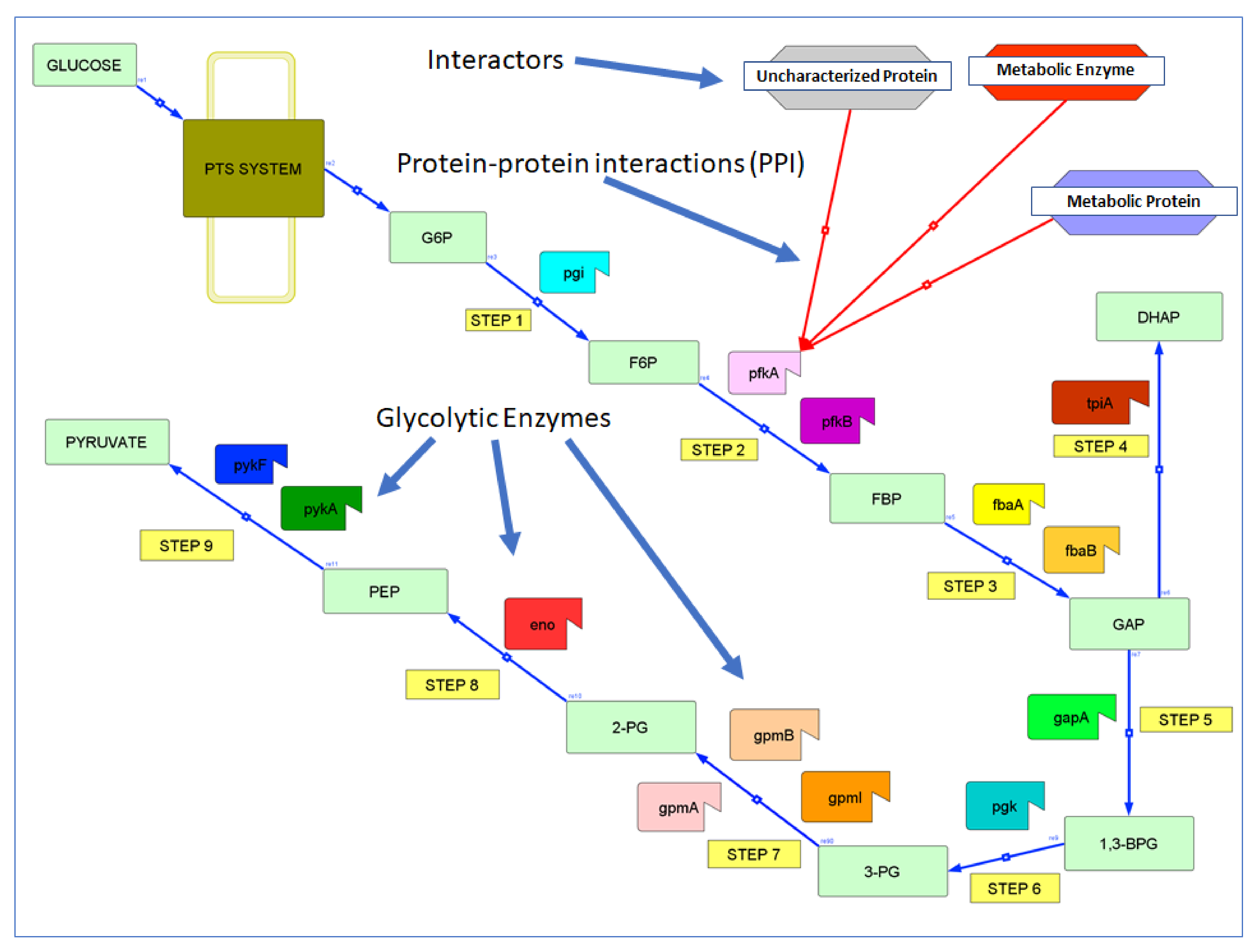
The Protein Interactome of Glycolysis in Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein and Interaction Data
2.1.1. PPIs from Intact
2.1.2. PPIs from String
2.2. Published Evidence for Protein-Protein Interactions Regulating Metabolism
2.3. Duplicate Interactions in Multiple Sources
2.4. Properties of Glycolytic Interactors
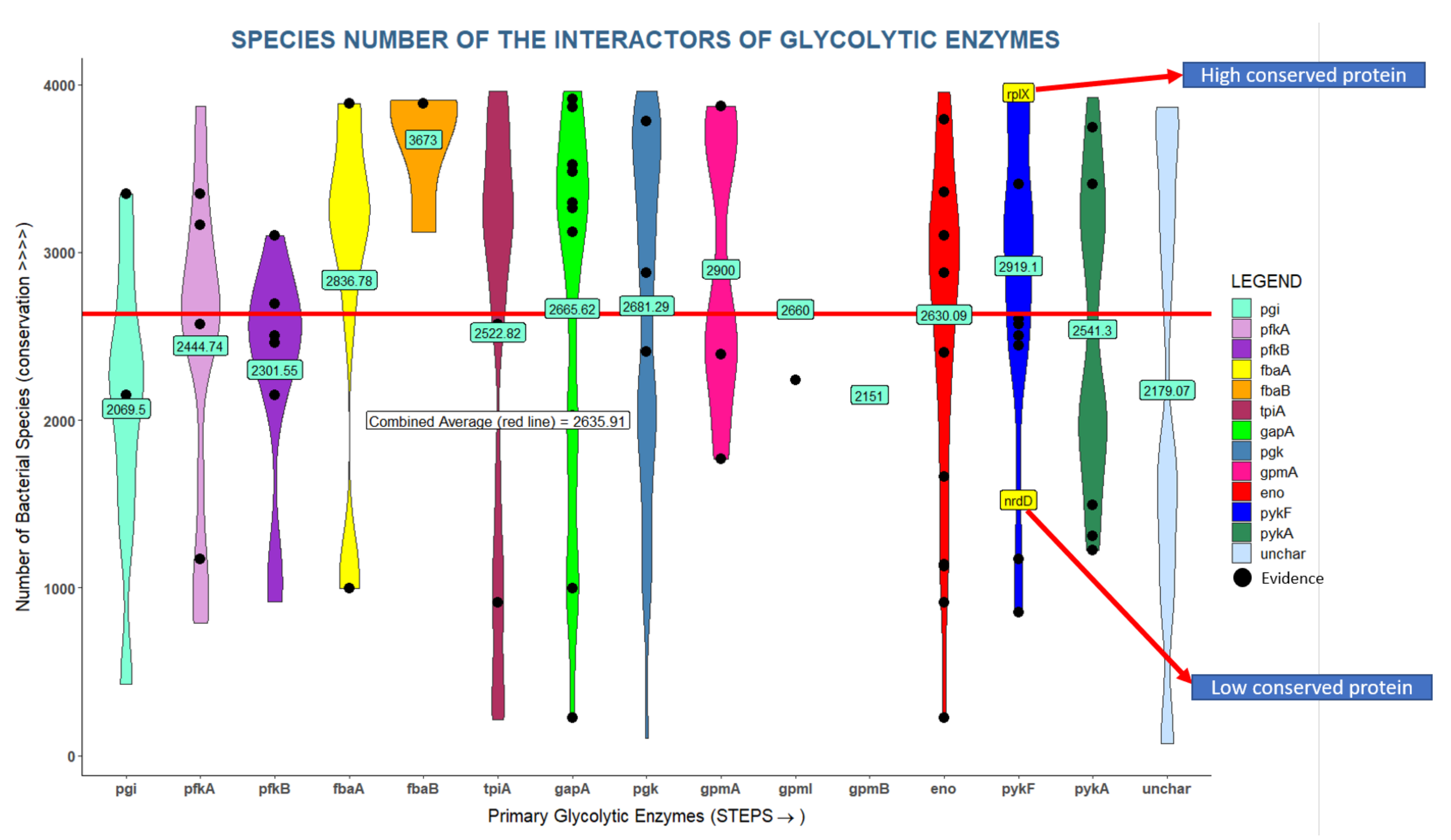
2.5. Conservation and Abundance of Glycolytic Interactors
3. Results
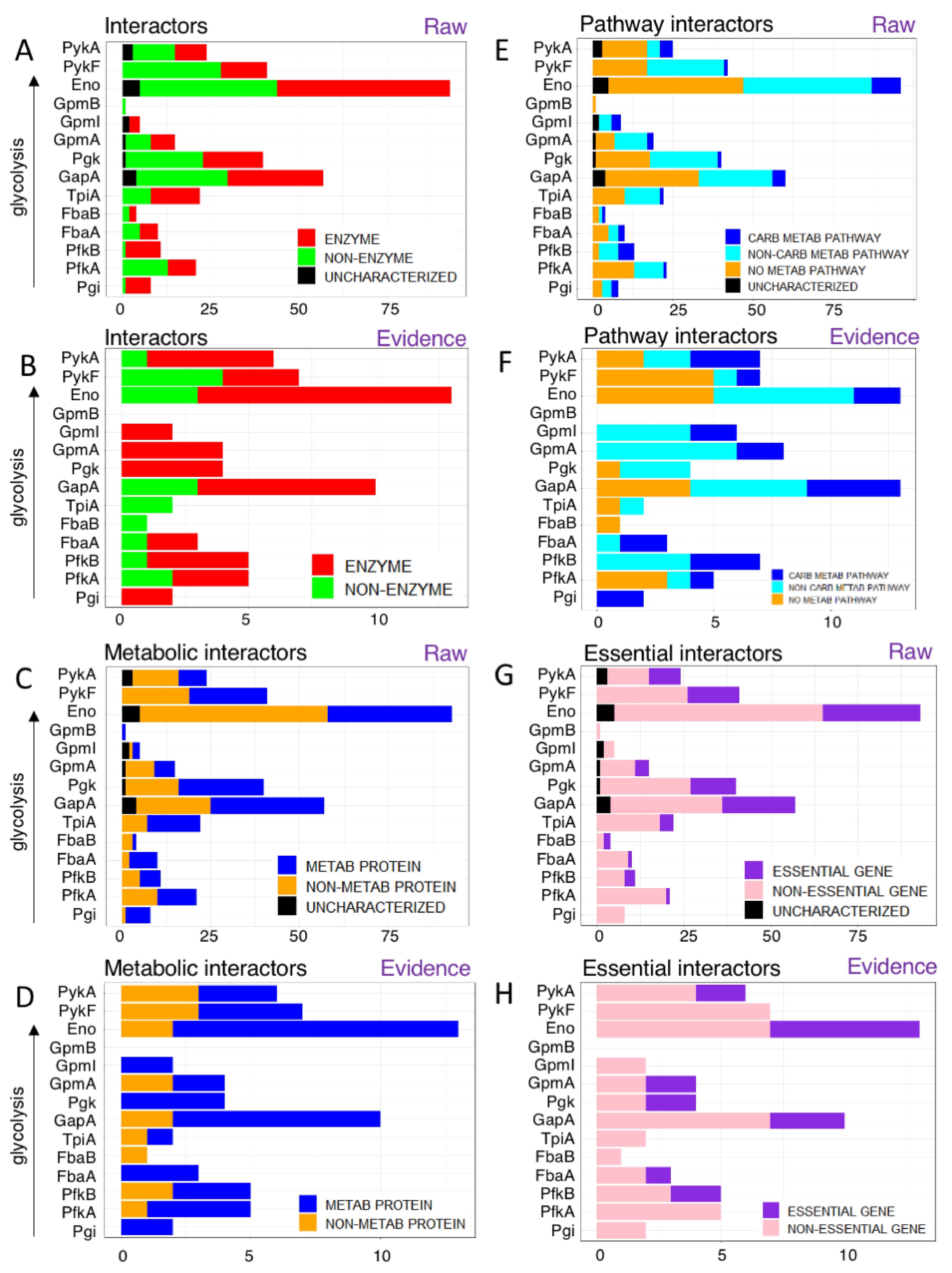
3.1. Glycolytic Enzymes Interact Very Specifically with Non-Glycolytic Proteins
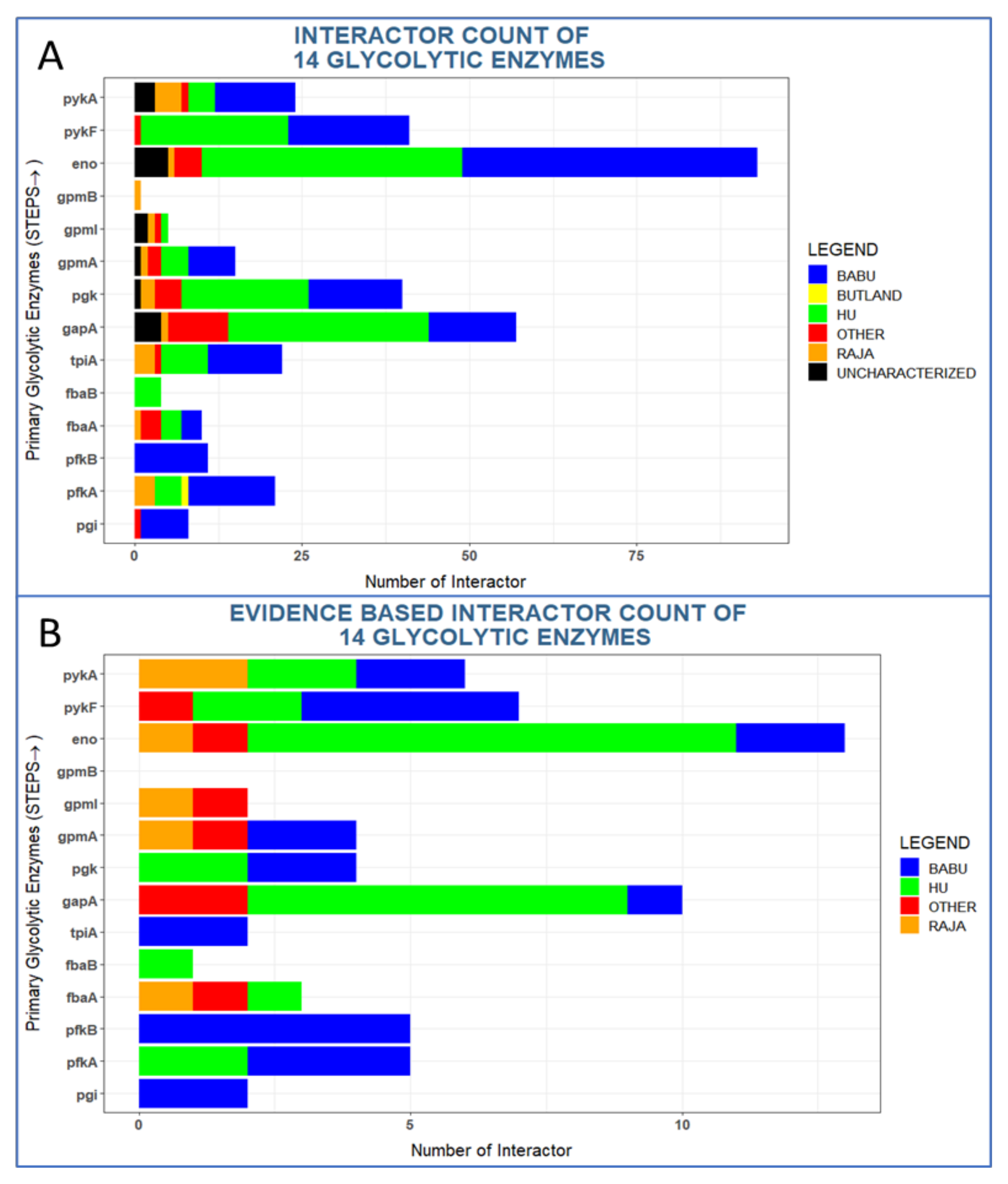
3.1.1. Glycolytic Enzymes Interact with Both Enzymatic and Non-Enzymatic Proteins
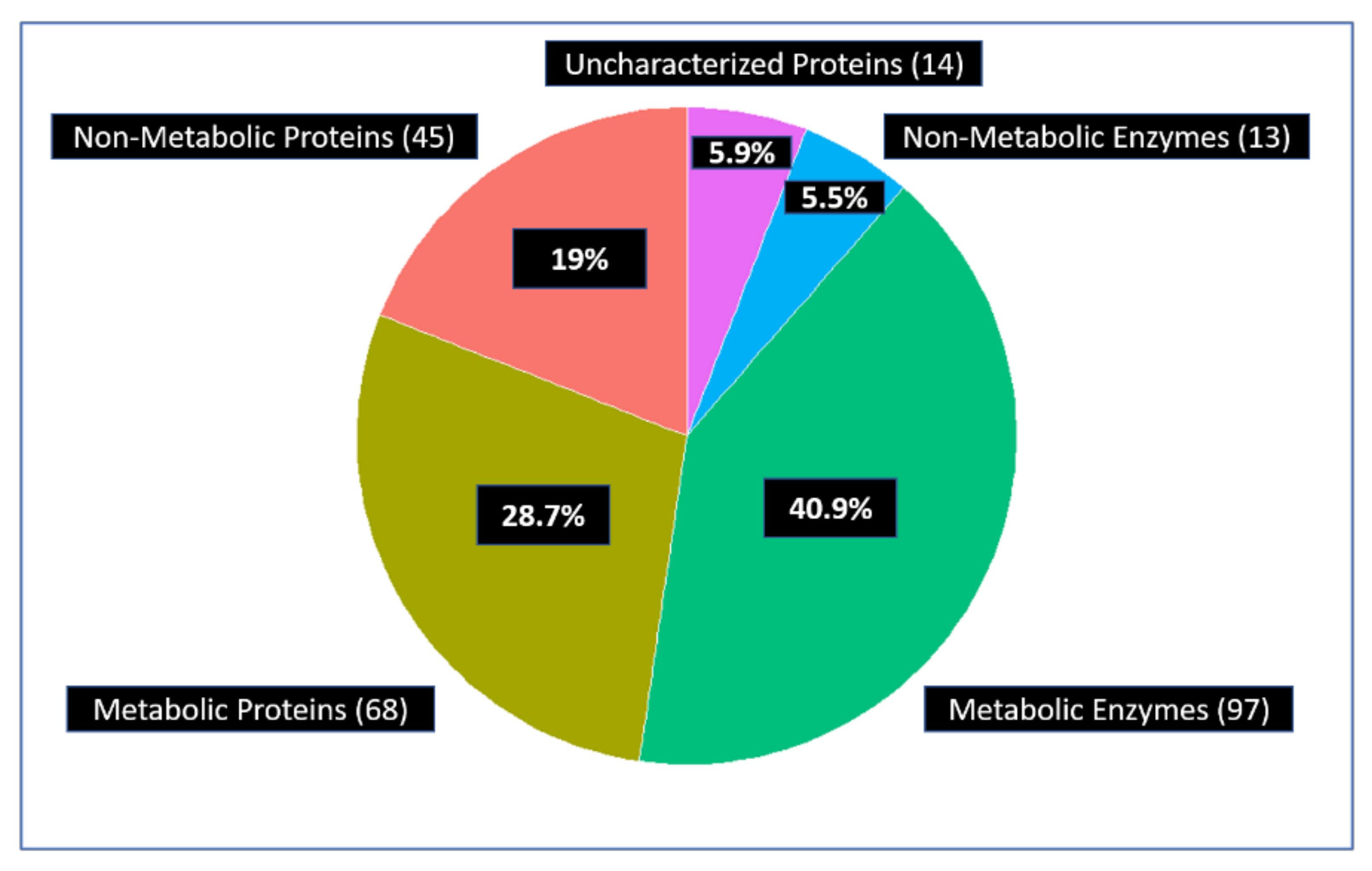
3.1.2. Glycolytic Enzymes Interact with Both Metabolic and Non-Metabolic Proteins
3.1.3. Glycolytic Enzymes Interact Primarily with Non-Essential Genes
3.1.4. Glycolytic Enzymes Interact Primarily with Metabolic Proteins
3.2. Many Interactors of Glycolytic Enzymes Are Highly Conserved
3.3. Most Glycolytic Enzymes Are More Abundant than Their Interactors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmidt, A.; Kochanowski, K.; Vedelaar, S.; Ahrné, E.; Volkmer, B.; Callipo, L.; Knoops, K.; Bauer, M.; Aebersold, R.; Heinemann, M. The Quantitative and Condition-Dependent Escherichia Coli Proteome. Nat. Biotechnol. 2016, 34, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Rajagopala, S.V.; Sikorski, P.; Kumar, A.; Mosca, R.; Vlasblom, J.; Arnold, R.; Franca-Koh, J.; Pakala, S.B.; Phanse, S.; Ceol, A.; et al. The Binary Protein-Protein Interaction Landscape of Escherichia Coli. Nat. Biotechnol. 2014, 32, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Janga, S.C.; Babu, M.; Díaz-Mejía, J.J.; Butland, G.; Yang, W.; Pogoutse, O.; Guo, X.; Phanse, S.; Wong, P.; et al. Global Functional Atlas of Escherichia Coli Encompassing Previously Uncharacterized Proteins. PLoS Biol. 2009, 7, e96. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, E.; Proudfoot, M.; Sanders, S.A.; Reinking, J.; Savchenko, A.; Arrowsmith, C.H.; Edwards, A.M.; Yakunin, A.F. Enzyme Genomics: Application of General Enzymatic Screens to Discover New Enzymes. FEMS Microbiol. Rev. 2005, 29, 263–279. [Google Scholar] [CrossRef]
