Histone Sample Preparation for Bottom-Up Mass Spectrometry: A Roadmap to Informed Decisions
Abstract
1. Introduction
2. Materials and Methods
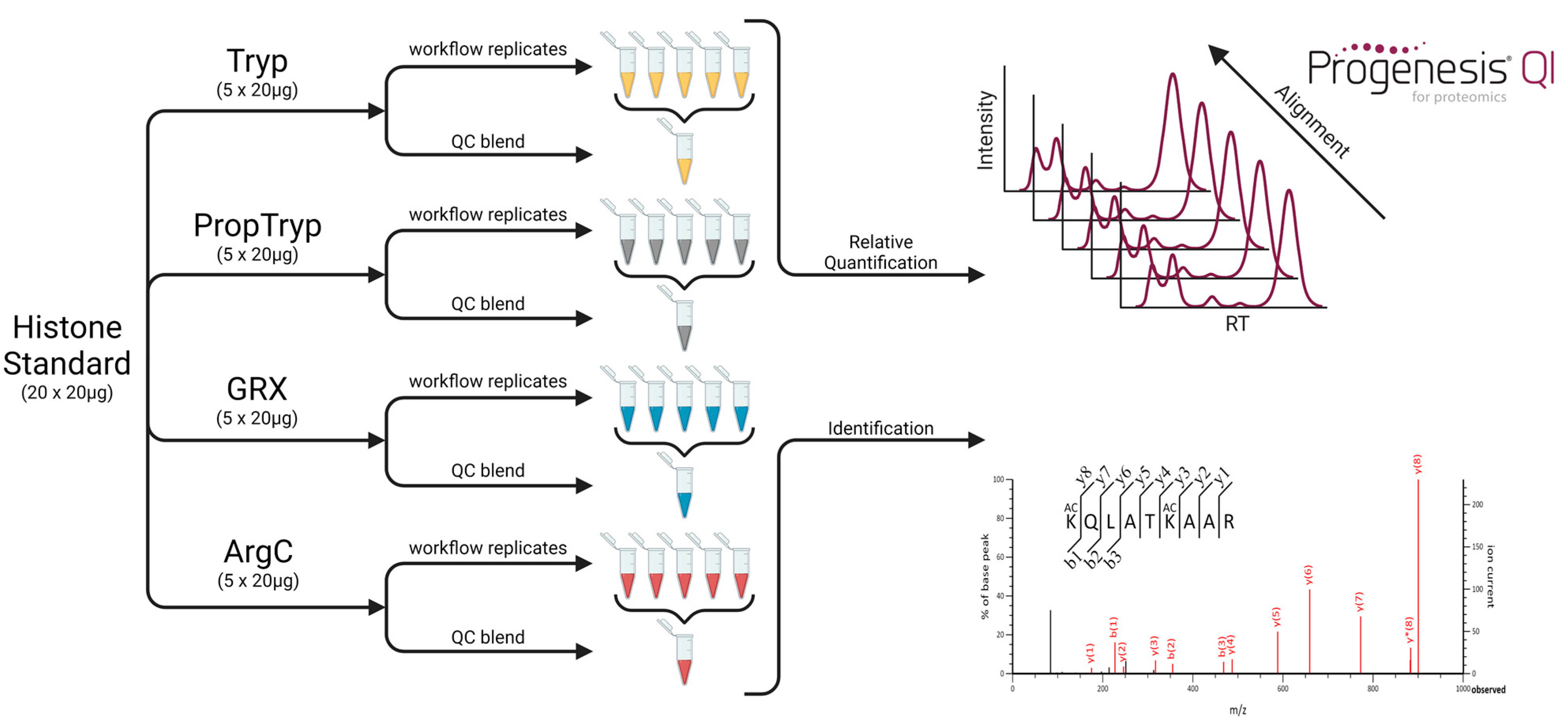
2.1. Sample Preparation
2.2. Sample Acquisition
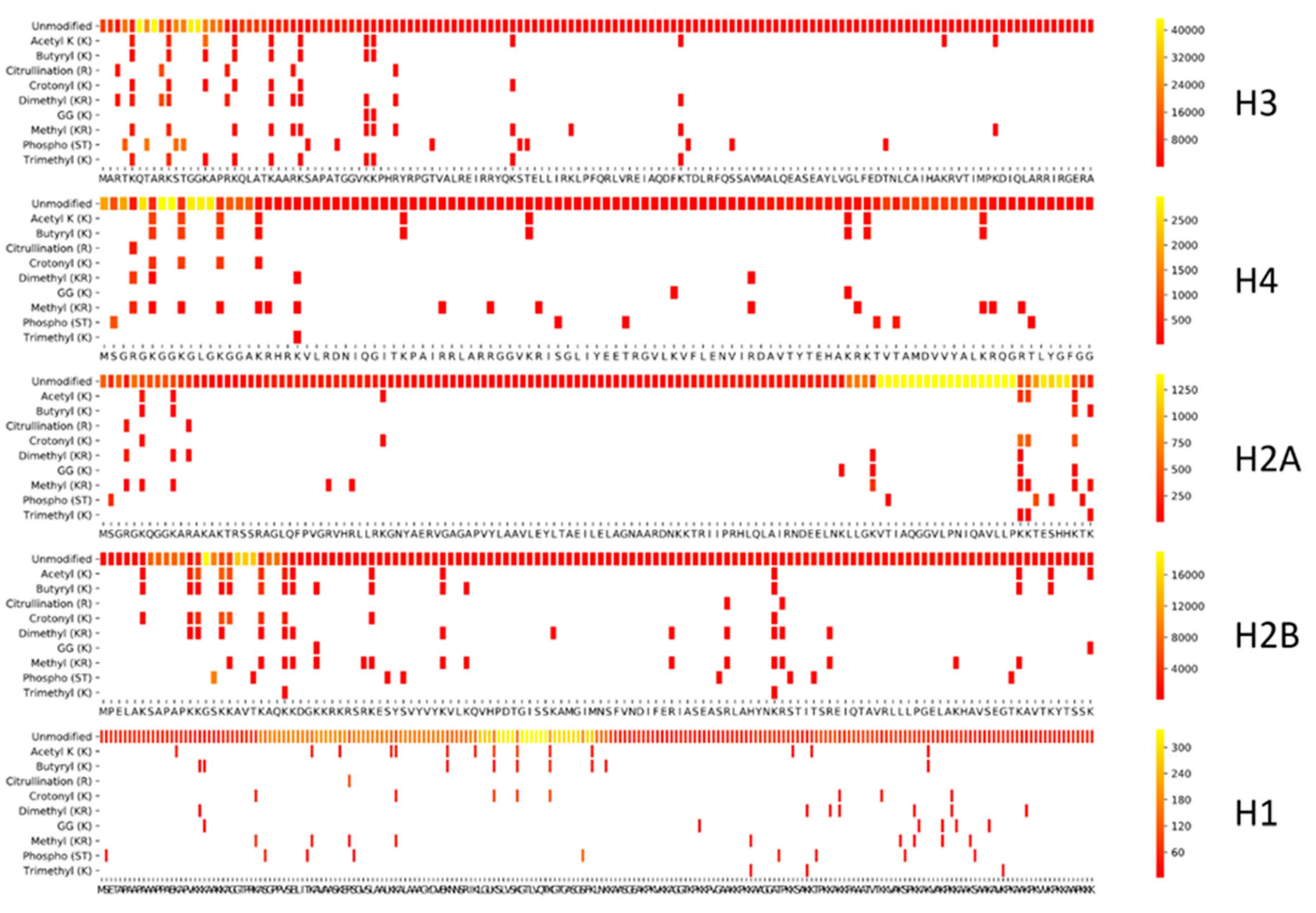
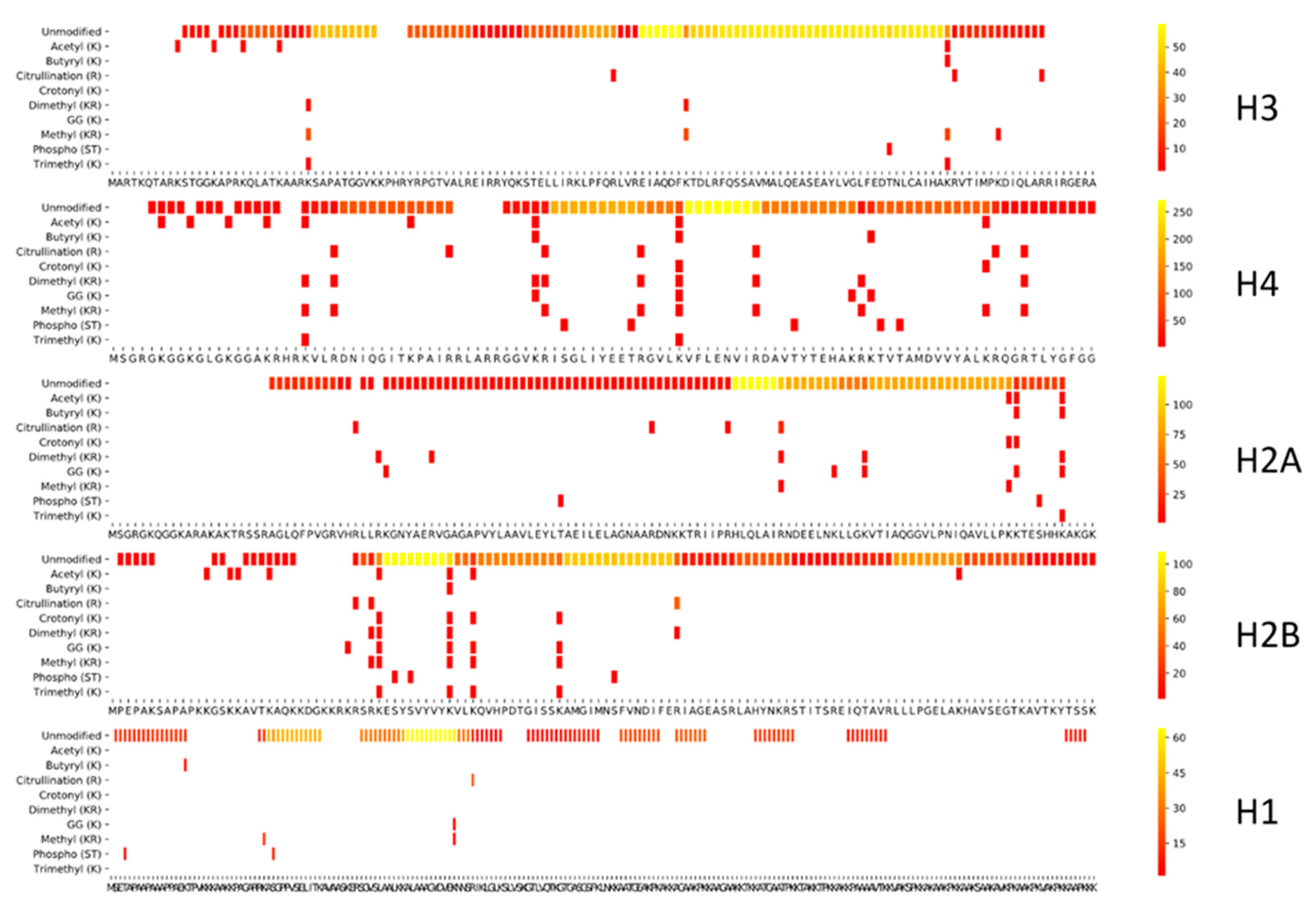
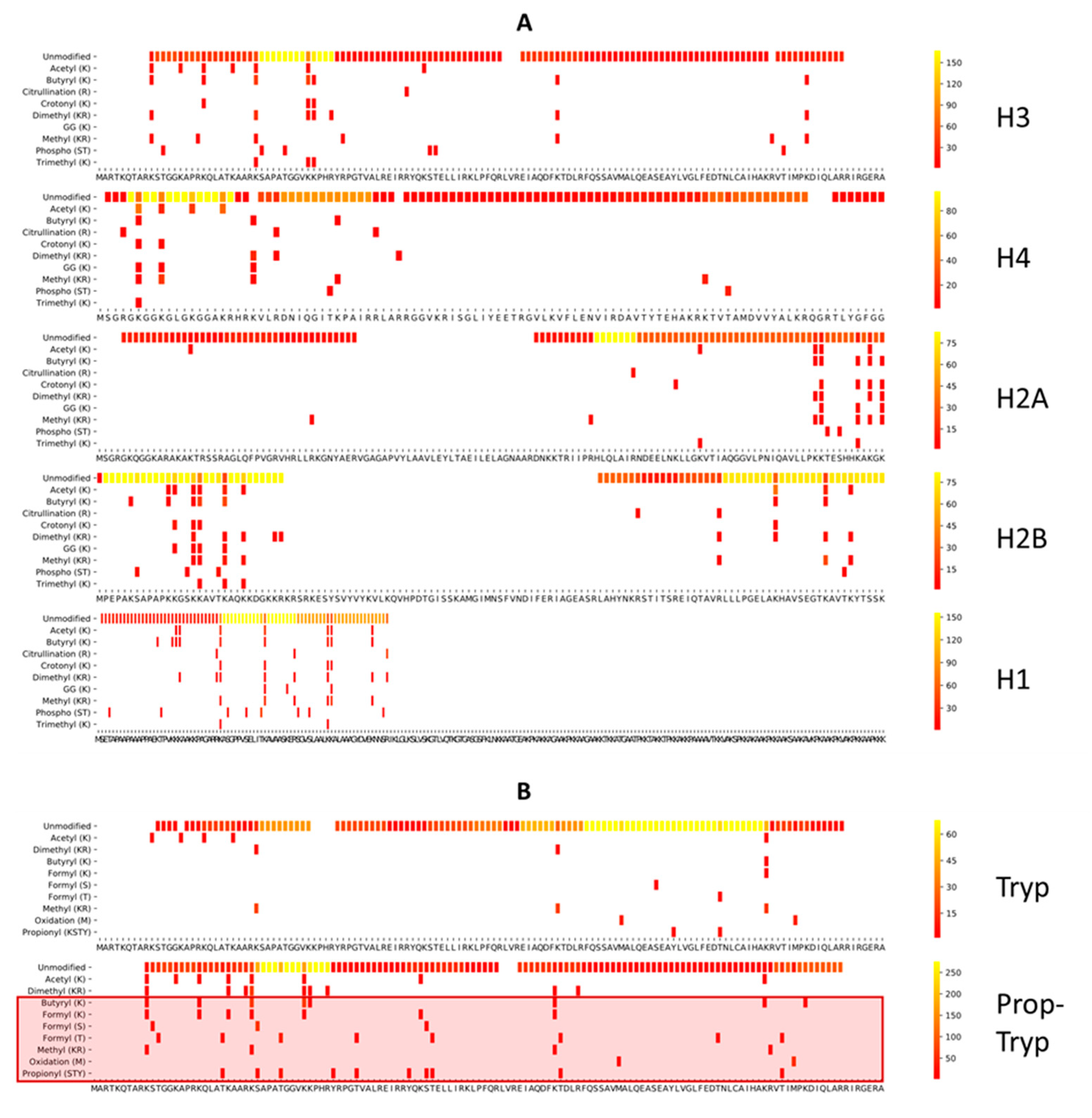
2.3. Coverage Plots
2.4. Quantitative Analysis with Progenesis QIP
2.5. Variant Calling
2.6. Data Availability
3. Results
3.1. The Histone Code Coverage with Tryptic Specificity
3.1.1. A Theoretical Tryptic Histone Snapshot
3.1.2. Theory Versus Practice
3.1.3. Exploring Other Options
3.2. Comparing Three Different Enzymatic Treatments with ArgC Specificity
3.2.1. Side Reactions and Chemical Noise
3.2.2. Histone Variant Calling
3.2.3. Feature Detectability
3.2.4. Enzyme Specificity and Efficiency
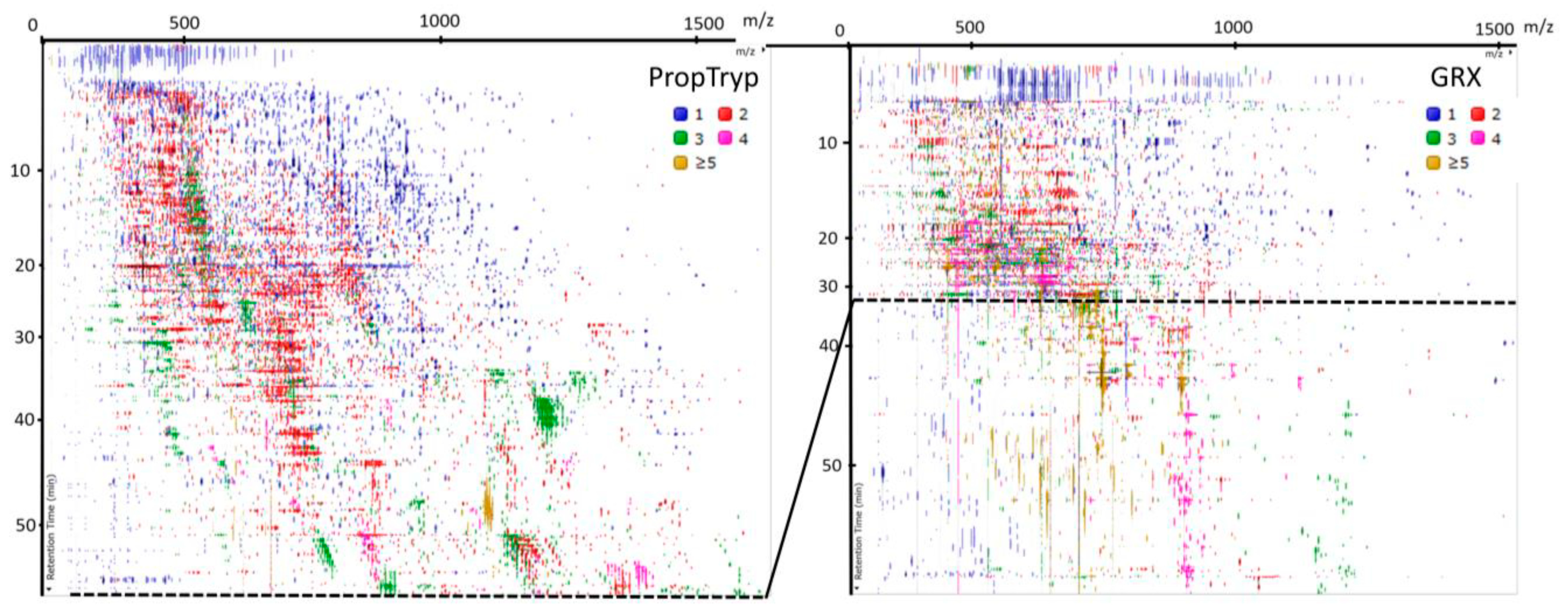
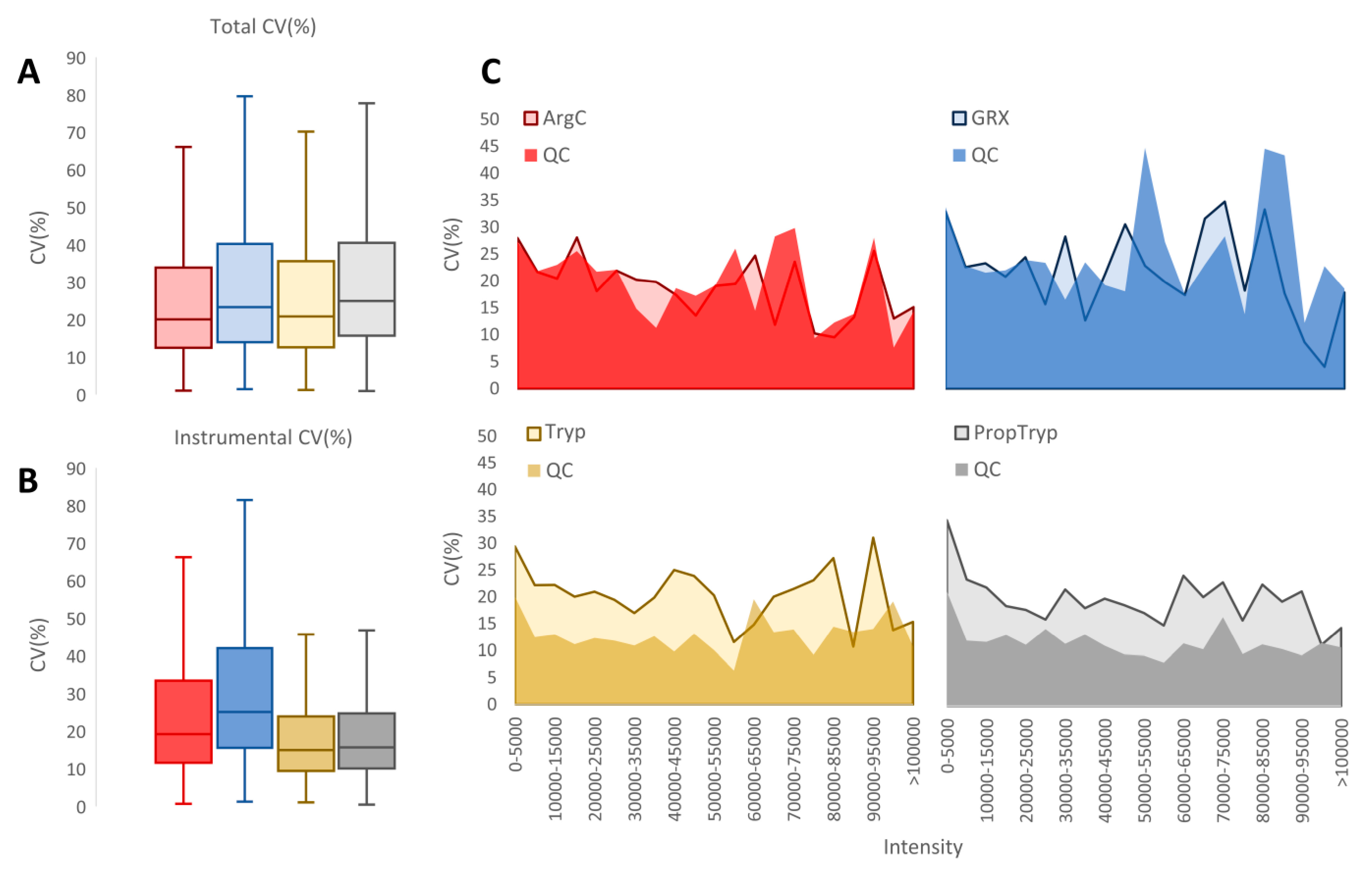
3.3. Workflow Variability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brunk, C.F.; Martin, W.F. Archaeal Histone Contributions to the Origin of Eukaryotes. Trends Microbiol. 2019, 27, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Henikoff, S. Phylogenomics of the nucleosome. Nat. Struct. Mol. Biol. 2003, 10, 882–891. [Google Scholar] [CrossRef]
