Mimicry in the Bite: Shared Sequences Between Aedes aegypti Salivary Proteins and Human Proteins
Abstract
1. Introduction
2. Materials and Methods
2.1. Determination of Occurrence of Identical Peptide Sequences in Proteins from Aedes aegypti Saliva and Humans
2.2. Prediction of Peptide Binding Affinity for MHC Molecules
2.3. Determination of the Presence of Sequences Shared Between Aedes aegypti Salivary Proteins and Human Proteins in Validated Peptides from the Immune Epitope Database
3. Results
3.1. Occurrence of Identical Peptide Sequences in Aedes aegypti Salivary Proteins and Human Proteins
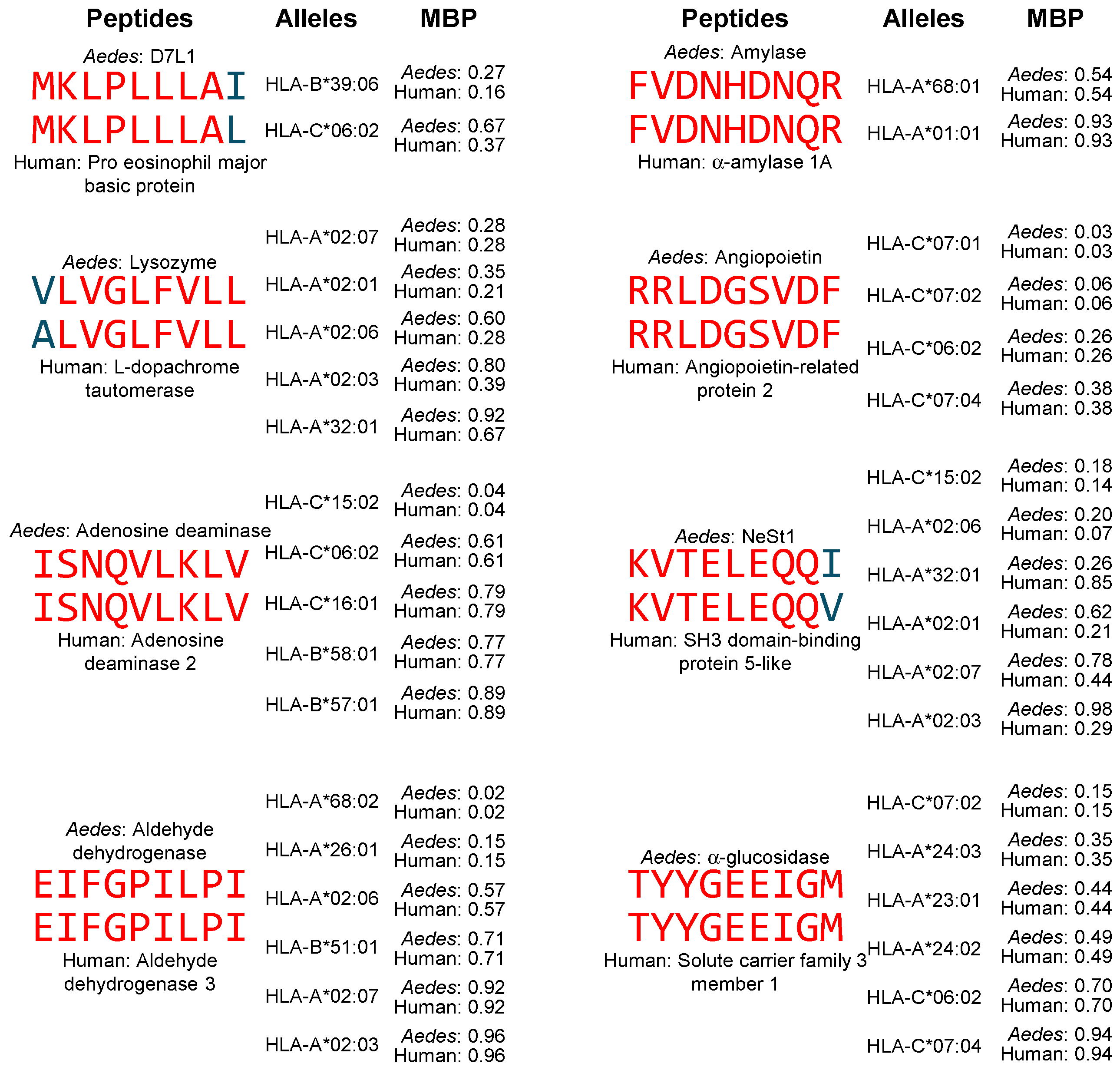
3.2. High-Affinity Binding of Peptides Containing Shared Sequences Between Aedes aegypti Salivary Proteins and Human Proteins to MHC-II Molecules
3.3. High-Affinity Binding of Peptides Containing Shared Sequences Between Aedes aegypti Salivary Proteins and Human Proteins to MHC-I Molecules
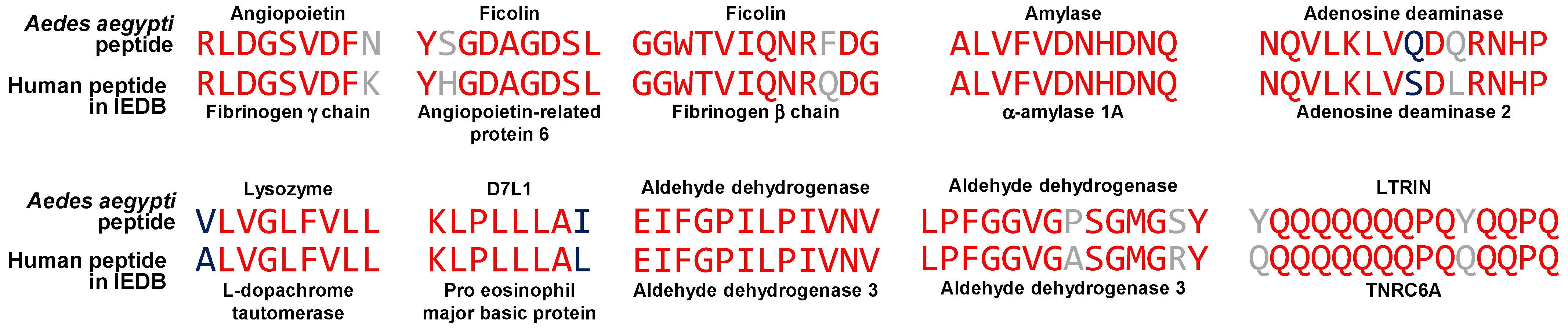
3.4. Peptides in the Immune Epitope Database Containing Sequences Shared Between Aedes aegypti Salivary Proteins and Human Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AaVA-1 | Aedes aegypti venom allergen-1 |
| AeMOPE | Aedes-specific modulatory peptide |
| AgBR | Aedes aegypti bacteria-responsive protein |
| AIH | Autoimmune hepatitis |
| CD | Crohn’s disease |
| CHD-1 | Chromodomain-helicase-DNA-binding protein-1 |
| CHIKV | Chikungunya virus |
| CLIP | Cytoplasmic linker protein |
| CNS | Central nervous system |
| DENV | Dengue virus |
| ECM | Extracellular matrix |
| EGFR | Epidermal growth factor receptor |
| ER | Endoplasmic reticulum |
| GD | Graves’ disease |
| GEF | Guanine nucleotide exchange factor |
| ITP | Immune thrombocytopenic purpura |
| LTRIN | Lymphotoxin β receptor inhibitor |
| MG | Myasthenia gravis |
| MIDEAS | Mitotic deacetylase-associated SANT domain protein |
| MS | Multiple sclerosis |
| NeSt | Neutrophil-stimulating factor |
| PAMP | Pathogen-associated molecular pattern |
| PE | Phosphatidylethanolamine |
| PI | Phosphatidylinositol |
| PR | Polymyalgia rheumatica |
| RA | Rheumatoid arthritis |
| RPTP | Receptor-type tyrosine-protein phosphatase |
| SCAPER | S phase cyclin A-associated protein in the endoplasmic reticulum |
| SLC3A1 | Solute carrier family 3 member 1 |
| SLE | Systemic lupus erythematosus |
| SS | Sjögren’s syndrome |
| T1D | Type 1 diabetes |
| TNRC6A | Trinucleotide repeat containing adaptor 6A |
| UC | Ulcerative colitis |
| ZIKV | Zika virus |
References
- Souza-Neto, J.A.; Powell, J.R.; Bonizzoni, M. Aedes aegypti vector competence studies: A review. Infect. Genet. Evol. 2019, 67, 191–209. [Google Scholar] [CrossRef]
- Alonso-Palomares, L.A.; Moreno-García, M.; Lanz-Mendoza, H.; Salazar, M.I. Molecular Basis for Arbovirus Transmission by Aedes aegypti Mosquitoes. Intervirology 2018, 61, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.A.; Yadav, M.K.; Lim, D.-W.; Kim, H.; Wang, J.-H.; Ansari, A. Viral-host molecular interactions and metabolic modulation: Strategies to inhibit flaviviruses pathogenesis. World J. Virol. 2024, 13, 99110. [Google Scholar] [CrossRef]
- Aguilar-Briseño, J.A.; Moser, J.; Rodenhuis-Zybert, I.A. Understanding immunopathology of severe dengue: Lessons learnt from sepsis. Curr. Opin. Virol. 2020, 43, 41–49. [Google Scholar] [CrossRef]
- Christian, K.M.; Song, H.; Ming, G.-L. Pathophysiology and Mechanisms of Zika Virus Infection in the Nervous System. Annu. Rev. Neurosci. 2019, 42, 249–269. [Google Scholar] [CrossRef] [PubMed]
- Cherie, T.J.J.; Choong, C.S.H.; Abid, M.B.; Weber, M.W.; Yap, E.S.; Seneviratne, S.L.; Abeysuriya, V.; de Mel, S. Immuno-Haematologic Aspects of Dengue Infection: Biologic Insights and Clinical Implications. Viruses 2024, 16, 1090. [Google Scholar] [CrossRef] [PubMed]
- Lardo, S.; Soesatyo, M.H.; Juffrie, J.; Umniyati, S.R. The Autoimmune Mechanism in Dengue Hemorrhagic Fever. Acta Med. Indones. 2018, 50, 70–79. Available online: https://pubmed.ncbi.nlm.nih.gov/29686179/ (accessed on 22 July 2025).