- Anton, B.P.; Chang, Y.-C.; Brown, P.; Choi, H.-P.; Faller, L.L.; Guleria, J.; Hu, Z.; Klitgord, N.; Levy-Moonshine, A.; Maksad, A.; et al. The COMBREX Project: Design, Methodology, and Initial Results. PLoS Biol. 2013, 11, e1001638. [Google Scholar] [CrossRef]
- Gerosa, L.; Sauer, U. Regulation and Control of Metabolic Fluxes in Microbes. Curr. Opin. Biotechnol. 2011, 22, 566–575. [Google Scholar] [CrossRef]
- Pisithkul, T.; Patel, N.M.; Amador-Noguez, D. Post-Translational Modifications as Key Regulators of Bacterial Metabolic Fluxes. Curr. Opin. Microbiol. 2015, 24, 29–37. [Google Scholar] [CrossRef]
- Rodionova, I.A.; Goodacre, N.; Babu, M.; Emili, A.; Uetz, P.; Saier, M.H. The Nitrogen Regulatory PII Protein (GlnB) and N -Acetylglucosamine 6-Phosphate Epimerase (NanE) Allosterically Activate Glucosamine 6-Phosphate Deaminase (NagB) in Escherichia Coli. J. Bacteriol. 2017, 200, e00691-17. [Google Scholar] [CrossRef]
- Rodionova, I.A.; Zhang, Z.; Mehla, J.; Goodacre, N.; Babu, M.; Emili, A.; Uetz, P.; Saier, M.H. The Phosphocarrier Protein HPr of the Bacterial Phosphotransferase System Globally Regulates Energy Metabolism by Directly Interacting with Multiple Enzymes in Escherichia Coli. J. Biol. Chem. 2017, 292, 14250–14257. [Google Scholar] [CrossRef]
- Rodionova, I.A.; Goodacre, N.; Do, J.; Hosseinnia, A.; Babu, M.; Uetz, P.; Saier, M.H. The Uridylyltransferase GlnD and TRNA Modification GTPase MnmE Allosterically Control Escherichia Coli Folylpoly-γ-Glutamate Synthase FolC. J. Biol. Chem. 2018, 293, 15725–15732. [Google Scholar] [CrossRef]
- Ashkavand, Z.; O’Flanagan, C.; Hennig, M.; Du, X.; Hursting, S.D.; Krupenko, S.A. Metabolic Reprogramming by Folate Restriction Leads to a Less Aggressive Cancer Phenotype. Mol. Cancer Res. 2017, 15, 189–200. [Google Scholar] [CrossRef]