- Henneman, B.; van Emmerik, C.; van Ingen, H.; Dame, R.T. Structure and Function of Archaeal Histones. PLoS Genet. 2018, 14, e1007582. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Katada, S.; Imhof, A.; Sassone-Corsi, P. Connecting Threads: Epigenetics and Metabolism. Cell 2012, 148, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.S.; Koutelou, E.; Schibler, A.C.; Dent, S.Y.R. Histone-modifying enzymes: Regulators of developmental decisions and driv-ers of human disease. Epigenomics 2012, 4, 163–177. [Google Scholar] [CrossRef]
- Huang, H.; Lin, S.; Garcia, B.A.; Zhao, Y. Quantitative Proteomic Analysis of Histone Modifications. Chem. Rev. 2015, 115, 2376–2418. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Huang, X.; Kelleher, N.L. Epiproteomics: Quantitative analysis of histone marks and codes by mass spectrometry. Curr. Opin. Chem. Biol. 2016, 33, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Janssen, K.; Sidoli, S.; Garcia, B. Recent Achievements in Characterizing the Histone Code and Approaches to Integrating Epigenomics and Systems Biology. Methods Enzymol. 2017, 586, 359–378. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.; Scotcher, J.; Wu, S.; Chu, R.K.; Tolić, N.; Ntai, I.; Thomas, P.M.; Fellers, R.T.; Early, B.P.; Zheng, Y.; et al. The first pilot project of the consortium for top-down proteomics: A status report. Proteomics 2014, 14, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Coradin, M.; Mendoza, M.R.; Sidoli, S.; Alpert, A.J.; Lu, C.; Garcia, B.A. Bullet points to evaluate the performance of the mid-dle-down proteomics workflow