- Martina, B.E.E.; Koraka, P.; Osterhaus, A.D.M.E. Dengue virus pathogenesis: An integrated view. Clin. Microbiol. Rev. 2009, 22, 564–581. [Google Scholar] [CrossRef]
- Srikiatkhachorn, A.; Mathew, A.; Rothman, A.L. Immune-mediated cytokine storm and its role in severe dengue. Semin. Immunopathol. 2017, 39, 563–574. [Google Scholar] [CrossRef]
- Lin, C.-F.; Lei, H.-Y.; Shiau, A.-L.; Liu, C.-C.; Liu, H.-S.; Yeh, T.-M.; Chen, S.-H.; Lin, Y.-S. Antibodies from dengue patient sera cross-react with endothelial cells and induce damage. J. Med. Virol. 2003, 69, 82–90. [Google Scholar] [CrossRef]
- Lin, C.-F.; Lei, H.-Y.; Shiau, A.-L.; Liu, H.-S.; Yeh, T.-M.; Chen, S.-H.; Liu, C.-C.; Chiu, S.-C.; Lin, Y.-S. Endothelial cell apoptosis induced by antibodies against dengue virus nonstructural protein 1 via production of nitric oxide. J. Immunol. 2002, 169, 657–664. [Google Scholar] [CrossRef]
- Chungue, E.; Poli, L.; Roche, C.; Gestas, P.; Glaziou, P.; Markoff, L.J. Correlation between detection of plasminogen cross-reactive antibodies and hemorrhage in dengue virus infection. J. Infect. Dis. 1994, 170, 1304–1307. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.-W.; Lin, C.-F.; Yeh, T.-M.; Liu, C.-C.; Liu, H.-S.; Wang, S.; Ling, P.; Anderson, R.; Lei, H.-Y.; Lin, Y.-S. Autoimmunity in dengue pathogenesis. J. Formos. Med. Assoc. 2013, 112, 3–11. [Google Scholar] [CrossRef]
- Falconar, A.K. The dengue virus nonstructural-1 protein (NS1) generates antibodies to common epitopes on human blood clotting, integrin/adhesin proteins and binds to human endothelial cells: Potential implications in haemorrhagic fever pathogenesis. Arch. Virol. 1997, 142, 897–916. [Google Scholar] [CrossRef]
- Cheng, H.-J.; Luo, Y.-H.; Wan, S.-W.; Lin, C.-F.; Wang, S.-T.; Hung, N.T.; Liu, C.-C.; Ho, T.-S.; Liu, H.-S.; Yeh, T.-M.; et al. Correlation between serum levels of anti-endothelial cell autoantigen and anti-dengue virus nonstructural protein 1 antibodies in dengue patients. Am. J. Trop. Med. Hyg. 2015, 92, 989–995. [Google Scholar] [CrossRef]
- Ghorai, T.; Sarkar, A.; Roy, A.; Bhowmick, B.; Nayak, D.; Das, S. Role of auto-antibodies in the mechanisms of dengue pathogenesis and its progression: A comprehensive review. Arch. Microbiol. 2024, 206, 214. [Google Scholar] [CrossRef]
- Rajadhyaksha, A.; Mehra, S. Dengue fever evolving into systemic lupus erythematosus and lupus nephritis: A case report. Lupus 2012, 21, 999–1002. [Google Scholar] [CrossRef]
- Jardim, D.L.F.; Tsukumo, D.M.L.; Angerami, R.N.; Carvalho Filho, M.A.; de Saad, M.J.A. Autoimmune features caused by dengue fever: A case report. Braz. J. Infect. Dis. 2012, 16, 92–95. [Google Scholar] [CrossRef][Green Version]
- Harris, V.K.; Danda, D.; Murali, N.S.; Das, P.K.; Abraham, M.; Cherian, A.M.; Chandy, M. Unusual association of Kikuchi’s disease and dengue virus infection evolving into systemic lupus erythematosus. J. Indian Med. Assoc. 2000, 98, 391–393. [Google Scholar] [PubMed]
- Puccioni-Sohler, M.; Ornelas, A.M.M.; de Souza, A.S.; Cabral-Castro, M.J.; Ramos, J.T.M.A.; Rosadas, C.; Salgado, M.C.F.; Castiglione, A.A.; Ferry, F.; Peralta, J.M.; et al. First report of persistent dengue-1-associated autoimmune neurological disturbance: Neuromyelitis optica spectrum disorder. J. Neurovirol. 2017, 23, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Correa, J.; de Siqueira, I.C.; Mota, S.; do Rosário, M.S.; Pereira de Jesus, P.A.; Alcantara, L.C.J.; Ernst, J.D.; Rodriguez, A. Anti-ganglioside antibodies in patients with Zika virus infection-associated Guillain-Barré Syndrome in Brazil. PLoS Negl. Trop. Dis. 2019, 13, e0007695. [Google Scholar] [CrossRef]
- Acosta-Ampudia, Y.; Monsalve, D.M.; Castillo-Medina, L.F.; Rodríguez, Y.; Pacheco, Y.; Halstead, S.; Willison, H.J.; Anaya, J.-M.; Ramírez-Santana, C. Autoimmune Neurological Conditions Associated with Zika Virus Infection. Front. Mol. Neurosci. 2018, 11, 116. [Google Scholar] [CrossRef]
- Anaya, J.-M.; Rodríguez, Y.; Monsalve, D.M.; Vega, D.; Ojeda, E.; González-Bravo, D.; Rodríguez-Jiménez, M.; Pinto-Díaz, C.A.; Chaparro, P.; Gunturiz, M.L.; et al. A comprehensive analysis and immunobiology of autoimmune neurological syndromes during the Zika virus outbreak in Cúcuta, Colombia. J. Autoimmun. 2017, 77, 123–138. [Google Scholar] [CrossRef]
- Payus, A.O.; Ibrahim, A.; Lin, C.L.S.; Jan, T.H. Sensory Predominant Guillain-Barré Syndrome Concomitant with Dengue Infection: A Case Report. Case Rep. Neurol. 2022, 14, 281–285. [Google Scholar] [CrossRef]
- Dalugama, C.; Shelton, J.; Ekanayake, M.; Gawarammana, I.B. Dengue fever complicated with Guillain-Barré syndrome: A case report and review of the literature. J. Med. Case Rep. 2018, 12, 137. [Google Scholar] [CrossRef] [PubMed]
- Sreelakshmi, V.; Pattanaik, A.; Marate, S.; Mani, R.S.; Pai, A.R.; Mukhopadhyay, C. Guillain-barré syndrome (GBS) with antecedent chikungunya infection: A case report and literature review. Neurol. Res. Pract. 2024, 6, 21. [Google Scholar] [CrossRef]
- Oehler, E.; Watrin, L.; Larre, P.; Leparc-Goffart, I.; Lastere, S.; Valour, F.; Baudouin, L.; Mallet, H.; Musso, D.; Ghawche, F. Zika virus infection complicated by Guillain-Barre syndrome—Case report, French Polynesia, December 2013. Euro. Surveill. 2014, 19, 20720. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Concepción, J.R.; Betancourt, J.P.; Cerra, J.; Reyes, E. The Zika Virus: An Association to Guillain-Barré Syndrome in the United States—A Case Report. P. R. Health Sci. J. 2018, 37, S93–S95. [Google Scholar]
- Mancera-Páez, O.; Román, G.C.; Pardo-Turriago, R.; Rodríguez, Y.; Anaya, J.-M. Concurrent Guillain-Barré syndrome, transverse myelitis and encephalitis post-Zika: A case report and review of the pathogenic role of multiple arboviral immunity. J. Neurol. Sci. 2018, 395, 47–53. [Google Scholar] [CrossRef]
- Bautista, L.E. Zika virus infection and risk of Guillain-Barré syndrome: A meta-analysis. J. Neurol. Sci. 2019, 403, 99–105. [Google Scholar] [CrossRef]
- Simmer, P.E.; Powell, V.; Hoch, V.; Noblett, C.; Eckert, P.; Abdelmaseeh, P. Ascending Trouble: Guillain-Barré-Like Syndrome Due to West Nile Virus. Cureus 2025, 17, e85240. [Google Scholar] [CrossRef] [PubMed]
- Beshai, R.; Bibawy, D.; Bibawy, J. Guillain-Barré Syndrome Secondary to West Nile Virus in New York City. Case Rep. Infect. Dis. 2020, 2020, 6501658. [Google Scholar] [CrossRef]
- Karagianni, P.; Alexopoulos, H.; Sourdi, A.; Papadimitriou, D.; Dimitrakopoulos, A.N.; Moutsopoulos, H.M. West Nile Virus infection triggering autoimmune encephalitis: Pathophysiological and therapeutic implications. Clin. Immunol. 2019, 207, 97–99. [Google Scholar] [CrossRef]
- Theofilopoulos, A.N.; Kono, D.H.; Baccala, R. The multiple pathways to autoimmunity. Nat. Immunol. 2017, 18, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Marson, A.; Housley, W.J.; Hafler, D.A. Genetic basis of autoimmunity. J. Clin. Investig. 2015, 125, 2234–2241. [Google Scholar] [CrossRef]
- Rosenblum, M.D.; Remedios, K.A.; Abbas, A.K. Mechanisms of human autoimmunity. J. Clin. Investig. 2015, 125, 2228–2233. [Google Scholar] [CrossRef] [PubMed]
- Cusick, M.F.; Libbey, J.E.; Fujinami, R.S. Molecular mimicry as a mechanism of autoimmune disease. Clin. Rev. Allergy Immunol. 2012, 42, 102–111. [Google Scholar] [CrossRef]
- Rojas, M.; Restrepo-Jiménez, P.; Monsalve, D.M.; Pacheco, Y.; Acosta-Ampudia, Y.; Ramírez-Santana, C.; Leung, P.S.C.; Ansari, A.A.; Gershwin, M.E.; Anaya, J.-M. Molecular mimicry and autoimmunity. J. Autoimmun. 2018, 95, 100–123. [Google Scholar] [CrossRef]
- Suliman, B.A. Potential clinical implications of molecular mimicry-induced autoimmunity. Immun. Inflamm. Dis. 2024, 12, e1178. [Google Scholar] [CrossRef]
- Kalil, J.; Guilherme, L. Rheumatic Fever: A Model of Autoimmune Disease due to Molecular Mimicry between Human and Pathogen Proteins. Crit. Rev. Immunol. 2020, 40, 419–422. [Google Scholar] [CrossRef]
- Shahrizaila, N.; Yuki, N. Guillain-barré syndrome animal model: The first proof of molecular mimicry in human autoimmune disorder. J. Biomed Biotechnol. 2011, 2011, 829129. [Google Scholar] [CrossRef]
- Poole, B.D.; Scofield, R.H.; Harley, J.B.; James, J.A. Epstein-Barr virus and molecular mimicry in systemic lupus erythematosus. Autoimmunity 2006, 39, 63–70. [Google Scholar] [CrossRef]
- Goh, L.; Kerkar, N. Hepatitis C Virus and Molecular Mimicry. Pathogens 2024, 13, 527. [Google Scholar] [CrossRef]
- Arévalo-Cortés, A.; Rodriguez-Pinto, D.; Aguilar-Ayala, L. Evidence for Molecular Mimicry between SARS-CoV-2 and Human Antigens: Implications for Autoimmunity in COVID-19. Autoimmune Dis. 2024, 2024, 8359683. [Google Scholar] [CrossRef]
- Hussein, H.M.; Rahal, E.A. The role of viral infections in the development of autoimmune diseases. Crit. Rev. Microbiol. 2019, 45, 394–412. [Google Scholar] [CrossRef]
- Cunningham, M.W. Molecular Mimicry, Autoimmunity, and Infection: The Cross-Reactive Antigens of Group A Streptococci and their Sequelae. Microbiol. Spectr. 2019, 7, GPP3-0045-2018. [Google Scholar] [CrossRef]
- Szymula, A.; Rosenthal, J.; Szczerba, B.M.; Bagavant, H.; Fu, S.M.; Deshmukh, U.S. T cell epitope mimicry between Sjögren’s syndrome Antigen A (SSA)/Ro60 and oral, gut, skin and vaginal bacteria. Clin. Immunol. 2014, 152, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Reichlin, M. A possible link between infection with burkholderia bacteria and systemic lupus erythematosus based on epitope mimicry. Clin. Dev. Immunol. 2008, 2008, 683489. [Google Scholar] [CrossRef] [PubMed]