- Janga, S.C.; Díaz-Mejía, J.J.; Moreno-Hagelsieb, G. Network-Based Function Prediction and Interactomics: The Case for Metabolic Enzymes. Metab. Eng. 2011, 13, 1–10. [Google Scholar] [CrossRef]
- Aho, T.; Almusa, H.; Matilainen, J.; Larjo, A.; Ruusuvuori, P.; Aho, K.-L.; Wilhelm, T.; Lähdesmäki, H.; Beyer, A.; Harju, M.; et al. Reconstruction and Validation of RefRec: A Global Model for the Yeast Molecular Interaction Network. PLoS ONE 2010, 5, e10662. [Google Scholar] [CrossRef]
- Osada-Oka, M.; Goda, N.; Saiga, H.; Yamamoto, M.; Takeda, K.; Ozeki, Y.; Yamaguchi, T.; Soga, T.; Tateishi, Y.; Miura, K.; et al. Metabolic Adaptation to Glycolysis Is a Basic Defense Mechanism of Macrophages for Mycobacterium Tuberculosis Infection. Int. Immunol. 2019, 31, 781–793. [Google Scholar] [CrossRef]
- Yakandawala, N.; Romeo, T.; Friesen, A.D.; Madhyastha, S. Metabolic Engineering of Escherichia Coli to Enhance Phenylalanine Production. Appl. Microbiol. Biotechnol. 2008, 78, 283–291. [Google Scholar] [CrossRef]
- Kawaguchi, H.; Miyagawa, H.; Nakamura-Tsuruta, S.; Takaya, N.; Ogino, C.; Kondo, A. Enhanced Phenyllactic Acid Production in Escherichia Coli Via Oxygen Limitation and Shikimate Pathway Gene Expression. Biotechnol. J. 2019, 14, e1800478. [Google Scholar] [CrossRef]
- Emmerling, M.; Bailey, J.E.; Sauer, U. Glucose Catabolism of Escherichia Coli Strains with Increased Activity and Altered Regulation of Key Glycolytic Enzymes. Metab. Eng. 1999, 1, 117–127. [Google Scholar] [CrossRef]
- Sabnis, N.A.; Yang, H.; Romeo, T. Pleiotropic Regulation of Central Carbohydrate Metabolism in Escherichia Coli via the Gene CsrA. J. Biol. Chem. 1995, 270, 29096–29104. [Google Scholar] [CrossRef]
- García-Contreras, R.; Vos, P.; Westerhoff, H.V.; Boogerd, F.C. Why In Vivo May Not Equal In Vitro—New Effectors Revealed by Measurement of Enzymatic Activities under the Same In Vivo-like Assay Conditions. FEBS J. 2012, 279, 4145–4159. [Google Scholar] [CrossRef]