for histone modification analysis. Methods 2020, 184, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Sidoli, S.; Garcia, B.A. Middle-down proteomics: A still unexploited resource for chromatin biology. Expert Rev. Proteom. 2017, 14, 617–626. [Google Scholar] [CrossRef]
- Moradian, A.; Kalli, A.; Sweredoski, M.J.; Hess, S. The top-down, middle-down, and bottom-up mass spectrometry approaches for characterization of histone variants and their post-translational modifications. Proteomics 2013, 14, 489–497. [Google Scholar] [CrossRef]
- Krueger, R.; Hobbs, T.; Mihal, K.; Tehrani, J.; Zeece, M.G. Analysis of endoproteinase Arg C action on adrenocortico-trophic hormone by capillary electrophoresis and reversed-phasse high-performance liquid chromatography. J. Chromatogr. A 1991, 543, 451–461. Available online: https://www.sciencedirect.com/science/article/pii/S0021967301957966 (accessed on 9 February 2021). [CrossRef]
- Garcia, B.; Mollah, S.; Ueberheide, B.M.; Busby, S.; Muratore, T.L.; Shabanowitz, J.; Hunt, D.F. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc. 2007, 2, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Meert, P.; Govaert, E.; Scheerlinck, E.; Dhaenens, M.; Deforce, D. Pitfalls in histone propionylation during bottom-up mass spec-trometry analysis. Proteomics 2015, 15, 2966–2971. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26010583 (accessed on 9 February 2021). [CrossRef]
