exp(s.be+s.the)/(1+exp(s.the)+exp(s.be+s.the))
term2=exp(s.be)/(1+exp(s.be))
## exp. terms in the derivative of ell w.r.t. theta
term3=(exp(s.the)+exp(s.be+s.the))/(1+exp(s.the)+exp(s.be+s.the))
term4=exp(s.the)/(1+exp(s.the))
return(c(term1, term2, term3, term4))
}
.
## evaluate the two first derivates
dell.be=as.vector(rep(0,p+1))
dell.the=rep(0,m+1)
for(i in 1:nrow(x)){
terms.f=term.f(x[i,], z[i,], be, the)
dell.be=dell.be+(y[i]+(1-y[i])*terms.f[1]-terms.f[2])*x[i,]
dell.the=dell.the+((1-y[i])*terms.f[3]-terms.f[4])*z[i,]
}
.
## find the vector of lamda
lam2.v=sign(be)*rep(lam2, p+1)
dell.be=-dell.be+(lam1*sum(be))+lam2.v
dell.the=-dell.the
## export log-likelood-lam*sum(be) and log-likelood-kap*sum(the)
return(list(dell.be=dell.be, dell.the=dell.the))
}
.
## udate the parameter one time
update.f<-function(be, the, gam1, gam2, lam1, lam2){
grad<-dbe_the(be, the, lam1, lam2)
be.new<-be-gam1*grad$dell.be
the.new<-the-gam2*grad$dell.the
return(list(be.new=be.new,the.new=the.new))
}
.
## searching the optimal estimates using momentum updating
grad.des<-function(be.o, the.o, gam1, gam2, lam1, lam2, M){
nu.t1=nu.t2=0
be.new=be.o+nu.t1
the.new=the.o+nu.t2
for (i in 1:M){
grad.para=dbe_the(be.new, the.new, lam1, lam2)
nu.t1=0.9*nu.t1-gam1*grad.para$dell.be
nu.t2=0.9*nu.t2-gam2*grad.para$dell.the
be.new=be.new+nu.t1
the.new=the.new+nu.t2
loglike.o=like.f(be.o, the.o, lam1, lam2)
loglike.n=like.f(be.new, the.new, lam1, lam2)
err<-abs(loglike.n-loglike.o)
## cat(i, err, "")
if (err<0.001){break} ## -log.like increases or err is smaller than threshold
else{
be.o<-be.new; the.o<-the.new
}
}
return(list(be.best=round(be.new,4),the.best=round(the.new,4),
iter.f=i,loglike.f=loglike.n))
}
.
## best cut prob. and error rate
errR.f=function(x.tra, y.tra, z.tra, be.est, the.est,
x.tes, z.tes, y.tes){
n=nrow(x.tra)
pp.est=array()
for(i in 1:n){
s.be=exp(sum(be.est*x.tra[i,]))
s.the=exp(sum(the.est*z.tra[i,]))
pp.est[i]=round(s.be/((1+s.be)*(1+s.the)),3)
}
.
errR=array()
seq.v=seq(0.1, 0.7, 0.05);
for (jj in 1:length(seq.v)){
y.est=rep(0,n); y.est[which(pp.est>=seq.v[jj])]=1
errR[jj]=length(which(y.est!=y))/n
}
## take the minimal one if multiple values are available
best.cut=min(seq.v[which(errR==min(errR))], na.rm=TRUE)
.
nt=nrow(x.tes)
pp.est=array()
for(i in 1:nt){
s.be=exp(sum(be.est*x.tes[i,]))
s.the=exp(sum(the.est*z.tes[i,]))
pp.est[i]=round(s.be/((1+s.be)*(1+s.the)),3)
}
.
y.est=rep(0,nt); y.est[which(pp.est>=best.cut)]=1
errR.tes=round(length(which(y.est!=y.tes))/nt,4)
return(list(bestCutP=best.cut, y.est=y.est, errR.tes=errR.tes))
}
.
## end function----------------------------------------------
.
data("PimaIndiansDiabetes2", package = "mlbench")
.
Diabetes2 <- na.omit(PimaIndiansDiabetes2)
Diabetes2.sta=apply(Diabetes2[,c(-9)],2,minmax)
diabetes<-factor(Diabetes2[,c(9)])
Diabetes3=data.frame(Diabetes2.sta,diabetes)
xx=Diabetes3[,c(-9)]; xx=apply(xx,2,minmax)
zz=Diabetes3[,c(3,6,8)]; zz=apply(zz,2,minmax)
y=ifelse(Diabetes3$diabetes=="neg", 0, 1)
.