- Muthye, V.; Wasmuth, J.D. Proteome-wide comparison of tertiary protein structures reveals molecular mimicry in Plasmodium-human interactions. Front. Parasitol. 2023, 2, 1162697. [Google Scholar] [CrossRef]
- Emiliani, Y.; Muzi, G.; Sánchez, A.; Sánchez, J.; Munera, M. Prediction of molecular mimicry between proteins from Trypanosoma sp. and human antigens associated with systemic lupus erythematosus. Microb. Pathog. 2022, 172, 105760. [Google Scholar] [CrossRef]
- Trier, N.H.; Houen, G. Antibody Cross-Reactivity in Auto-Immune Diseases. Int. J. Mol. Sci. 2023, 24, 13609. [Google Scholar] [CrossRef]
- Kammer, A.R.; van der Burg, S.H.; Grabscheid, B.; Hunziker, I.P.; Kwappenberg, K.M.; Reichen, J.; Melief, C.J.; Cerny, A. Molecular mimicry of human cytochrome P450 by hepatitis C virus at the level of cytotoxic T cell recognition. J. Exp. Med. 1999, 190, 169–176. [Google Scholar] [CrossRef]
- Johnston, A.; Gudjonsson, J.E.; Sigmundsdottir, H.; Love, T.J.; Valdimarsson, H. Peripheral blood T cell responses to keratin peptides that share sequences with streptococcal M proteins are largely restricted to skin-homing CD8+ T cells. Clin. Exp. Immunol. 2004, 138, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Liu, I.-J.; Chiu, C.-Y.; Chen, Y.-C.; Wu, H.-C. Molecular mimicry of human endothelial cell antigen by autoantibodies to nonstructural protein 1 of dengue virus. J. Biol. Chem. 2011, 286, 9726–9736. [Google Scholar] [CrossRef]
- Cheng, H.-J.; Lin, C.-F.; Lei, H.-Y.; Liu, H.-S.; Yeh, T.-M.; Luo, Y.-H.; Lin, Y.-S. Proteomic analysis of endothelial cell autoantigens recognized by anti-dengue virus nonstructural protein 1 antibodies. Exp. Biol. Med. 2009, 234, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-J.; Lei, H.-Y.; Lin, C.-F.; Luo, Y.-H.; Wan, S.-W.; Liu, H.-S.; Yeh, T.-M.; Lin, Y.-S. Anti-dengue virus nonstructural protein 1 antibodies recognize protein disulfide isomerase on platelets and inhibit platelet aggregation. Mol. Immunol. 2009, 47, 398–406. [Google Scholar] [CrossRef]
- Chang, H.-H.; Shyu, H.-F.; Wang, Y.-M.; Sun, D.-S.; Shyu, R.-H.; Tang, S.-S.; Huang, Y.-S. Facilitation of cell adhesion by immobilized dengue viral nonstructural protein 1 (NS1): Arginine-glycine-aspartic acid structural mimicry within the dengue viral NS1 antigen. J. Infect. Dis. 2002, 186, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Chang, B.I.; Lei, H.Y.; Liu, H.S.; Liu, C.C.; Wu, H.L.; Yeh, T.M. Antibodies against dengue virus E protein peptide bind to human plasminogen and inhibit plasmin activity. Clin. Exp. Immunol. 1997, 110, 35–40. [Google Scholar] [CrossRef]
- Lin, Y.-S.; Yeh, T.-M.; Lin, C.-F.; Wan, S.-W.; Chuang, Y.-C.; Hsu, T.-K.; Liu, H.-S.; Liu, C.-C.; Anderson, R.; Lei, H.-Y. Molecular mimicry between virus and host and its implications for dengue disease pathogenesis. Exp. Biol. Med. 2011, 236, 515–523. [Google Scholar] [CrossRef]
- Lucchese, G.; Kanduc, D. Zika virus and autoimmunity: From microcephaly to Guillain-Barré syndrome, and beyond. Autoimmun. Rev. 2016, 15, 801–808. [Google Scholar] [CrossRef]
- França, L.C.; Fontes-Dantas, F.L.; Garcia, D.G.; de Araújo, A.D.; da Costa Gonçalves, J.P.; da Silav Rêgo, C.C.; da Silva, E.V.; do Nascimento, O.J.M.; Lopes, F.C.R.; Herlinger, A.L.; et al. Molecular mimicry between Zika virus and central nervous system inflammatory demyelinating disorders: The role of NS5 Zika virus epitope and PLP autoantigens. Arq. Neuropsiquiatr. 2023, 81, 357–368. [Google Scholar] [CrossRef]
- Nasirudeen, A.M.A.; Wong, H.H.; Thien, P.; Xu, S.; Lam, K.-P.; Liu, D.X. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl. Trop. Dis. 2011, 5, e926. [Google Scholar] [CrossRef]
- Ye, S.; Liang, Y.; Chang, Y.; Lai, B.; Zhong, J. Dengue Virus Replicative-Form dsRNA Is Recognized by Both RIG-I and MDA5 to Activate Innate Immunity. J. Med. Virol. 2025, 97, e70194. [Google Scholar] [CrossRef] [PubMed]
- Suthar, M.S.; Aguirre, S.; Fernandez-Sesma, A. Innate Immune Sensing of Flaviviruses. PLoS Pathog. 2013, 9, e1003541. [Google Scholar] [CrossRef] [PubMed]
- Pingen, M.; Schmid, M.A.; Harris, E.; McKimmie, C.S. Mosquito Biting Modulates Skin Response to Virus Infection. Trends Parasitol. 2017, 33, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Demeure, C.E.; Brahimi, K.; Hacini, F.; Marchand, F.; Péronet, R.; Huerre, M.; St.-Mezard, P.; Nicolas, J.-F.; Brey, P.; Delespesse, G.; et al. Anopheles Mosquito Bites Activate Cutaneous Mast Cells Leading to a Local Inflammatory Response and Lymph Node Hyperplasia1. J. Immunol. 2005, 174, 3932–3940. [Google Scholar] [CrossRef]
- Pingen, M.; Bryden, S.R.; Pondeville, E.; Schnettler, E.; Kohl, A.; Merits, A.; Fazakerley, J.K.; Graham, G.J.; McKimmie, C.S. Host Inflammatory Response to Mosquito Bites Enhances the Severity of Arbovirus Infection. Immunity 2016, 44, 1455–1469. [Google Scholar] [CrossRef]
- Guerrero, D.; Cantaert, T.; Missé, D. Aedes Mosquito Salivary Components and Their Effect on the Immune Response to Arboviruses. Front. Cell. Infect. Microbiol. 2020, 10, 407. Available online: https://www.frontiersin.org/journals/cellular-and-infection-microbiology/articles/10.3389/fcimb.2020.00407/full (accessed on 23 July 2025). [CrossRef]
- Ader, D.B.; Celluzzi, C.; Bisbing, J.; Gilmore, L.; Gunther, V.; Peachman, K.K.; Rao, M.; Barvir, D.; Sun, W.; Palmer, D.R. Modulation of Dengue Virus Infection of Dendritic Cells by Aedes aegypti Saliva. Viral Immunol. 2004, 17, 252–265. [Google Scholar] [CrossRef]
- Schneider, B.S.; Soong, L.; Zeidner, N.S.; Higgs, S. Aedes aegypti Salivary Gland Extracts Modulate Anti-Viral and TH1/TH2 Cytokine Responses to Sindbis Virus Infection. Viral Immunol. 2004, 17, 565–573. [Google Scholar] [CrossRef]
- Schmid, M.A.; Glasner, D.R.; Shah, S.; Michlmayr, D.; Kramer, L.D.; Harris, E. Mosquito Saliva Increases Endothelial Permeability in the Skin, Immune Cell Migration, and Dengue Pathogenesis during Antibody-Dependent Enhancement. PLoS Pathog. 2016, 12, e1005676. [Google Scholar] [CrossRef]
- Martin-Martin, I.; Kern, O.; Brooks, S.; Smith, L.B.; Valenzuela-Leon, P.C.; Bonilla, B.; Ackerman, H.; Calvo, E. Biochemical characterization of AeD7L2 and its physiological relevance in blood feeding in the dengue mosquito vector, Aedes aegypti. FEBS J. 2021, 288, 2014–2029. [Google Scholar] [CrossRef]
- Martin-Martin, I.; Valenzuela Leon, P.C.; Amo, L.; Shrivastava, G.; Iniguez, E.; Aryan, A.; Brooks, S.; Kojin, B.B.; Williams, A.E.; Bolland, S.; et al. Aedes aegypti sialokinin facilitates mosquito blood feeding and modulates host immunity and vascular biology. Cell Rep. 2022, 39, 110648. [Google Scholar] [CrossRef]
- Ribeiro, J.M. Characterization of a vasodilator from the salivary glands of the yellow fever mosquito Aedes aegypti. J. Exp. Biol. 1992, 165, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Rasic, N.; Liu, Y.; Simons, F.E.R. Mosquito saliva–specific IgE and IgG antibodies in 1059 blood donors. J. Allergy Clin. Immunol. 2002, 110, 816–817. [Google Scholar] [CrossRef] [PubMed]
- Londoño-Rentería, B.; Cárdenas, J.C.; Giovanni, J.E.; Cárdenas, L.; Villamizar, P.; Rolón, J.; Chisenhall, D.M.; Christofferson, R.C.; Carvajal, D.J.; Pérez, O.G.; et al. Aedes aegypti anti-salivary gland antibody concentration and dengue virus exposure history in healthy individuals living in an endemic area in Colombia. Biomedica 2015, 35, 572–581. [Google Scholar] [CrossRef]
- Chea, S.; Willen, L.; Nhek, S.; Ly, P.; Tang, K.; Oristian, J.; Salas-Carrillo, R.; Ponce, A.; Leon, P.C.V.; Kong, D.; et al. Antibodies to Aedes aegypti D7L salivary proteins as a new serological tool to estimate human exposure to Aedes mosquitoes. Front. Immunol. 2024, 15, 1368066. [Google Scholar] [CrossRef]
- Mathieu-Daudé, F.; Claverie, A.; Plichart, C.; Boulanger, D.; Mphande, F.A.; Bossin, H.C. Specific human antibody responses to Aedes aegypti and Aedes polynesiensis saliva: A new epidemiological tool to assess human exposure to disease vectors in the Pacific. PLoS Negl. Trop. Dis. 2018, 12, e0006660. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Aoki, V.; Liu, Z.; Prisayanh, P.; Valenzuela, J.G.; Diaz, L.A. From Insect Bites to a Skin Autoimmune Disease: A Conceivable Pathway to Endemic Pemphigus Foliaceus. Front. Immunol. 2022, 13, 907424. Available online: https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2022.907424/full (accessed on 11 October 2025). [CrossRef]
- Ribeiro, J.M.C.; Martin-Martin, I.; Arcà, B.; Calvo, E. A Deep Insight into the Sialome of Male and Female Aedes aegypti Mosquitoes. PLoS ONE 2016, 11, e0151400. [Google Scholar] [CrossRef]
- Ribeiro, J.M.C.; Arcà, B.; Lombardo, F.; Calvo, E.; Phan, V.M.; Chandra, P.K.; Wikel, S.K. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genom. 2007, 8, 6. [Google Scholar] [CrossRef]
- Wasinpiyamongkol, L.; Patramool, S.; Luplertlop, N.; Surasombatpattana, P.; Doucoure, S.; Mouchet, F.; Séveno, M.; Remoue, F.; Demettre, E.; Brizard, J.-P.; et al. Blood-feeding and immunogenic Aedes aegypti saliva proteins. Proteomics 2010, 10, 1906–1916. [Google Scholar] [CrossRef]
- Gavor, E.; Choong, Y.K.; Liu, Y.; Pompon, J.; Ooi, E.E.; Mok, Y.K.; Liu, H.; Kini, R.M.; Sivaraman, J. Identification of Aedes aegypti salivary gland proteins interacting with human immune receptor proteins. PLoS Negl. Trop. Dis. 2022, 16, e0010743. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; De Bruijn, M.L.; Vernie, L.N.; Kast, W.M.; Melief, C.J.; Neefjes, J.J.; Ploegh, H.L. Peptide selection by MHC class I molecules. Nature 1991, 350, 703–706. [Google Scholar] [CrossRef]