- Meert, P.; Dierickx, S.; Govaert, E.; De Clerck, L.; Willems, S.; Dhaenens, M.; Deforce, D. Tackling aspecific side reactions during histone propionylation: The promise of reversing overpropionylation. Proteomics 2016, 16, 1970–1974. [Google Scholar] [CrossRef] [PubMed]
- Sidoli, S.; Garcia, B.A. Properly reading the histone code by MS-based proteomics. Proteomics 2015, 15, 2901–2902. [Google Scholar] [CrossRef]
- Paternoster, V.; Edhager, A.V.; Sibbersen, C.; Nielsen, A.L.; Børglum, A.D.; Christensen, J.H.; Palmfeldt, J. Quantitative assessment of methyl-esterification and other side reactions in a standard propionylation protocol for detection of histone modifications. Proteomics 2016, 16, 2059–2063. [Google Scholar] [CrossRef] [PubMed]
- Soldi, M.; Cuomo, A.; Bonaldi, T. Quantitative assessment of chemical artefacts produced by propionylation of histones prior to mass spectrometry analysis. Proteomics 2016, 16, 1952–1954. [Google Scholar] [CrossRef]
- Banbula, A.; Mak, P.; Smoluch, M.; Travis, J.; Potempa, J. Arginine-specific cysteine proteinase from Porphyromanas gingivalis as a convenient tool in protein chemistry. Biol. Chem. 2001, 382, 1399–1404. Available online: https://www.degruyter.com/document/doi/10.1515/BC.2001.172/html (accessed on 9 February 2021). [CrossRef] [PubMed]