## x=xxtra; z=zztra; y=ytra
n=length(y)
cnst=rep(1,n)
x=data.frame(cnst, xx); z=data.frame(cnst, zz)
.
## parameters
## be: beta
## the: theta
## m: the number of randomly generated combinations of (lam1, lam2)
## gam1, gam2: constant learning rate
## M: max. number for the gradient decent algorithm
## iter: the repeatition of simulations
## seed.r: used for set.seed
.
gam1=0.01
gam2=0.01
.
M=500
k=150
p=ncol(x)-1
m=ncol(z)-1
.
be0=rep(1,ncol(x))
the0=rep(1,ncol(z))
.
## update parameters by gradient decent method
.
## Initial solutions of the parameters (random initial values)
## lasso, ridge, ENR
set.seed=123
lam=sample(seq(0, 1, 0.001), k)
seq1=sample(1:k, round(k/3))
seq2=sample(1:k, round(k/3))
seq3=sample(1:k, k-2*round(k/3))
lam.1=lam[seq1]
lam.2=lam[seq2]
lam.3=lam[seq3]
.
alp=0; penCoef1=cbind(lam.1*(1-alp)/2, lam.1*alp) ## lasso
alp=1; penCoef2=cbind(lam.2*(1-alp)/2, lam.2*alp) ## ridge
alp=0.5; penCoef3=cbind(lam.3*(1-alp)/2, lam.3*alp) ## ENR
.
penCoef=rbind(penCoef1,penCoef2,penCoef3)
.
## MLEs as initial values
## be.o=rnorm(length(be0)); the.o=rnorm(length(the0))
be.o=c(-7.562, 2.665, 6.931, 1.99, -0.19, 3.34,
5.246, 5.313, -2.863);
the.o=c(0.005, 2.232, -1.94, -10.494)
.
sink("Diabetes_ENR-ZIBerk150.lst")
iterBest=logLikeli=errR.v=bestCut_P=array()
beEst=array(dim=c(k, length(be0)))
theEst=array(dim=c(k, length(the0)))
for (i in 1:k){
aaa=grad.des(be.o, the.o, gam1, gam2, penCoef[i,1], penCoef[i,2], M)
.
be.best=as.numeric(aaa$be.best)
the.best=as.numeric(aaa$the.best)
.
iterBest[i]=aaa$iter.f
beEst[i,]=be.best
theEst[i,]=the.best
logLikeli[i]=aaa$loglike.f
errR.v[i]=errR.f(x, y, z, aaa$be.best, aaa$the.best, x, z, y)$errR.tes
cat(c(i, penCoef[i,1], penCoef[i,2], aaa$iter.f, be.best, the.best, aaa$loglike.n, errR.v[i]), "\n")
}
.
ind.errR=which(errR.v==min(errR.v)) ## using error rate
ind2=which(penCoef[ind.errR,2]==max(penCoef[ind.errR,2]))
ind=ind2
.
lam1.f=penCoef[ind,1]; lam2.f=penCoef[ind,2];
be.h=beEst[ind,]; the.h=theEst[ind,]
iter.best=iterBest[ind]
errR.best=errR.v[ind]
cat("\n")
cat(iter.best, lam1.f, lam2.f, be.h, the.h, errR.best, "\n")
cat("\n")
sink()
.