- Rammensee, H.G.; Friede, T.; Stevanoviíc, S. MHC ligands and peptide motifs: First listing. Immunogenetics 1995, 41, 178–228. [Google Scholar] [CrossRef]
- Renaudineau, Y.; Charras, A.; Natoli, V.; Congy-Jolivet, N.; Haldenby, S.; Liu, X.; Fang, Y.; Smith, E.M.; Beresford, M.W.; Hedrich, C.M.; et al. Across ancestries, HLA-B∗08:01∼DRB1∗03:01 (DR3) and HLA-DQA∗01:02 (DR2) increase the risk to develop juvenile-onset systemic lupus erythematosus through low complement C4 levels. J. Transl. Autoimmun. 2025, 10, 100268. [Google Scholar] [CrossRef]
- Butler-Laporte, G.; Farjoun, J.; Nakanishi, T.; Lu, T.; Abner, E.; Chen, Y.; Hultström, M.; Metspalu, A.; Milani, L.; Mägi, R.; et al. HLA allele-calling using multi-ancestry whole-exome sequencing from the UK Biobank identifies 129 novel associations in 11 autoimmune diseases. Commun. Biol. 2023, 6, 1113. [Google Scholar] [CrossRef] [PubMed]
- Al Naqbi, H.; Mawart, A.; Alshamsi, J.; Al Safar, H.; Tay, G.K. Major histocompatibility complex (MHC) associations with diseases in ethnic groups of the Arabian Peninsula. Immunogenetics 2021, 73, 131–152. [Google Scholar] [CrossRef]
- Fernando, M.M.A.; Stevens, C.R.; Walsh, E.C.; De Jager, P.L.; Goyette, P.; Plenge, R.M.; Vyse, T.J.; Rioux, J.D. Defining the role of the MHC in autoimmunity: A review and pooled analysis. PLoS Genet. 2008, 4, e1000024. [Google Scholar] [CrossRef]
- Erlich, H.; Valdes, A.M.; Noble, J.; Carlson, J.A.; Varney, M.; Concannon, P.; Mychaleckyj, J.C.; Todd, J.A.; Bonella, P.; Fear, A.L.; et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: Analysis of the type 1 diabetes genetics consortium families. Diabetes 2008, 57, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Pirie, F.J.; Hammond, M.G.; Motala, A.A.; Omar, M.A. HLA class II antigens in South African Blacks with type I diabetes. Tissue Antigens 2001, 57, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Heward, J.M.; Allahabadia, A.; Daykin, J.; Carr-Smith, J.; Daly, A.; Armitage, M.; Dodson, P.M.; Sheppard, M.C.; Barnett, A.H.; Franklyn, J.A.; et al. Linkage disequilibrium between the human leukocyte antigen class II region of the major histocompatibility complex and Graves’ disease: Replication using a population case control and family-based study. J. Clin. Endocrinol. Metab. 1998, 83, 3394–3397. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hernández-Doño, S.; Jakez-Ocampo, J.; Márquez-García, J.E.; Ruiz, D.; Acuña-Alonzo, V.; Lima, G.; Llorente, L.; Tovar-Méndez, V.H.; García-Silva, R.; Granados, J.; et al. Heterogeneity of Genetic Admixture Determines SLE Susceptibility in Mexican. Front. Genet. 2021, 12, 701373. [Google Scholar] [CrossRef]
- Al-Harbi, E.M.; Abbassi, A.-J.; Tamim, H.; al-Jenaidi, F.; Kooheji, M.; Kamal, M.; al-Mahroos, S.; al-Nasir, F.; Motala, A.A.; Almawi, W.Y. Specific HLA-DRB and -DQB alleles and haplotypes confer disease susceptibility or resistance in Bahraini type 1 diabetes patients. Clin. Diagn. Lab. Immunol. 2004, 11, 292–296. [Google Scholar] [CrossRef]
- Varade, J.; Wang, N.; Lim, C.K.; Zhang, T.; Zhang, Y.; Liu, X.; Piehl, F.; Matell, R.; Cao, H.; Xu, X.; et al. Novel genetic loci associated HLA-B*08:01 positive myasthenia gravis. J. Autoimmun. 2018, 88, 43–49. [Google Scholar] [CrossRef]
- Le, T.T.V.; Vuong, T.T.B.; Ong, T.P.; Do, M.D. Allele frequency and the associations of HLA-DRB1 and HLA-DQB1 polymorphisms with pemphigus subtypes and disease severity. Medicine 2022, 101, e28855. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Li, W.; Song, J.; Liu, X.; Gu, Y.; Yan, C.; Wu, H.; Xi, J.; Zhou, S.; Zhao, C. HLA typing using next-generation sequencing for Chinese juvenile- and adult-onset myasthenia gravis patients. J. Clin. Neurosci. 2019, 59, 179–184. [Google Scholar] [CrossRef]
- de Almeida, D.E.; Ling, S.; Holoshitz, J. New insights into the functional role of the rheumatoid arthritis shared epitope. FEBS Lett. 2011, 585, 3619–3626. [Google Scholar] [CrossRef]
- van Drongelen, V.; Holoshitz, J. Human Leukocyte Antigen-Disease Associations in Rheumatoid Arthritis. Rheum. Dis. Clin. N. Am. 2017, 43, 363–376. [Google Scholar] [CrossRef]
- Oliveira, L.C.; Porta, G.; Marin, M.L.C.; Bittencourt, P.L.; Kalil, J.; Goldberg, A.C. Autoimmune hepatitis, HLA and extended haplotypes. Autoimmun. Rev. 2011, 10, 189–193. [Google Scholar] [CrossRef]
- Jun, K.R.; Choi, S.E.; Cha, C.H.; Oh, H.B.; Heo, Y.S.; Ahn, H.Y.; Lee, K.J. Meta-analysis of the association between HLA-DRB1 allele and rheumatoid arthritis susceptibility in Asian populations. J. Korean Med. Sci. 2007, 22, 973–980. [Google Scholar] [CrossRef]
- Gough, S.C.L.; Simmonds, M.J. The HLA Region and Autoimmune Disease: Associations and Mechanisms of Action. Curr. Genom. 2007, 8, 453–465. [Google Scholar] [CrossRef]
- Patsopoulos, N.A.; Barcellos, L.F.; Hintzen, R.Q.; Schaefer, C.; van Duijn, C.M.; Noble, J.A.; Raj, T.; IMSGC; ANZgene; Gourraud, P.-A.; et al. Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS Genet. 2013, 9, e1003926. [Google Scholar] [CrossRef] [PubMed]
- Hachicha, H.; Kammoun, A.; Mahfoudh, N.; Marzouk, S.; Feki, S.; Fakhfakh, R.; Fourati, H.; Haddouk, S.; Frikha, F.; Gaddour, L.; et al. Human leukocyte antigens-DRB1*03 is associated with systemic lupus erythematosus and anti-SSB production in South Tunisia. Int. J. Health Sci. 2018, 12, 21–27. [Google Scholar]
- Jean, S.; Quelvennec, E.; Alizadeh, M.; Guggenbuhl, P.; Birebent, B.; Perdriger, A.; Grosbois, B.; Pawlotsky, P.Y.; Semana, G. DRB1*15 and DRB1*03 extended haplotype interaction in primary Sjögren’s syndrome genetic susceptibility. Clin. Exp. Rheumatol. 1998, 16, 725–728. [Google Scholar] [PubMed]
- Yeo, T.W.; De Jager, P.L.; Gregory, S.G.; Barcellos, L.F.; Walton, A.; Goris, A.; Fenoglio, C.; Ban, M.; Taylor, C.J.; Goodman, R.S.; et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann. Neurol. 2007, 61, 228–236. [Google Scholar] [CrossRef]
- Kikili, C.İ.; Kivanç, D.; Ortaboz, D.; Şentürk Çiftçi, H.; Özbalak, M.M.; Yenerel, M.N.; Nalçaci, M.; Ar, M.C.; Oğuz, F.S.; Beşişik, S.K. Identification of HLA alleles involved in immune thrombotic thrombocytopenic purpura patients from Turkey. Blood Coagul. Fibrinolysis. 2024, 35, 307–315. [Google Scholar] [CrossRef]
- Stasiak, M.; Zawadzka-Starczewska, K.; Tymoniuk, B.; Stasiak, B.; Lewiński, A. Significance of HLA in the development of Graves’ orbitopathy. Genes Immun. 2023, 24, 32–38. [Google Scholar] [CrossRef]
- Sirikong, M.; Tsuchiya, N.; Chandanayingyong, D.; Bejrachandra, S.; Suthipinittharm, P.; Luangtrakool, K.; Srinak, D.; Thongpradit, R.; Siriboonrit, U.; Tokunaga, K. Association of HLA-DRB1*1502-DQB1*0501 haplotype with susceptibility to systemic lupus erythematosus in Thais. Tissue Antigens 2002, 59, 113–117. [Google Scholar] [CrossRef]
- Prinz, J.C. Human Leukocyte Antigen-Class I Alleles and the Autoreactive T Cell Response in Psoriasis Pathogenesis. Front. Immunol. 2018, 9, 954. [Google Scholar] [CrossRef] [PubMed]
- Noble, J.A.; Valdes, A.M. Genetics of the HLA region in the prediction of type 1 diabetes. Curr. Diab. Rep. 2011, 11, 533–542. [Google Scholar] [CrossRef]
- Bugawan, T.L.; Klitz, W.; Alejandrino, M.; Ching, J.; Panelo, A.; Solfelix, C.M.; Petrone, A.; Buzzetti, R.; Pozzilli, P.; Erlich, H.A. The association of specific HLA class I and II alleles with type 1 diabetes among Filipinos. Tissue Antigens 2002, 59, 452–469. [Google Scholar] [CrossRef] [PubMed]
- Prinz, J.C. Melanocytes: Target Cells of an HLA-C*06:02-Restricted Autoimmune Response in Psoriasis. J. Investig. Dermatol. 2017, 137, 2053–2058. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.J.; Bridges, S.L., Jr.; Ahmed, S. HLA-C: An Accomplice in Rheumatic Diseases. ACR Open Rheumatol. 2019, 1, 571–579. [Google Scholar] [CrossRef]
- Kovjazin, R.; Carmon, L. The use of signal peptide domains as vaccine candidates. Hum. Vaccin Immunother. 2014, 10, 2733–2740. [Google Scholar] [CrossRef] [PubMed]
- Smith-Garvin, J.E.; Koretzky, G.A.; Jordan, M.S. T cell activation. Annu. Rev. Immunol. 2009, 27, 591–619. [Google Scholar] [CrossRef]
- Messaoudi, I.; Guevara Patiño, J.A.; Dyall, R.; LeMaoult, J.; Nikolich-Zugich, J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science 2002, 298, 1797–1800. [Google Scholar] [CrossRef]
- Engels, B.; Engelhard, V.H.; Sidney, J.; Sette, A.; Binder, D.C.; Liu, R.B.; Kranz, D.M.; Meredith, S.C.; Rowley, D.A.; Schreiber, H. Relapse or eradication of cancer is predicted by peptide-MHC affinity. Cancer Cell 2013, 23, 516–526. [Google Scholar] [CrossRef]
- Harndahl, M.; Rasmussen, M.; Roder, G.; Dalgaard Pedersen, I.; Sørensen, M.; Nielsen, M.; Buus, S. Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur. J. Immunol. 2012, 42, 1405–1416. [Google Scholar] [CrossRef]
- Vecchio, F.; Carré, A.; Korenkov, D.; Zhou, Z.; Apaolaza, P.; Tuomela, S.; Burgos-Morales, O.; Snowhite, I.; Perez-Hernandez, J.; Brandao, B.; et al. Coxsackievirus infection induces direct pancreatic β cell killing but poor antiviral CD8+ T cell responses. Sci. Adv. 2024, 10, eadl1122. [Google Scholar] [CrossRef]
- Zheng, Z.; Mergaert, A.M.; Fahmy, L.M.; Bawadekar, M.; Holmes, C.L.; Ong, I.M.; Bridges, A.J.; Newton, M.A.; Shelef, M.A. Disordered Antigens and Epitope Overlap Between Anti-Citrullinated Protein Antibodies and Rheumatoid Factor in Rheumatoid Arthritis. Arthritis Rheumatol. 2020, 72, 262–272. [Google Scholar] [CrossRef]