- Veillard, F.; Potempa, B.; Guo, Y.; Ksiazek, M.; Sztukowska, M.N.; Houston, J.A.; Koneru, L.; Nguyen, K.A.; Potempa, J. Purification and characterisation of recombi-nant His-tagged RgpB gingipain from Porphymonas gingivalis. Biol. Chem. 2015, 396, 377–384. Available online: https://www.degruyter.com/document/doi/10.1515/hsz-2014-0304/html (accessed on 9 February 2021). [CrossRef] [PubMed]
- Helm, D.; Vissers, J.P.C.; Hughes, C.J.; Hahne, H.; Ruprecht, B.; Pachl, F.; Grzyb, A.; Richardson, K.; Wildgoose, J.; Maier, S.K.; et al. Ion Mobility Tandem Mass Spectrometry Enhances Performance of Bottom-up Proteomics. Mol. Cell. Proteom. 2014, 13, 3709–3715. [Google Scholar] [CrossRef] [PubMed]
- Van Puyvelde, B.; Willems, S.; Gabriels, R.; Daled, S.; De Clerck, L.; Casteele, S.V.; Staes, A.; Impens, F.; Deforce, D.; Martens, L.; et al. Removing the Hidden Data Dependency of DIA with Predicted Spectral Libraries. Proteomics 2020, 20. [Google Scholar] [CrossRef] [PubMed]
- Gabriels, R.; Martens, L.; Degroeve, S. Updated MS²PIP web server delivers fast and accurate MS² peak intensity prediction for multiple fragmentation methods, instruments and labeling techniques. Nucleic Acids Res. 2019, 47, W295–W299. [Google Scholar] [CrossRef]
- Bouwmeester, R.; Gabriels, R.; Hulstaert, N.; Martens, L.; Degroeve, S. DeepLC can predict retention times for peptides that carry as-yet unseen modifications. bioRxiv 2020. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.S.; Bouwmeester, R.; Martens, L.; Degroeve, S. Accurate peptide fragmentation predictions allow data driven ap-proaches to replace and improve upon proteomics search engine scoring functions. Bioinformatics 2019, 35, 5243–5248. Available online: http://www.ncbi.nlm.nih.gov/pubmed/31077310 (accessed on 9 February 2021). [CrossRef]