write.csv(iterBest, file="iterBestk150.csv", row.names = FALSE)
write.csv(beEst, file="beEstk150.csv", row.names = FALSE)
write.csv(theEst, file="theEstk150.csv", row.names = FALSE)
write.csv(errR.v, file="errR150.csv", row.names = FALSE)
References
- Diop, A.; Diop, A.; Dupuy, J.-F. Simulation-based inference in a zero-inflated Bernoulli regression model. Commun. Stat.-Simul. Comput. 2016, 45, 3597–3614. [Google Scholar] [CrossRef]
- Ridout, M.; Demétrio, C.G.B.; Hinde, J. Models for counts data with many zeros. In Proceedings of the XIXth International Biometric Conference, Cape Town, South Africa, 14–18 December 1998; Invited Papers. International Biometric Society: Cape Town, South Africa, 1998; pp. 179–192. [Google Scholar]
- Lambert, D. Zero-inflated Poisson regression, with an application to defects in manufacturing. Technometrics 1992, 34, 1–14. [Google Scholar] [CrossRef]
- Hall, D.B. Zero-inflated Poisson and binomial regression with random effects: A case study. Biometrics 2000, 56, 1030–1039. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.B. Zero-inflated models for regression analysis of count data: A study of growth and development. Stat. Med. 2002, 21, 1461–1469. [Google Scholar] [CrossRef] [PubMed]
- Gelfand, A.E.; Citron-Pousty, S. Zero-inflated models with application to spatial count data. Environ. Ecol. Stat. 2002, 9, 341–355. [Google Scholar]
- Rodrigues, J. Bayesian analysis of zero-inflated distributions. Commun.-Stat.-Theory Methods 2003, 32, 281–289. [Google Scholar] [CrossRef]
- Ghosh, S.K.; Mukhopadhyay, P.; Lu, J.C. Bayesian analysis of zero-inflated regression models. J. Stat. Plan. Inference 2006, 136, 1360–1375. [Google Scholar] [CrossRef]
- Harris, M.N.; Zhao, X. A zero-inflated ordered probit model, with an application to modelling tobacco consumption. J. Econom. 2007, 141, 1073–1099. [Google Scholar] [CrossRef]
- Loeys, T.; Moerkerke, B.; De Smet, O.; Buysse, A. The analysis of zero-inflated count data: Beyond zero-inflated Poisson regression. Br. J. Math. Stat. Psychol. 2011, 65, 163–180. [Google Scholar] [CrossRef]
- Diop, A.; Diop, A.; Dupuy, J.F. Maximum likelihood estimation in the logistic regression model with a cure fraction. Electron. J. Stat. 2011, 5, 460–483. [Google Scholar] [CrossRef]
- Staub, K.E.; Winkelmann, R. Consistent estimation of zero-inflated count models. Health Econ. 2012, 22, 673–686. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Tang, W.; Wang, W.; Crits-Christoph, P. Structural zeroes and zero-inflated models. Shanghai Arch. Psychiatry 2014, 26, 236–242. [Google Scholar] [PubMed]
- Zuur, A.F.; Ieno, E.N. Beginner’s Guide to Zero-Inflated Models with R; Highland Statistics Limited: Newburgh, NY, USA, 2016. [Google Scholar]
- Lee, S.M.; Pho, K.H.; Li, C.S. Validation likelihood estimation method for a zero-inflated Bernoulli regression model with missing covariates. J. Stat. Plan. Inference 2021, 214, 105–127. [Google Scholar] [CrossRef]
- Pho, K.H. Goodness of fit test for a zero-inflated Bernoulli regression model. Commun.-Stat.-Simul. Comput. 2024, 53, 756–771. [Google Scholar] [CrossRef]
- Li, C.S.; Lu, M. Semiparametric zero-inflated Bernoulli regression with applications. J. Appl. Stat. 2022, 49, 2845–2869. [Google Scholar] [CrossRef]
- Pho, K.H. Zero-inflated probit Bernoulli model: A new model for binary data. In Communications in Statistics-Simulation and Computation; Taylor & Francis: Abingdon, UK, 2023; pp. 1–21. [Google Scholar]
- Lu, M.; Li, C.S.; Wagner, K.D. Penalised estimation of partially linear additive zero-inflated Bernoulli regression models. J. Nonparametric Stat. 2024, 36, 863–890. [Google Scholar] [CrossRef]
- Chiang, J.-Y.; Lio, Y.L.; Hsu, C.-Y.; Ho, C.-L.; Tsai, T.R. Binary classification with imbalanced data. Entropy 2023, 26, 15. [Google Scholar] [CrossRef]
- Yu, X.-H.; Chen, G.A. Efficient backpropagation learning using optimal learning rate and momentum. Neural Netw. 1997, 10, 517–527. [Google Scholar] [CrossRef]
- Attoh-Okine, N.O. Analysis of learning rate and momentum term in backpropagation neural network algorithm trained to predict pavement performance. Adv. Eng. Softw. 1999, 30, 291–302. [Google Scholar] [CrossRef]
- Qian, N. On the momentum term in gradient descent learning algorithms. Neural Netw. 1999, 12, 145–151. [Google Scholar] [CrossRef]
- Wang, J.; Yang, J.; Wu, W. Convergence of cyclic and almost-cyclic learning with momentum for feedforward neural networks. IEEE Trans. Neural Netw. 2011, 22, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, Y.; Yin, W. An improved analysis of stochastic gradient descent with momentum. Adv. Neural Inf. Process. Syst. 2020, 33, 18261–18271. [Google Scholar]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Simon, N.; Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J. Stat. Softw. 2011, 39, 1–13. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).