- Doan, T.; Martin, L.; Zorrilla, S.; Chaix, D.; Aymerich, S.; Labesse, G.; Declerck, N. A Phospho-Sugar Binding Domain Homologous to NagB Enzymes Regulates the Activity of the Central Glycolytic Genes Repressor. Proteins 2008, 71, 2038–2050. [Google Scholar] [CrossRef]
- van der Oost, J.; Huynen, M.A.; Verhees, C.H. Molecular Characterization of Phosphoglycerate Mutase in Archaea. FEMS Microbiol. Lett. 2002, 212, 111–120. [Google Scholar] [CrossRef]
- Johnsen, U.; Schönheit, P. Characterization of Cofactor-Dependent and Cofactor-Independent Phosphoglycerate Mutases from Archaea. Extremophiles 2007, 11, 647–657. [Google Scholar] [CrossRef]
- Bardey, V.; Vallet, C.; Robas, N.; Charpentier, B.; Thouvenot, B.; Mougin, A.; Hajnsdorf, E.; Régnier, P.; Springer, M.; Branlant, C. Characterization of the Molecular Mechanisms Involved in the Differential Production of Erythrose-4-Phosphate Dehydrogenase, 3-Phosphoglycerate Kinase and Class II Fructose-1,6-Bisphosphate Aldolase in Escherichia Coli. Mol. Microbiol. 2005, 57, 1265–1287. [Google Scholar] [CrossRef]
- Wang, M.; Herrmann, C.J.; Simonovic, M.; Szklarczyk, D.; von Mering, C. Version 4.0 of PaxDb: Protein Abundance Data, Integrated across Model Organisms, Tissues, and Cell-Lines. Proteomics 2015, 15, 3163–3168. [Google Scholar] [CrossRef]
- Butland, G.; Peregrín-Alvarez, J.M.; Li, J.; Yang, W.; Yang, X.; Canadien, V.; Starostine, A.; Richards, D.; Beattie, B.; Krogan, N.; et al. Interaction Network Containing Conserved and Essential Protein Complexes in Escherichia Coli. Nature 2005, 433, 531–537. [Google Scholar] [CrossRef]
- Babu, M.; Bundalovic-Torma, C.; Calmettes, C.; Phanse, S.; Zhang, Q.; Jiang, Y.; Minic, Z.; Kim, S.; Mehla, J.; Gagarinova, A.; et al. Global Landscape of Cell Envelope Protein Complexes in Escherichia Coli. Nat. Biotechnol. 2018, 36, 103–112. [Google Scholar] [CrossRef]