- Mayer, R.L.; Verbeke, R.; Asselman, C.; Aernout, I.; Gul, A.; Eggermont, D.; Boucher, K.; Thery, F.; Maia, T.M.; Demol, H.; et al. Immunopeptidomics-based design of mRNA vaccine formulations against Listeria monocytogenes. Nat. Commun. 2022, 13, 6075. [Google Scholar] [CrossRef]
- Marcu, A.; Bichmann, L.; Kuchenbecker, L.; Kowalewski, D.J.; Freudenmann, L.K.; Backert, L.; Mühlenbruch, L.; Szolek, A.; Lübke, M.; Wagner, P.; et al. HLA Ligand Atlas: A benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy. J. Immunother. Cancer 2021, 9, e002071. [Google Scholar] [CrossRef]
- Ritz, D.; Gloger, A.; Weide, B.; Garbe, C.; Neri, D.; Fugmann, T. High-sensitivity HLA class I peptidome analysis enables a precise definition of peptide motifs and the identification of peptides from cell lines and patients’ sera. Proteomics 2016, 16, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Sarkizova, S.; Klaeger, S.; Le, P.M.; Li, L.W.; Oliveira, G.; Keshishian, H.; Hartigan, C.R.; Zhang, W.; Braun, D.A.; Ligon, K.L.; et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 2020, 38, 199–209. [Google Scholar] [CrossRef]
- Fujiwara, K.; Shao, Y.; Niu, N.; Zhang, T.; Herbst, B.; Henderson, M.; Muth, S.; Zhang, P.; Zheng, L. Direct identification of HLA class I and class II-restricted T cell epitopes in pancreatic cancer tissues by mass spectrometry. J. Hematol. Oncol. 2022, 15, 154. [Google Scholar] [CrossRef]
- Solleder, M.; Guillaume, P.; Racle, J.; Michaux, J.; Pak, H.-S.; Müller, M.; Coukos, G.; Bassani-Sternberg, M.; Gfeller, D. Mass Spectrometry Based Immunopeptidomics Leads to Robust Predictions of Phosphorylated HLA Class I Ligands. Mol. Cell. Proteom. 2020, 19, 390–404. [Google Scholar] [CrossRef]
- Sudhir, P.-R.; Lin, T.-D.; Zhang, Q. HLA Allele-Specific Quantitative Profiling of Type 1 Diabetic B Lymphocyte Immunopeptidome. J. Proteome Res. 2022, 21, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Caron, E.; Espona, L.; Kowalewski, D.J.; Schuster, H.; Ternette, N.; Alpízar, A.; Schittenhelm, R.B.; Ramarathinam, S.H.; Lindestam Arlehamn, C.S.; Chiek Koh, C.; et al. An open-source computational and data resource to analyze digital maps of immunopeptidomes. eLife 2015, 4, e07661. [Google Scholar] [CrossRef] [PubMed]
- Klatt, M.G.; Mack, K.N.; Bai, Y.; Aretz, Z.E.H.; Nathan, L.I.; Mun, S.S.; Dao, T.; Scheinberg, D.A. Solving an MHC allele-specific bias in the reported immunopeptidome. JCI Insight 2020, 5, 141264. [Google Scholar] [CrossRef]
- Bassani-Sternberg, M.; Chong, C.; Guillaume, P.; Solleder, M.; Pak, H.; Gannon, P.O.; Kandalaft, L.E.; Coukos, G.; Gfeller, D. Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLoS Comput. Biol. 2017, 13, e1005725. [Google Scholar] [CrossRef] [PubMed]
- Shraibman, B.; Barnea, E.; Kadosh, D.M.; Haimovich, Y.; Slobodin, G.; Rosner, I.; López-Larrea, C.; Hilf, N.; Kuttruff, S.; Song, C.; et al. Identification of Tumor Antigens Among the HLA Peptidomes of Glioblastoma Tumors and Plasma. Mol. Cell. Proteom. 2019, 18, 1255–1268. [Google Scholar] [CrossRef] [PubMed]
- Yarmarkovich, M.; Marshall, Q.F.; Warrington, J.M.; Premaratne, R.; Farrel, A.; Groff, D.; Li, W.; di Marco, M.; Runbeck, E.; Truong, H.; et al. Targeting of intracellular oncoproteins with peptide-centric CARs. Nature 2023, 623, 820–827. [Google Scholar] [CrossRef]
- Nicholas, B.; Bailey, A.; Staples, K.J.; Wilkinson, T.; Elliott, T.; Skipp, P. Immunopeptidomic analysis of influenza A virus infected human tissues identifies internal proteins as a rich source of HLA ligands. PLoS Pathog. 2022, 18, e1009894. [Google Scholar] [CrossRef]
- Parkhurst, M.R.; Fitzgerald, E.B.; Southwood, S.; Sette, A.; Rosenberg, S.A.; Kawakami, Y. Identification of a shared HLA-A*0201-restricted T-cell epitope from the melanoma antigen tyrosinase-related protein 2 (TRP2). Cancer Res. 1998, 58, 4895–4901. [Google Scholar] [PubMed]
- Wang, J.; Jelcic, I.; Mühlenbruch, L.; Haunerdinger, V.; Toussaint, N.C.; Zhao, Y.; Cruciani, C.; Faigle, W.; Naghavian, R.; Foege, M.; et al. HLA-DR15 Molecules Jointly Shape an Autoreactive T Cell Repertoire in Multiple Sclerosis. Cell 2020, 183, 1264–1281.e20. [Google Scholar] [CrossRef]
- Moser, J.J.; Chan, E.K.L.; Fritzler, M.J. An SNP in the trinucleotide repeat region of the TNRC6A gene maps to a major TNGW1 autoepitope in patients with autoantibodies to GW182. Adv. Exp. Med. Biol. 2013, 768, 243–259. [Google Scholar] [CrossRef]
- Rosen, A.; Casciola-Rosen, L. Autoantigens in systemic autoimmunity: Critical partner in pathogenesis. J. Intern. Med. 2009, 265, 625–631. [Google Scholar] [CrossRef]
- Pedersen, A.E. The potential for induction of autoimmune disease by a randomly-mutated self-antigen. Med. Hypotheses 2007, 68, 1240–1246. [Google Scholar] [CrossRef]
- Xiang, Y.; Zhang, M.; Jiang, D.; Su, Q.; Shi, J. The role of inflammation in autoimmune disease: A therapeutic target. Front. Immunol. 2023, 14, 1267091. Available online: https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2023.1267091/full (accessed on 10 September 2025). [CrossRef]
- Muñoz, L.E.; Lauber, K.; Schiller, M.; Manfredi, A.A.; Herrmann, M. The role of defective clearance of apoptotic cells in systemic autoimmunity. Nat. Rev. Rheumatol. 2010, 6, 280–289. [Google Scholar] [CrossRef] [PubMed]
- UniProt. UniProt-P54253·ATX1_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P54253/entry (accessed on 3 September 2025).
- UniProt. UniProt-O14646·CHD1_HUMAN. Available online: https://www.uniprot.org/uniprotkb/O14646/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q9UJU5·FOXD3_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q9UJU5/entry (accessed on 3 September 2025).
- UniProt. UniProt-O14686·KMT2D_HUMAN. Available online: https://www.uniprot.org/uniprotkb/O14686/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q8NBZ0·IN80E_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q8NBZ0/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q7Z3Z5·Q7Z3Z5_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q7Z3Z5/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q6PJG2·MDEAS_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q6PJG2/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q8TBK2·SETD6_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q8TBK2/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q9Y618·NCOR2_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q9Y618/entry (accessed on 3 September 2025).
- UniProt. UniProt-P06748·NPM_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P06748/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q10571·MN1_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q10571/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q9H3D4·P63_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q9H3D4/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q15911·ZFHX3_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q15911/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q5VZL5·ZMYM4_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q5VZL5/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q96KR1·ZFR_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q96KR1/entry (accessed on 3 September 2025).
- UniProt. UniProt-A0A2R8Y5A6·A0A2R8Y5A6_HUMAN. Available online: https://www.uniprot.org/uniprotkb/A0A2R8Y5A6/entry (accessed on 3 September 2025).
- UniProt. UniProt-E7ENM8·E7ENM8_HUMAN. Available online: https://www.uniprot.org/uniprotkb/E7ENM8/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q6Y7W6·GGYF2_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q6Y7W6/entry (accessed on 3 September 2025).
- UniProt. UniProt-P42858·HD_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P42858/entry (accessed on 3 September 2025).
- UniProt. UniProt-P40126·TYRP2_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P40126/entry (accessed on 3 September 2025).
- UniProt. UniProt-C9JE40·PATL2_HUMAN. Available online: https://www.uniprot.org/uniprotkb/C9JE40/entry (accessed on 3 September 2025).
- UniProt. UniProt-A1IGU5·ARH37_HUMAN. Available online: https://www.uniprot.org/uniprotkb/A1IGU5/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q7L8J4·3BP5L_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q7L8J4/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q8NDV7·TNR6A_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q8NDV7/entry (accessed on 3 September 2025).
- UniProt. UniProt-P51648·AL3A2_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P51648/entry (accessed on 3 September 2025).
- UniProt. UniProt-O75460·ERN1_HUMAN. Available online: https://www.uniprot.org/uniprotkb/O75460/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q9BY12·SCAPE_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q9BY12/entry (accessed on 3 September 2025).
- UniProt. UniProt-P54098·DPOG1_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P54098/entry (accessed on 3 September 2025).
- UniProt. UniProt-P03891·NU2M_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P03891/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q2M2I8·AAK1_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q2M2I8/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q5T1A1·DCST2_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q5T1A1/entry (accessed on 3 September 2025).
- UniProt. UniProt-P21917·DRD4_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P21917/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q13492·PICAL_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q13492/entry (accessed on 3 September 2025).
- UniProt. UniProt-B7Z2A4·B7Z2A4_HUMAN. Available online: https://www.uniprot.org/uniprotkb/B7Z2A4/entry (accessed on 3 September 2025).