- Willems, S.; Dhaenens, M.; Govaert, E.; De Clerck, L.; Meert, P.; Van Neste, C.; Van Nieuwerburgh, F.; Deforce, D. Flagging False Positives Following Untargeted LC–MS Characterization of Histone Post-Translational Modification Combinations. J. Proteome Res. 2016, 16, 655–664. [Google Scholar] [CrossRef]
- Lin, S.; Wein, S.; Gonzales-Cope, M.; Otte, G.L.; Yuan, Z.-F.; Afjehi-Sadat, L.; Maile, T.; Berger, S.L.; Rush, J.; Lill, J.R.; et al. Stable-isotope-labeled Histone Peptide Library for Histone Post-translational Modification and Variant Quantification by Mass Spectrometry. Mol. Cell. Proteom. 2014, 13, 2450–2466. [Google Scholar] [CrossRef]
- Dhaenens, M.; Glibert, P.; Meert, P.; Vossaert, L.; Deforce, D. Histone proteolysis: A proposal for categorization into ‘clipping’ and ‘degradation’. BioEssays 2015, 37, 70–79. [Google Scholar] [CrossRef]
- Dhaenens, M.; Glibert, P.; Lambrecht, S.; Vossaert, L.; Van Steendam, K.; Elewaut, D.; Deforce, D. Neutrophil Elastase in the capacity of the “H2A-specific protease”. Int. J. Biochem. Cell Biol. 2014, 51, 39–44. Available online: https://linkinghub.elsevier.com/retrieve/pii/S1357272514000922 (accessed on 10 February 2021). [CrossRef] [PubMed]