- Shatsky, M.; Allen, S.; Gold, B.L.; Liu, N.L.; Juba, T.R.; Reveco, S.A.; Elias, D.A.; Prathapam, R.; He, J.; Yang, W.; et al. Bacterial Interactomes: Interacting Protein Partners Share Similar Function and Are Validated in Independent Assays More Frequently Than Previously Reported. Mol. Cell Proteomics 2016, 15, 1539–1555. [Google Scholar] [CrossRef]
- van Bloois, E.; Dekker, H.L.; Fröderberg, L.; Houben, E.N.G.; Urbanus, M.L.; de Koster, C.G.; de Gier, J.-W.; Luirink, J. Detection of Cross-Links between FtsH, YidC, HflK/C Suggests a Linked Role for These Proteins in Quality Control upon Insertion of Bacterial Inner Membrane Proteins. FEBS Lett. 2008, 582, 1419–1424. [Google Scholar] [CrossRef]
- Houry, W.A.; Frishman, D.; Eckerskorn, C.; Lottspeich, F.; Hartl, F.U. Identification of In Vivo Substrates of the Chaperonin GroEL. Nature 1999, 402, 147–154. [Google Scholar] [CrossRef]
- Kumar, J.K.; Tabor, S.; Richardson, C.C. Proteomic Analysis of Thioredoxin-Targeted Proteins in Escherichia Coli. Proc. Natl. Acad. Sci. USA 2004, 101, 3759–3764. [Google Scholar] [CrossRef]
- Häuser, R.; Ceol, A.; Rajagopala, S.V.; Mosca, R.; Siszler, G.; Wermke, N.; Sikorski, P.; Schwarz, F.; Schick, M.; Wuchty, S.; et al. A Second-Generation Protein-Protein Interaction Network of Helicobacter Pylori. Mol. Cell Proteomics 2014, 13, 1318–1329. [Google Scholar] [CrossRef] [PubMed]
- Lasserre, J.-P.; Beyne, E.; Pyndiah, S.; Lapaillerie, D.; Claverol, S.; Bonneu, M. A Complexomic Study of Escherichia Coli Using Two-Dimensional Blue Native/SDS Polyacrylamide Gel Electrophoresis. Electrophoresis 2006, 27, 3306–3321. [Google Scholar] [CrossRef] [PubMed]
- Le, H.-T.; Gautier, V.; Kthiri, F.; Malki, A.; Messaoudi, N.; Mihoub, M.; Landoulsi, A.; An, Y.J.; Cha, S.-S.; Richarme, G. YajL, Prokaryotic Homolog of Parkinsonism-Associated Protein DJ-1, Functions as a Covalent Chaperone for Thiol Proteome. J. Biol. Chem. 2012, 287, 5861–5870. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Zhang, R.; Ou, H.-Y.; Zhang, C.-T. DEG: A Database of Essential Genes. Nucleic Acids Res. 2004, 32, D271–D272. [Google Scholar] [CrossRef]