- UniProt. UniProt-A0A087X0R9·A0A087X0R9_HUMAN. Available online: https://www.uniprot.org/uniprotkb/A0A087X0R9/entry (accessed on 3 September 2025).
- UniProt. UniProt-A0A2R8Y4T1·A0A2R8Y4T1_HUMAN. Available online: https://www.uniprot.org/uniprotkb/A0A2R8Y4T1/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q9NZK5·ADA2_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q9NZK5/entry (accessed on 3 September 2025).
- UniProt. UniProt-P0DUB6·AMY1A_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P0DUB6/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q9Y264·ANGP4_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q9Y264/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q9UKU9·ANGL2_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q9UKU9/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q9Y5C1·ANGL3_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q9Y5C1/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q9BY76·ANGL4_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q9BY76/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q8NI99·ANGL6_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q8NI99/entry (accessed on 3 September 2025).
- UniProt. UniProt-P49747·COMP_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P49747/entry (accessed on 3 September 2025).
- UniProt. UniProt-P36222·CH3L1_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P36222/entry (accessed on 3 September 2025).
- UniProt. UniProt-P02675·FIBB_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P02675/entry (accessed on 3 September 2025).
- UniProt. UniProt-P02679·FIBG_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P02679/entry (accessed on 3 September 2025).
- UniProt. UniProt-Q08830·FGL1_HUMAN. Available online: https://www.uniprot.org/uniprotkb/Q08830/entry (accessed on 3 September 2025).
- UniProt. UniProt-O00602·FCN1_HUMAN. Available online: https://www.uniprot.org/uniprotkb/O00602/entry (accessed on 3 September 2025).
- UniProt. UniProt-P13727·PRG2_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P13727/entry (accessed on 3 September 2025).
- UniProt. UniProt-P13611·CSPG2_HUMAN. Available online: https://www.uniprot.org/uniprotkb/P13611/entry (accessed on 3 September 2025).
- Malavige, G.N.; Jeewandara, C.; Ogg, G.S. Dysfunctional Innate Immune Responses and Severe Dengue. Front. Cell. Infect. Microbiol. 2020, 10, 590004. Available online: https://www.frontiersin.org/journals/cellular-and-infection-microbiology/articles/10.3389/fcimb.2020.590004/full (accessed on 10 September 2025). [CrossRef]
- Khandia, R.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Malik, Y.S.; Singh, R.K.; Chaicumpa, W. Modulation of Dengue/Zika Virus Pathogenicity by Antibody-Dependent Enhancement and Strategies to Protect Against Enhancement in Zika Virus Infection. Front. Immunol. 2018, 9, 597. [Google Scholar] [CrossRef]
- Maucourant, C.; Queiroz, G.A.N.; Samri, A.; Grassi, M.F.R.; Yssel, H.; Vieillard, V. Zika virus in the eye of the cytokine storm. Eur. Cytokine Netw. 2019, 30, 74–81. [Google Scholar] [CrossRef]
- Nanaware, N.; Banerjee, A.; Mullick Bagchi, S.; Bagchi, P.; Mukherjee, A. Dengue Virus Infection: A Tale of Viral Exploitations and Host Responses. Viruses 2021, 13, 1967. [Google Scholar] [CrossRef] [PubMed]
- Clé, M.; Desmetz, C.; Barthelemy, J.; Martin, M.-F.; Constant, O.; Maarifi, G.; Foulongne, V.; Bolloré, K.; Glasson, Y.; De Bock, F.; et al. Zika Virus Infection Promotes Local Inflammation, Cell Adhesion Molecule Upregulation, and Leukocyte Recruitment at the Blood-Brain Barrier. mBio 2020, 11, e01183-20. [Google Scholar] [CrossRef] [PubMed]
- Pavesi, A.; Tiecco, G.; Rossi, L.; Sforza, A.; Ciccarone, A.; Compostella, F.; Lovatti, S.; Tomasoni, L.R.; Castelli, F.; Quiros-Roldan, E. Inflammatory Response Associated with West Nile Neuroinvasive Disease: A Systematic Review. Viruses 2024, 16, 383. [Google Scholar] [CrossRef]
- Lazarski, C.A.; Chaves, F.A.; Jenks, S.A.; Wu, S.; Richards, K.A.; Weaver, J.M.; Sant, A.J. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity 2005, 23, 29–40. [Google Scholar] [CrossRef]
- Rasmussen, M.; Fenoy, E.; Harndahl, M.; Kristensen, A.B.; Nielsen, I.K.; Nielsen, M.; Buus, S. Pan-specific prediction of peptide-MHC-I complex stability; a correlate of T cell immunogenicity. J. Immunol. 2016, 197, 1517–1524. [Google Scholar] [CrossRef]
- George, A.J.T.; Stark, J.; Chan, C. Understanding specificity and sensitivity of T-cell recognition. Trends Immunol. 2005, 26, 653–659. [Google Scholar] [CrossRef]
- Fox, D.A. Citrullination: A Specific Target for the Autoimmune Response in Rheumatoid Arthritis. J. Immunol. 2015, 195, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Xu, W.W.; Sham, Y.; Lam, H.; Sun, D.; Li, C.; Rasic, N.F.; Guan, Q.; James, A.A.; Simons, F.E.R. Mosquito salivary allergen Aed a 3: Cloning, comprehensive molecular analysis, and clinical evaluation. Allergy 2016, 71, 621–628. [Google Scholar] [CrossRef]
- Conway, M.J. Type I hypersensitivity promotes Aedes aegypti blood feeding. Sci. Rep. 2021, 11, 14891. [Google Scholar] [CrossRef]
- Wang, Z.; Liang, Y.; Zeng, F.; Li, T.; Cheng, G. A capture enzyme-linked immunosorbent assay for detection of mosquito salivary protein-specific immunoglobulin E. PLoS Neglected Trop. Dis. 2025, 19, e0013468. [Google Scholar] [CrossRef]
- Pandey, R.K.; Dahiya, S.; Mahita, J.; Sowdhamini, R.; Prajapati, V.K. Vaccination and immunization strategies to design Aedes aegypti salivary protein based subunit vaccine tackling Flavivirus infection. Int. J. Biol. Macromol. 2019, 122, 1203–1211. [Google Scholar] [CrossRef] [PubMed]
- Manning, J.E.; Morens, D.M.; Kamhawi, S.; Valenzuela, J.G.; Memoli, M. Mosquito Saliva: The Hope for a Universal Arbovirus Vaccine? J. Infect. Dis. 2018, 218, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ling, L.; Jiang, L.; Marin-Lopez, A. Research progress toward arthropod salivary protein vaccine development for vector-borne infectious diseases. PLoS Neglected Trop. Dis. 2024, 18, e0012618. [Google Scholar] [CrossRef]

| Protein | GenBank Accession Number | Length (Amino Acids) | Octapeptides Probed |
|---|---|---|---|
| AaVA-1 | A0A1S4EWW7.1 | 255 | 248 |
| Adenosine deaminase | XP_021698106.1 | 530 | 523 |
| Aegyptin | O01949.2 | 273 | 266 |
| AeMOPE-1 | Q8T9V8.1 | 95 | 88 |
| AgBR1 | ABF18180.1 | 439 | 432 |
| Aldehyde dehydrogenase | AGI96742.1 | 494 | 487 |
| α-glucosidase | P13080.1 | 579 | 572 |
| Amylase | AAB60935.1 | 737 | 730 |
| Angiopoietin | ABF18152.1 | 354 | 347 |
| Apyrase | P50635.2 | 562 | 555 |
| CLIPA3 | A0A6I8TBG6.1 | 472 | 465 |
| C-type lectin | ABF18475.1 | 163 | 156 |
| D7L1 | P18153.2 | 321 | 314 |
| D7L2 | Q0IF93.1 | 332 | 325 |
| Ficolin | JAN95606.1 | 283 | 276 |
| LTRIN | J9HGJ1.1 | 210 | 203 |
| Lysozyme | AAU09087.1 | 148 | 141 |
| Mucin 2 | ABF18030.1 | 281 | 274 |
| Mucin 3 | JAN95076.1 | 292 | 285 |
| Mucin 4 | JAN95079.1 | 574 | 567 |
| Mucin 6 | JAN95078.1 | 130 | 123 |
| NeSt1 | Q17F11.1 | 316 | 309 |
| PE-binding protein | ABF18501.1 | 224 | 217 |
| Proline-rich mucin | ABF18067.1 | 100 | 93 |
| Purine hydrolase | AAL76010.1 | 338 | 331 |
| Serpin | ABF18509.1 | 417 | 410 |
| SG34 | A0A1S4F550.1 | 312 | 305 |
| Sialokinin | P42634.2 | 85 | 78 |
| Venom allergen 5 | NP_001395334.1 | 200 | 193 |
| Total | 9516 | 9513 |
| MHC-II Allele | Associated Autoimmune Disease | References | |
|---|---|---|---|
| HLA-DQA1* | 01:02 | SLE; MS | [86,87] |
| 02:01 | Psoriasis; T1D | [88] | |
| 04:01 | T1D; SLE | [87,89] | |
| 04:02 | SLE | [88] | |
| 05:01 | T1D; GD | [89,90,91,92] | |
| HLA-DQB1* | 02:01 | T1D; celiac disease; SLE; MG | [87,90,91,93,94,95] |
| 03:02 | RA; T1D; PR; UC; pemphigus | [87,90,91,94,96] | |
| 03:03 | MG; psoriasis | [88,97] | |
| HLA-DRB1* | 01:01 | RA; AIH | [98,99,100,101] |
| 01:02 | RA | [102] | |
| 03:01 | SLE; T1D; MS; RA; AIH; SS; MG | [86,89,90,91,93,94,95,99,100,103,104,105] | |
| 03:04 | GD; MS | [92,106] | |
| 03:07 | RA | [88] | |
| 03:08 | RA | [88] | |
| 04:01 | RA; PR; MS; T1D; AIH | [87,94,98,99,100,101,103] | |
| 04:02 | Pemphigus | [96] | |
| 04:04 | T1D; PR; MS; RA | [87,90,98,99,103] | |
| 04:05 | T1D; RA | [90,98,101] | |
| 04:08 | RA | [98] | |
| 04:10 | RA | [101] | |
| 07:01 | AIH; RA; CD | [89,100,102] | |
| 08:02 | SLE | [88] | |
| 09:01 | RA; T1D; MG | [89,97,99] | |
| 10:01 | RA | [99,101] | |
| 13:03 | MS | [103] | |
| 14:01 | ITP; MS; GD | [103,107,108] | |