- Glibert, P.; Vossaert, L.; Van Steendam, K.; Lambrecht, S.; Van Nieuwerburgh, F.; Offner, F.; Kipps, T.; Dhaenens, M.; Deforce, D. Quantitative proteomics to char-acterize specific histone H2A proteolysis in chronic lymphocytic leukemia and the myeloid THP-1 cell line. Int. J. Mol. Sci. 2014, 15, 9407–9421. Available online: http://www.ncbi.nlm.nih.gov/pubmed/24871368 (accessed on 10 February 2021). [CrossRef] [PubMed]
- Sidoli, S.; Yuan, Z.-F.; Lin, S.; Karch, K.; Wang, X.; Bhanu, N.; Arnaudo, A.M.; Britton, L.-M.; Cao, X.-J.; Gonzales-Cope, M.; et al. Drawbacks in the use of unconventional hydrophobic anhydrides for histone derivatization in bottom-up proteomics PTM analysis. Proteomics 2015, 15, 1459–1469. [Google Scholar] [CrossRef] [PubMed]
- Lenčo, J.; Khalikova, M.A.; Švec, F. Dissolving Peptides in 0.1% Formic Acid Brings Risk of Artificial Formylation. J. Proteome Res. 2020, 19, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, W.M. Cleavage at arginine residues by clostripain. Methods Enzymol. 1977, 47, 165–170. [Google Scholar] [CrossRef]
- Chen, Z.; Potempa, J.; Polanowski, A.; Wikstrom, M.; Travis, J. Purification and characterization of a 50-kDa cysteine proteinase (gingipain) from Porphyromonas gingivalis. J. Biol. Chem. 1992, 267, 18896–18901. [Google Scholar] [CrossRef]
- De Clerck, L.; Willems, S.; Noberini, R.; Restellini, C.; Van Puyvelde, B.; Daled, S.; Bonaldi, T.; Deforce, D.; Dhaenens, M. hSWATH: Unlocking SWATH’s Full Poten-tial for an Untargeted Histone Perspective. J. Proteome Res. 2019, 18, 3840–3849. Available online: https://pubs.acs.org/sharingguidelines (accessed on 6 April 2021).