- Funahashi, A.; Morohashi, M.; Kitano, H.; Tanimura, N. CellDesigner: A Process Diagram Editor for Gene-Regulatory and Biochemical Networks. BIOSILICO 2003, 1, 159–162. [Google Scholar] [CrossRef]
- Mora, C.; Tittensor, D.P.; Adl, S.; Simpson, A.G.B.; Worm, B. How Many Species Are There on Earth and in the Ocean? PLoS Biol. 2011, 9, e1001127. [Google Scholar] [CrossRef]
- Schwikowski, B.; Uetz, P.; Fields, S. A Network of Protein-Protein Interactions in Yeast. Nat. Biotechnol. 2000, 18, 1257–1261. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Boscá, L.; Corredor, C. Is Phosphofructokinase the Rate-Limiting Step of Glycolysis? Trends Biochem. Sci. 1984, 9, 372–373. [Google Scholar] [CrossRef]

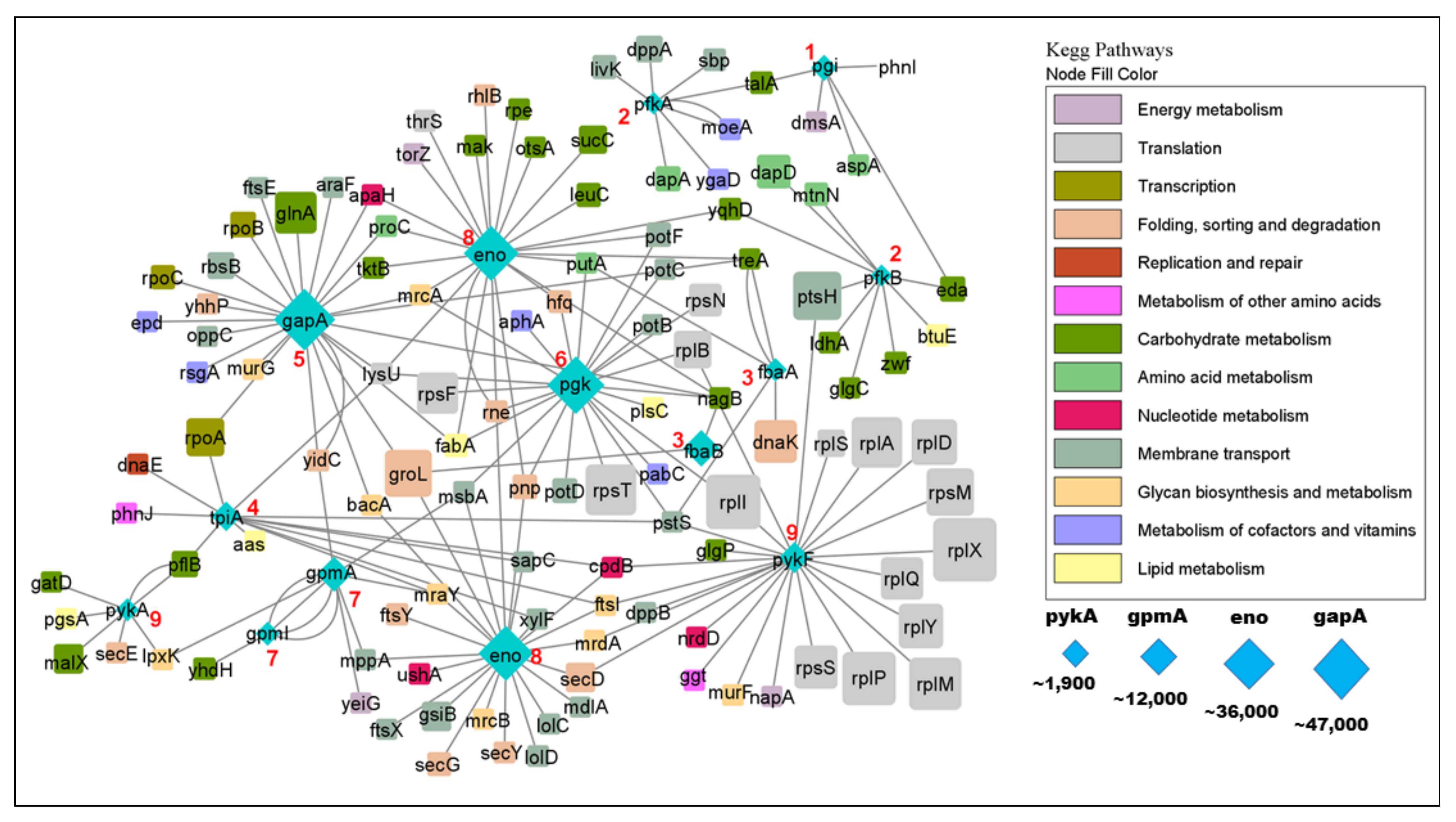
| Step | Enzyme | Uniprot | Full Name | Substrate | Char. IPs | Unchar. IPs |
|---|---|---|---|---|---|---|
| 1 | Pgi | P0A6T1 | Glucose-6-phosphate isomerase | Glucose-6-phosphate (G6P) | 8 | 0 |
| 2 | PfkA | P0A796 | ATP-dependent 6-phosphofructokinase 1 | Fructose-6-phosphate (F6P) | 19 | 0 |
| 2 | PfkB | P06999 | ATP-dependent 6-phosphofructokinase 2 | Fructose-6-phosphate (F6P) | 11 | 0 |
| 3 | FbaA | P0AB71 | Fructose-bisphosphate aldolase class 2 | Fructose-1,6-bisphosphate (FBP) | 9 | 0 |
| 3 | FbaB | P0A991 | Fructose-bisphosphate aldolase class 1 | Fructose-1,6-bisphosphate (FBP) | 4 | 0 |
| 4 | TpiA | P0A858 | Triosephosphate isomerase | Glyceraldehyde-3-phosphate (GAP) | 22 | 0 |
| 5 | GapA | P0A9B2 | Glyceraldehyde-3-phosphate dehydrogenase A | Glyceraldehyde-3-phosphate (GAP) | 52 | 4 |
| 6 | Pgk | P0A799 | Phosphoglycerate kinase | 1,3-bisphosphoglycerate (1,3-BPG) | 38 | 1 |
| 7 | GpmA | P62707 | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | 3-phosphoglycerate (3-PG) | 13 | 1 |
| 7 | GpmI | P37689 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | 3-phosphoglycerate (3-PG) | 1 | 2 |
| 7 | GpmB | P0A7A2 | Probable phosphoglycerate mutase | 3-phosphoglycerate (3-PG) | 1 | 0 |
| 8 | Eno | P0A6P9 | Enolase | 2-phosphoglycerate (2-PG) | 85 | 5 |