| 14:02 | RA | [98,99] | |
| 15:01 | SLE; thyroid autoimmune disease; MS; RA | [86,87,89,102,103] | |
| 15:02 | SLE; UC | [89,109] | |
| 16:02 | SLE | [88] | |
| MHC-I Allele | Associated Autoimmune Disease | References | |
|---|---|---|---|
| HLA-A* | 02:07 | Psoriasis; MG | [97,110] |
| 24:02 | T1D; MG | [97,102,111,112] | |
| 24:03 | T1D | [102,112] | |
| HLA-B* | 07:02 | T1D | [111] |
| 08:01 | MG | [95] | |
| 18:01 | T1D | [102,111] | |
| 35:02 | T1D | [111] | |
| 37:01 | MS; GD | [103,108] | |
| 39:06 | T1D | [102,111] | |
| 44:03 | T1D | [111] | |
| 46:01 | MG | [97] | |
| HLA-C* | 01:02 | MG; T1D | [97,112] |
| 03:02 | T1D | [112] | |
| 03:03 | T1D | [111] | |
| 06:02 | Psoriasis | [110,113,114] | |
| 07:01 | Psoriasis; GD | [102,110] | |
| 07:02 | SLE; psoriasis; GD | [86,102,110] | |
| 07:04 | Psoriasis; GD | [102,110] | |
| 08:02 | GD; T1D | [88,108] | |
| 12:02 | Psoriasis | [110] | |
| 15:02 | MS | [102] | |
| 16:01 | Psoriasis | [114] | |
| Protein | Peptides with Match | Matches in Human Proteome | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 18-mer | 13-mer | 12-mer | 11-mer | 10-mer | 9-mer | 8-mer | Total | ||
| LTRIN | 0 | 0 | 0 | 0 | 1 | 2 | 6 | 9 | 18 |
| Aldehyde dehydrogenase | 0 | 0 | 1 | 1 | 0 | 1 | 4 | 7 | 7 |
| Ficolin | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 5 | 5 |
| Angiopoietin | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 4 | 7 |
| α-glucosidase | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 3 | 3 |
| AgBR1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 |
| Lysozyme | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 2 |
| Mucin 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 |
| NeSt1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| Adenosine deaminase | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| Aegyptin | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 |
| Amylase | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Apyrase | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| D7L1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| Mucin 4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| Mucin 6 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| Proline-rich mucin | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| Purine hydrolase | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| Serpin | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| SG34 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| Venom allergen 5 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| Total | 1 | 1 | 1 | 1 | 4 | 7 | 32 | 47 | 60 |
| Aedes aegypti Protein | Peptide | Length (Amino Acids) | Human Proteins |
|---|---|---|---|
| Amylase Angiopoietin | ALVFVDNHDNQRGHGAGG | 18 | α-amylase 1A |
| GWTVIQRRLDGSV | 13 | Angiopoietin-related protein 2 | |
| GWTVIQRR | 8 | Angiopoietin-related protein 4 | |
| Angiopoietin-related protein 6 | |||
| Fibrinogen-like 1 | |||
| GEYWLGLE | 8 | Angiopoietin-related protein 6 | |
| Angiopoietin-related protein 2 | |||
| RLDGSVDF | 8 | Fibrinogen γ chain | |
| Aldehyde dehydrogenase | EIFGPILPIVNV | 12 | Aldehyde dehydrogenase 3 |
| TLELGGKSPCY | 11 | Aldehyde dehydrogenase 3 | |
| GQTCIAPDY | 9 | Aldehyde dehydrogenase 3 | |
| LELGGKSP | 8 | Cytosolic 10-formyltetrahydrofolate dehydrogenase | |
| KFLQEARS | 8 | Tyrosine-protein kinase | |
| SLPFGGVG | 8 | Aldehyde dehydrogenase 3 | |
| LASSRYPP | 8 | INO80 complex subunit E | |
| Adenosine deaminase | PISNQVLKLV | 10 | Adenosine deaminase |
| Mucin 6 | STPSSSNSTS | 10 | PI-binding clathrin assembly protein |
| LTRIN | QQQQQQHQQP | 10 | GRB10-interacting GYF protein 2 |
| QQQQQQHQQ | 9 | Mixed-lineage leukemia 2 | |
| Mediator of RNA polymerase II transcription | |||
| Histone-lysine N-methyltransferase | |||
| QQQQQQQPQ | 9 | Zinc finger homeobox protein 3 | |
| TNRC6A | |||
| QQQQQQHQ | 8 | CHD-1 | |
| Ataxin 1 | |||
| Tumor protein 63 | |||
| PQQQQQQH | 8 | Transcriptional activator MN1 | |
| QQQQQQPQ | 8 | DNA polymerase subunit γ-1 | |
| MIDEAS | |||
| QQQQQQQP | 8 | Ataxin 2 | |
| Huntingtin | |||
| Nuclear receptor corepressor 2 | |||
| Tensin-1 | |||
| YQQQQQQQ | 8 | AP2-associated protein kinase 1 isoform 4 | |
| Zinc finger RNA-binding protein | |||
| APHHGQPQ | 8 | Forkhead box protein D3 | |
| α-glucosidase | KDSDGDGIGD | 10 | Cartilage oligomeric matrix protein |
| TYYGEEIGM | 9 | SLC3A1 | |
| YPRSFKDS | 8 | SLC3A1 | |
| Lysozyme | LVGLFVLLA | 9 | L-dopachrome tautomerase |
| Ficolin | GGWTVIQNR | 9 | Fibrinogen β chain |
| FSTLDSDND | 9 | Angiopoietin-4 | |
| DGEFWLGL | 8 | Angiopoietin-related protein 3 | |
| GDAGDSLS | 8 | Angiopoietin-related protein 6 | |
| SNLNGLYL | 8 | Ficolin | |
| Aegyptin | EEENEGEE | 8 | PAT1 homolog 2 |
| Tubulin α-8 chain | |||
| AgBR1 | VSANNATT | 8 | RPTP-α |
| FDGLDLAW | 8 | Chitinase-3-like protein 1 | |
| Mucin 3 | LLIGAVLA | 8 | Dopamine receptor D4 |
| ELDSSDEE | 8 | DC-STAMP domain-containing protein 2 | |
| D7L1 | MKLPLLLA | 8 | Pro eosinophil major basic protein |
| NeSt1 | KVTELEQQ | 8 | SH3 domain-binding protein 5-like |
| Apyrase | KLTVGKRK | 8 | Zinc finger MYM-type protein 4 |
| Lysozyme | CSLAKALL | 8 | Rho GEF 37 |
| Mucin 4 | DDDGDDFD | 8 | Nucleophosmin |
| Proline-rich mucin | SLLLILSI | 8 | NADH dehydrogenase |
| Purine hydrolase | DQDGGGDD | 8 | N-lysine methyltransferase SETD6 |
| Serpin | DVLSKLKE | 8 | SCAPER |
| SG34 | EVQLLRES | 8 | ER to nucleus signaling 1 |
| Venom allergen 5 | QMVSDRTT | 8 | Versican |
| Aedes aegypti Protein | Matches | Aedes aegypti Peptides | Human Proteins | Alleles | ≥80% Identity |
|---|---|---|---|---|---|
| Angiopoietin | 113 | 23 | Angiopoietin-related protein 2; angiopoietin-related protein 4; angiopoietin-related protein 6 | HLA-DRB1*08:02 | 79 |
| HLA-DRB1*13:03 | |||||
| HLA-DRB1*14:02 | |||||
| Ficolin | 67 | 14 | Angiopoietin-4; fibrinogen β chain; angiopoietin-related protein 6 | HLA-DQA1*02:01/DQB1*02:01; HLA-DQA1*02:01/DQB1*02:02; HLA-DQA1*02:01/DQB1*02:04; HLA-DQA1*02:01/DQB1*02:05; HLA-DQA1*02:01/DQB1*02:06 | 36 |
| HLA-DQA1*04:01/DQB1*02:01; HLA-DQA1*04:01/DQB1*02:02; HLA-DQA1*04:01/DQB1*02:04; HLA-DQA1*04:01/DQB1*02:05; HLA-DQA1*04:01/DQB1*02:06; HLA-DQA1*04:01/DQB1*03:02 | |||||
| HLA-DQA1*04:02/DQB1*02:01; HLA-DQA1*04:02/DQB1*02:02; HLA-DQA1*04:02/DQB1*02:04; HLA-DQA1*04:02/DQB1*02:05; HLA-DQA1*04:02/DQB1*02:06; HLA-DQA1*04:02/DQB1*03:02 | |||||
| HLA-DQA1*05:01/DQB1*02:01; HLA-DQA1*05:01/DQB1*02:02; HLA-DQA1*05:01/DQB1*02:04; HLA-DQA1*05:01/DQB1*02:05; HLA-DQA1*05:01/DQB1*02:06 | |||||
| HLA-DRB1*04:05 | |||||
| HLA-DRB1*04:08 | |||||
| HLA-DRB1*04:10 | |||||
| LTRIN | 33 | 6 | Forkhead box protein D3 | HLA-DQA1*01:02/DQB1*03:01 | 0 |
| HLA-DQA1*02:01/DQB1*03:01 | |||||
| HLA-DQA1*04:01/DQB1*03:01; HLA-DQA1*04:01/DQB1*03:03 | |||||
| HLA-DQA1*04:02/DQB1*03:01; HLA-DQA1*04:02/DQB1*03:03; HLA-DQA1*04:02/DQB1*03:04 | |||||
| HLA-DQA1*05:01/DQB1*03:01; HLA-DQA1*05:01/DQB1*03:03; HLA-DQA1*05:01/DQB1*03:04 | |||||
| Aldehyde dehydrogenase | 25 | 16 | Aldehyde dehydrogenase 3 | HLA-DRB1*04:01 | 14 |
| HLA-DRB1*04:02 | |||||
| HLA-DRB1*04:04 | |||||
| HLA-DRB1*04:08 | |||||
| HLA-DRB1*04:10 | |||||
| HLA-DRB1*10:01 | |||||
| Amylase | 12 | 7 | α-amylase 1A | HLA-DRB1*03:01 | 12 |
| HLA-DRB1*03:04 | |||||
| HLA-DRB1*03:08 | |||||
| HLA-DRB1*04:02 | |||||
| HLA-DRB1*14:02 | |||||
| Venom allergen 5 | 9 | 3 | Versican | HLA-DRB1*03:01 | 3 |
| HLA-DRB1*03:04 | |||||
| HLA-DRB1*03:07 | |||||
| HLA-DRB1*03:08 | |||||
| 6 | 259 | 69 | 10 | 46 | 144 |
| Matches | MHC-II Alleles | Total Alleles |
|---|---|---|
| 58 | HLA-DRB1*13:03 | 1 |
| 37 | HLA-DRB1*08:02 | 1 |
| 21 | HLA-DRB1*14:02 | 1 |
| 13 | HLA-DRB1*04:05 | 1 |
| 10 | HLA-DRB1*10:01 | 1 |
| 7 | HLA-DRB1*04:02 | 1 |
| 6 | HLA-DQA1*04:01/DQB1*03:01; HLA-DQA1*04:02/DQB1*03:01; HLA-DRB1*04:04 | 3 |
| 5 | HLA-DRB1*03:04; HLA-DRB1*03:08 | 2 |
| 4 | HLA-DQA1*02:01/DQB1*02:05; HLA-DQA1*04:01/DQB1*02:05; HLA-DQA1*04:02/DQB1*02:05; HLA-DQA1*04:02/DQB1*03:04; HLA-DQA1*05:01/DQB1*02:05; HLA-DQA1*05:01/DQB1*03:01; HLA-DRB1*04:08; HLA-DRB1*04:10; HLA-DRB1*03:01 | 9 |
| 3 | HLA-DQA1*01:02/DQB1*03:01; HLA-DQA1*02:01/DQB1*02:01; HLA-DQA1*02:01/DQB1*02:02; HLA-DQA1*02:01/DQB1*02:04; HLA-DQA1*02:01/DQB1*02:06; HLA-DQA1*05:01/DQB1*02:01; HLA-DQA1*05:01/DQB1*02:02; HLA-DQA1*05:01/DQB1*02:04; HLA-DQA1*05:01/DQB1*02:06 | 9 |
| 2 | HLA-DQA1*02:01/DQB1*03:01; HLA-DQA1*04:01/DQB1*03:03; HLA-DQA1*04:02/DQB1*03:03; HLA-DQA1*05:01/DQB1*03:03; HLA-DQA1*05:01/DQB1*03:04 | 5 |