- Levin, Y. The role of statistical power analysis in quantitative proteomics. Proteomics 2011, 11, 2565–2567. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daled, S.; Willems, S.; Van Puyvelde, B.; Corveleyn, L.; Verhelst, S.; De Clerck, L.; Deforce, D.; Dhaenens, M. Histone Sample Preparation for Bottom-Up Mass Spectrometry: A Roadmap to Informed Decisions. Proteomes 2021, 9, 17. https://doi.org/10.3390/proteomes9020017
Daled S, Willems S, Van Puyvelde B, Corveleyn L, Verhelst S, De Clerck L, Deforce D, Dhaenens M. Histone Sample Preparation for Bottom-Up Mass Spectrometry: A Roadmap to Informed Decisions. Proteomes. 2021; 9(2):17. https://doi.org/10.3390/proteomes9020017
Chicago/Turabian StyleDaled, Simon, Sander Willems, Bart Van Puyvelde, Laura Corveleyn, Sigrid Verhelst, Laura De Clerck, Dieter Deforce, and Maarten Dhaenens. 2021. "Histone Sample Preparation for Bottom-Up Mass Spectrometry: A Roadmap to Informed Decisions" Proteomes 9, no. 2: 17. https://doi.org/10.3390/proteomes9020017
APA StyleDaled, S., Willems, S., Van Puyvelde, B., Corveleyn, L., Verhelst, S., De Clerck, L., Deforce, D., & Dhaenens, M. (2021). Histone Sample Preparation for Bottom-Up Mass Spectrometry: A Roadmap to Informed Decisions. Proteomes, 9(2), 17. https://doi.org/10.3390/proteomes9020017