| 9 | PykA | P21599 | Pyruvate kinase II | Phosphoenolpyruvate (PEP) | 20 | 2 |
| 9 | PykF | P0AD61 | Pyruvate kinase I | Phosphoenolpyruvate (PEP) | 41 | 0 |
| Scope | Source | PPIs (Count) | PPIs (%) | REF |
|---|---|---|---|---|
| E. coli complexes | Butland et al. | 1 | 0.3 | [25] |
| E. coli complexes | Hu et al. | 148 | 42 | [3] |
| Cell envelope complexes | Babu et al. | 156 | 44.3 | [26] |
| Binary PPIs (Y2H) | Rajagopala et al. | 20 | 5.7 | [2] |
| Bacterial Interactome comparison | Shatsky et al. | 6 | 1.7 | [27] |
| Bacterial inner membrane proteins | Bloois et al. | 1 | 0.3 | [28] |
| Chaperonin GroEL substrates | Houry et al. | 2 | 0.6 | [29] |
| Thioredoxin-targeted proteins | Kumar et al. | 5 | 1.4 | [30] |
| Helicobacter pylori/E. coli PPIs | Hauser et al. | 8 | 2.3 | [31] |
| E. coli complexes native/SDS-PAGE | Lasserre et al. | 3 | 0.9 | [32] |
| YajL and the thiol proteome | Le et al. | 2 | 0.6 | [33] |
| Step | Enzyme | Uniprot ID | Interactor | Uniprot ID | Literature Source |
|---|---|---|---|---|---|
| 2 | PfkA | P0A796 | MoeA | P12281 | Butland/Rajagopala [25,2] |
| 2 | PfkA | P0A796 | UcpA | P37440 | Hu/Rajagopala [3,2] |
| 3 | FbaA | P0AB71 | TreA | P13482 | Hu/Shatsky [3,27] |
| 5 | GapA | P0A9B2 | YidC | P25714 | Babu/Bloois [26,28] |
| 6 | Pgk | P0A799 | Usg | P08390 | Rajagopala/Lasserre [2,32] |
| 7 | GpmA | P62707 | NagC | P0AF20 | Hu/Shatsky [3,27] |
| 7 | GpmA | P62707 | GpmI | P37689 | Rajagopala/Lasserre [2,32] |
| 7 | GpmI | P37689 | GpmA | P62707 | Rajagopala/Lasserre [2,32] |
| 8 | Eno | P0A6P9 | Rne | P21513 | Hu/Rajagopala/Shatsky [3,2,27] |
| 8 | Eno | P0A6P9 | Pnp | P05055 | Hu/Shatsky [3,27] |
| 9 | PykA | P21599 | PflB | P09373 | Hu/Rajagopala [3,2] |
| 9 | PykA | P21599 | YggR (UP) | P52052 | Hu/Rajagopala [3,2] |
| Parameter | COUNT |
|---|---|
| Glycolytic enzymes | 14 |
| Unique protein-protein interactions (PPI) | 339 |
| Evidence based protein-protein interactions | 58 (note A) |
| Unique protein interactors | 237 |