| 1 | HLA-DQA1*04:01/DQB1*02:01; HLA-DQA1*04:01/DQB1*02:02; HLA-DQA1*04:01/DQB1*02:04; HLA-DQA1*04:01/DQB1*02:06; HLA-DQA1*04:01/DQB1*03:02; HLA-DQA1*04:02/DQB1*02:01; HLA-DQA1*04:02/DQB1*02:02; HLA-DQA1*04:02/DQB1*02:04; HLA-DQA1*04:02/DQB1*02:06; HLA-DQA1*04:02/DQB1*03:02; HLA-DRB1*04:01; HLA-DRB1*03:07 | 12 |
| Aedes aegypti Protein | Matches | Aedes aegypti Peptides | Human Proteins | Alleles | Identical |
|---|---|---|---|---|---|
| Aldehyde dehydrogenase | 10 | 3 | Aldehyde dehydrogenase 3; INO80 complex subunit E | HLA-A*02:07; HLA-A*24:02; HLA-A*24:03; HLA-B*07:02; HLA-B*35:02; HLA-B*46:01; HLA-C*03:02; HLA-C*07:04; HLA-C*16:01 | 7 |
| Ficolin | 6 | 4 | Angiopoietin-related protein 3; angiopoietin-related protein 6; ficolin | HLA-B*37:01; HLA-B*44:03; HLA-C*03:03; HLA-C*08:02; HLA-C*15:02; HLA-C*16:01 | 0 |
| Angiopoietin | 6 | 3 | Angiopoietin-related protein 2; fibrinogen γ chain | HLA-A*24:03; HLA-C*06:02; HLA-C*07:01; HLA-C*07:02; HLA-C*07:04; HLA-C*08:02 | 2 |
| Adenosine deaminase | 6 | 2 | Adenosine deaminase 2 | HLA-C*03:03; HLA-C*06:02; HLA-C*15:02; HLA-C*16:01 | 6 |
| α-glucosidase | 6 | 1 | SLC3A1 | HLA-A*24:02; HLA-A*24:03; HLA-C*06:02; HLA-C*07:01; HLA-C*07:02; HLA-C*07:04 | 6 |
| D7L1 | 3 | 2 | Pro eosinophil major basic protein | HLA-B*39:06; HLA-C*01:02; HLA-C*06:02 | 0 |
| NeSt1 | 2 | 1 | SH3 domain-binding protein 5-like | HLA-A*02:07; HLA-C*15:02 | 0 |
| AgBR1 | 1 | 1 | Chitinase-3-like protein 1 | HLA-A*24:02 | 0 |
| Lysozyme | 1 | 1 | L-dopachrome tautomerase | HLA-A*02:07 | 0 |
| Mucin 6 | 1 | 1 | PI-binding clathrin assembly protein | HLA-C*01:02 | 1 |
| 10 | 43 | 19 | 14 | 19 | 22 |
| Matches | MHC-II Alleles | Total Alleles |
|---|---|---|
| 4 | HLA-A*02:07; HLA-C*06:02; HLA-C*15:02; HLA-C*16:01 | 4 |
| 3 | HLA-A*24:02; HLA-A*24:03; HLA-C*07:04 | 3 |
| 2 | HLA-C*01:02; HLA-C*03:03; HLA-C*07:01; HLA-C*07:02; HLA-C*08:02 | 5 |
| 1 | HLA-B*07:02; HLA-B*35:02; HLA-B*37:01; HLA-B*39:06; HLA-B*44:03; HLA-B*46:01; HLA-C*03:02 | 7 |
| Human Protein | IEDB Peptide | Peptide ID | Validation | Reference |
|---|---|---|---|---|
| Fibrinogen β chain | GGWTVIQNRQDG | 961381 | B cell activation; association to RA | [121] |
| GWTVIQNRQDGS | 962133 | |||
| WTVIQNRQDGSV | 968636 | |||
| Aldehyde dehydrogenase 3 | EIFGPILPIVNV | 2244697 | Presentation bound to MHC-I | [122,123,124] |
| QDEIFGPILP | 1266040 | |||
| LPFGGVGASGMGRY | 621205 | |||
| Fibrinogen γ chain | LDGSVDFK | 1028523 | Processing from tumor antigen; presentation bound to MHC-I alleles HLA-C*16:01 and HLA-A*34:02 | [125,126,127] |
| RLDGSVDFK | 991140 | |||
| Angiopoietin-related protein 6 | YHGDAGDSL | 450124 | Presentation bound to MHC-I alleles HLA-B*15:10, HLA-B*38:01 and HLA-B*38:02; association to T1D | [125,128,129,130,131,132] |
| α-amylase 1A | ALVFVDNHDNQ | 2185722 | Presentation bound to MHC-I | [133] |
| Adenosine deaminase 2 | NQVLKLVSDLRNHP | 2038620 | Presentation bound to MHC-II | [134] |
| L-dopachrome tautomerase | ALVGLFVLL | 2961 | Processing from tumor antigen; presentation bound to MHC-I allele HLA-A*02:01 | [135] |
| Pro eosinophil major basic protein | KLPLLLAL | 1393024 | Presentation bound to MHC-II allele HLA-DRB1*15:01; association to MS | [136] |
| TNRC6A | QQQQQQQQPQQQQPQ | 177936 | B cell activation; association to SLE | [137] |
| Localization | Protein | Tissue/Cell Type Distribution | Function | Reference |
|---|---|---|---|---|
| Nucleus | Ataxin 1 | Neurons, oligodendrocytes | Transcriptional regulation | [142] |
| CHD-1 | Ubiquitous | Chromatin remodeling | [143] | |
| Forkhead box protein D3 | Schwann cells, melanocytes | Transcriptional regulation | [144] | |
| Histone-lysine N-methyltransferase 2D | Ubiquitous | Chromatin remodeling | [145] | |
| INO80 complex subunit E | Ubiquitous | Chromatin remodeling | [146] | |
| Mediator complex subunit 12 | Ubiquitous | Transcriptional regulation | [147] | |
| MIDEAS | Ubiquitous | Histone deacetylation | [148] | |
| N-lysine methyltransferase SETD6 | Ubiquitous | Transcriptional regulation | [149] | |
| Nuclear receptor corepressor 2 | Ubiquitous | Transcriptional repression | [150] | |
| Nucleophosmin | Ubiquitous | Ribosome nuclear export | [151] | |
| Transcriptional activator MN1 | Ubiquitous | Transcriptional activation | [152] | |
| Tumor protein 63 | Epithelial cells | Transcriptional regulation | [153] | |
| Zinc finger homeobox protein 3 | Ubiquitous | Transcriptional regulation | [154] | |
| Zinc finger MYM-type protein 4 | Ubiquitous | Transcriptional regulation | [155] | |
| Zinc finger RNA binding protein | Ubiquitous | Regulation of RNA metabolism | [156] | |
| Cytoplasm | Ataxin 2 | CNS | Regulation of EGFR endocytosis | [157] |
| Tyrosine-protein kinase | Ubiquitous | Regulation of cytoskeleton function | [158] | |
| GRB10-interacting GYF protein 2 | Ubiquitous | Translational repression | [159] | |
| Huntingtin | Brain | Scaffolding in vesicle transport | [160] | |
| L-dopachrome tautomerase | Melanocytes | Enzyme in melanin synthesis | [161] | |
| PAT1 homolog 2 | Oocytes | Translational repression | [162] | |
| Rho GEF 37 | Ubiquitous | Signal transduction | [163] | |
| SH3 domain-binding protein 5-like | Ubiquitous | Signal transduction | [164] | |
| TNRC6A | Ubiquitous | Translational repression | [165] | |
| Tubulin α-8 | Constituent of microtubules | |||
| ER | Aldehyde dehydrogenase 3 | Liver | Enzyme in lipid metabolism | [166] |
| ER to nucleus signaling 1 | Ubiquitous | Unfolded protein response | [167] | |
| SCAPER | Ubiquitous | Cell cycle regulation | [168] | |
| Mitochondria | DNA polymerase subunit γ-1 | Ubiquitous | Replication of mitochondrial DNA | [169] |
| NADH dehydrogenase subunit 2 | Brain, heart muscle | Enzyme in respiratory chain | [170] | |
| Cell membrane | AP2-associated protein kinase 1 | Neurons, T cells, NK cells | Regulation of endocytosis | [171] |
| DC-STAMP domain-containing protein 2 | Sperm cells | Sperm–egg fusion | [172] | |
| Dopamine receptor D4 | Retina, neurons | Neuronal signaling | [173] | |
| PI-binding clathrin assembly protein | Ubiquitous | Adapter protein in endocytosis | [174] | |
| RPTP-α | Ubiquitous | Focal adhesion formation | [175] | |
| SLC3A1 | Kidney and small intestine | Amino acid transporter | [176] | |
| Tensin-1 | Ubiquitous | Fibrillar adhesion formation | [177] | |
| Secreted | Adenosine deaminase 2 | Blood | Regulation of adenosine function | [178] |
| α-amylase 1A | Saliva | Carbohydrate digestion | [179] | |
| Angiopoietin-4 | Blood | Regulation of angiogenesis | [180] | |
| Angiopoietin-related protein 2 | Blood | Promotes inflammation | [181] | |
| Angiopoietin-related protein 3 | Blood | Regulation of lipid and glucose metabolism | [182] | |
| Angiopoietin-related protein 4 | Blood, ECM | Regulation of lipid metabolism and endothelial cell function | [183] | |
| Angiopoietin-related protein 6 | Blood | Wound healing | [184] | |
| Cartilage oligomeric matrix protein | ECM | Structural integrity of cartilage | [185] | |
| Chitinase-3-like protein 1 | Blood | Promotes inflammation | [186] | |
| Fibrinogen β chain | Blood | Clot formation | [187] | |
| Fibrinogen γ chain | Blood | Clot formation | [188] | |
| Fibrinogen-like 1 | Blood | Inhibition of T cell activation | [189] | |
| Ficolin | Blood | Pattern recognition receptor | [190] | |
| Pro eosinophil major basic protein | Blood | Cytotoxin and helminthotoxin | [191] | |
| Versican | ECM | Regulation of cell migration | [192] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arévalo-Cortés, A.; Rodriguez-Pinto, D. Mimicry in the Bite: Shared Sequences Between Aedes aegypti Salivary Proteins and Human Proteins. Proteomes 2025, 13, 56. https://doi.org/10.3390/proteomes13040056
Arévalo-Cortés A, Rodriguez-Pinto D. Mimicry in the Bite: Shared Sequences Between Aedes aegypti Salivary Proteins and Human Proteins. Proteomes. 2025; 13(4):56. https://doi.org/10.3390/proteomes13040056
Chicago/Turabian StyleArévalo-Cortés, Andrea, and Daniel Rodriguez-Pinto. 2025. "Mimicry in the Bite: Shared Sequences Between Aedes aegypti Salivary Proteins and Human Proteins" Proteomes 13, no. 4: 56. https://doi.org/10.3390/proteomes13040056
APA StyleArévalo-Cortés, A., & Rodriguez-Pinto, D. (2025). Mimicry in the Bite: Shared Sequences Between Aedes aegypti Salivary Proteins and Human Proteins. Proteomes, 13(4), 56. https://doi.org/10.3390/proteomes13040056