| Reproducible protein-protein interactions | 13 (note B) |
| Uncharacterized proteins | 14 |
| PPIs involving uncharacterized proteins (UP)s | 15 (note C) |
| Classification | Enzyme | Metabolic Protein |
|---|---|---|
| Metabolic Enzyme (ME) | X | X |
| Metabolic Protein (MP) | X | |
| Non-Metabolic Enzyme (NME) | X | |
| Non-Metabolic Protein (NMP) |
| Glycolytic Enzyme | Uncharacterized Interactor | Uniprot ID | Filtering Criteria |
|---|---|---|---|
| PykA | YodC (60 aa peptide) | P64517 | Top 10% abundance |
| Eno | YbcJ (70 aa peptide) | P0AAS7 | Top 10% abundance |
| GapA | YiiD (an acyl transferase—may initiate fatty acid biosynthesis | P0ADQ2 | Top 10% conservation |
| Pgk | YhhS (an MFS transporter-like YfeJ, an exporter of arabinose and herbicides | P37621 | Top 10% conservation |
| Eno | YadG (part of an ABC exporter (TC# 3.A.1.105.17)ss | P36879 | Top 10% conservation |
| GpmI | YegV (a sugar kinase) | P76419 | Top 10% conservation |
| GapA | YdjL (a zinc ADH) | P77539 | Multiple glycolytic interactor |
| Eno | YdjL (a zinc ADH) | P77539 | Multiple glycolytic interactor |
| PykA | YggR a pilus biogenesis ATPase) | P52052 | Found in two studies |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chowdhury, S.; Hepper, S.; Lodi, M.K.; Saier, M.H., Jr.; Uetz, P. The Protein Interactome of Glycolysis in Escherichia coli. Proteomes 2021, 9, 16. https://doi.org/10.3390/proteomes9020016
Chowdhury S, Hepper S, Lodi MK, Saier MH Jr., Uetz P. The Protein Interactome of Glycolysis in Escherichia coli. Proteomes. 2021; 9(2):16. https://doi.org/10.3390/proteomes9020016
Chicago/Turabian StyleChowdhury, Shomeek, Stephen Hepper, Mudassir K. Lodi, Milton H. Saier, Jr., and Peter Uetz. 2021. "The Protein Interactome of Glycolysis in Escherichia coli" Proteomes 9, no. 2: 16. https://doi.org/10.3390/proteomes9020016
APA StyleChowdhury, S., Hepper, S., Lodi, M. K., Saier, M. H., Jr., & Uetz, P. (2021). The Protein Interactome of Glycolysis in Escherichia coli. Proteomes, 9(2), 16. https://doi.org/10.3390/proteomes9020016