Identification of Protein Markers for Chronic Ischemic Heart Disease Through Integrated Analysis of the Human Plasma Proteome and Genome-Wide Association Data
Abstract
1. Introduction
2. Materials and Methods
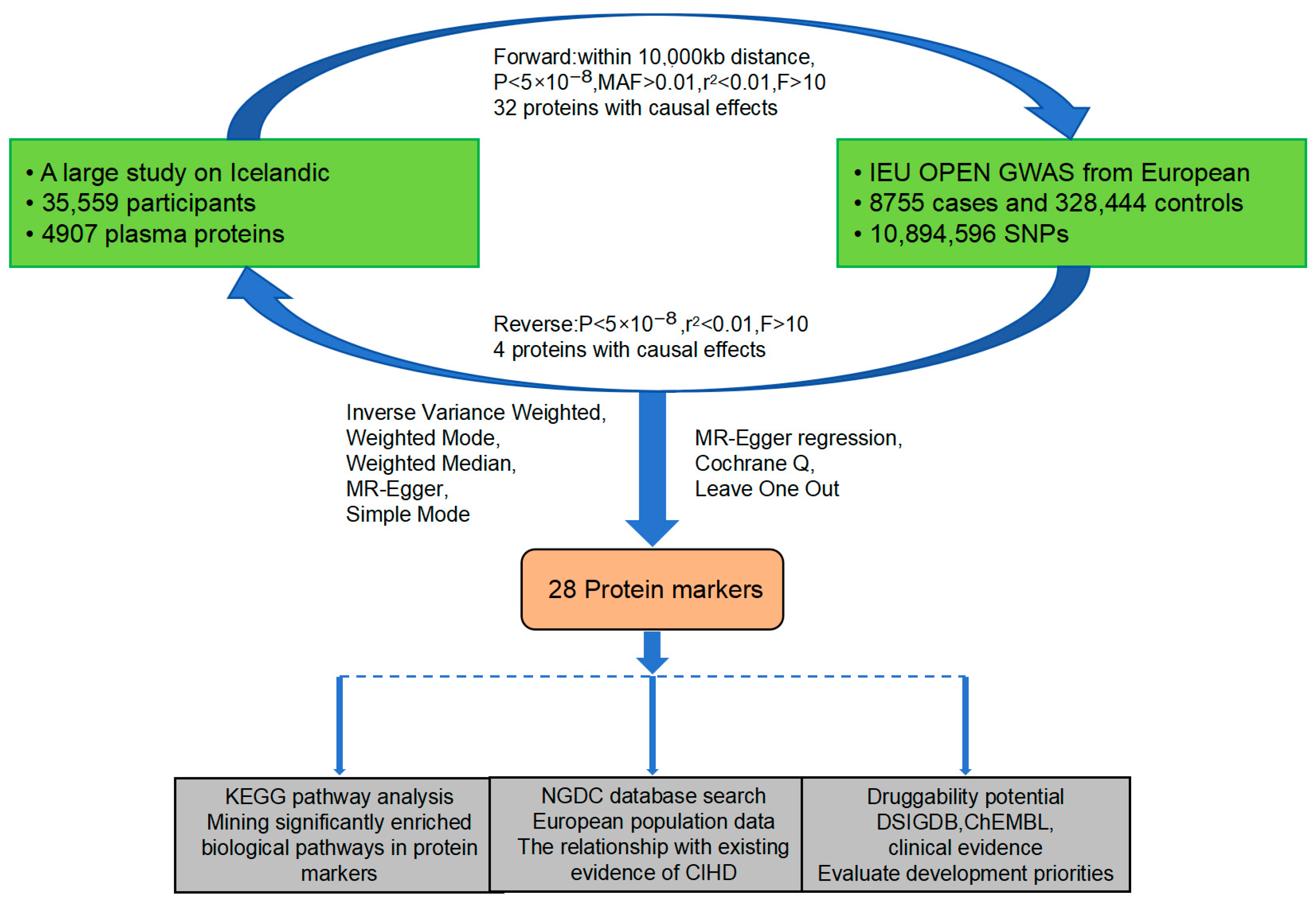
2.1. Research Design and Datasets
2.2. Choosing Genetic Instruments and Multiple Testing Correction
2.3. Bidirectional MR Analysis and Screening
2.3.1. Forward MR Analysis and Screening
2.3.2. Reverse MR
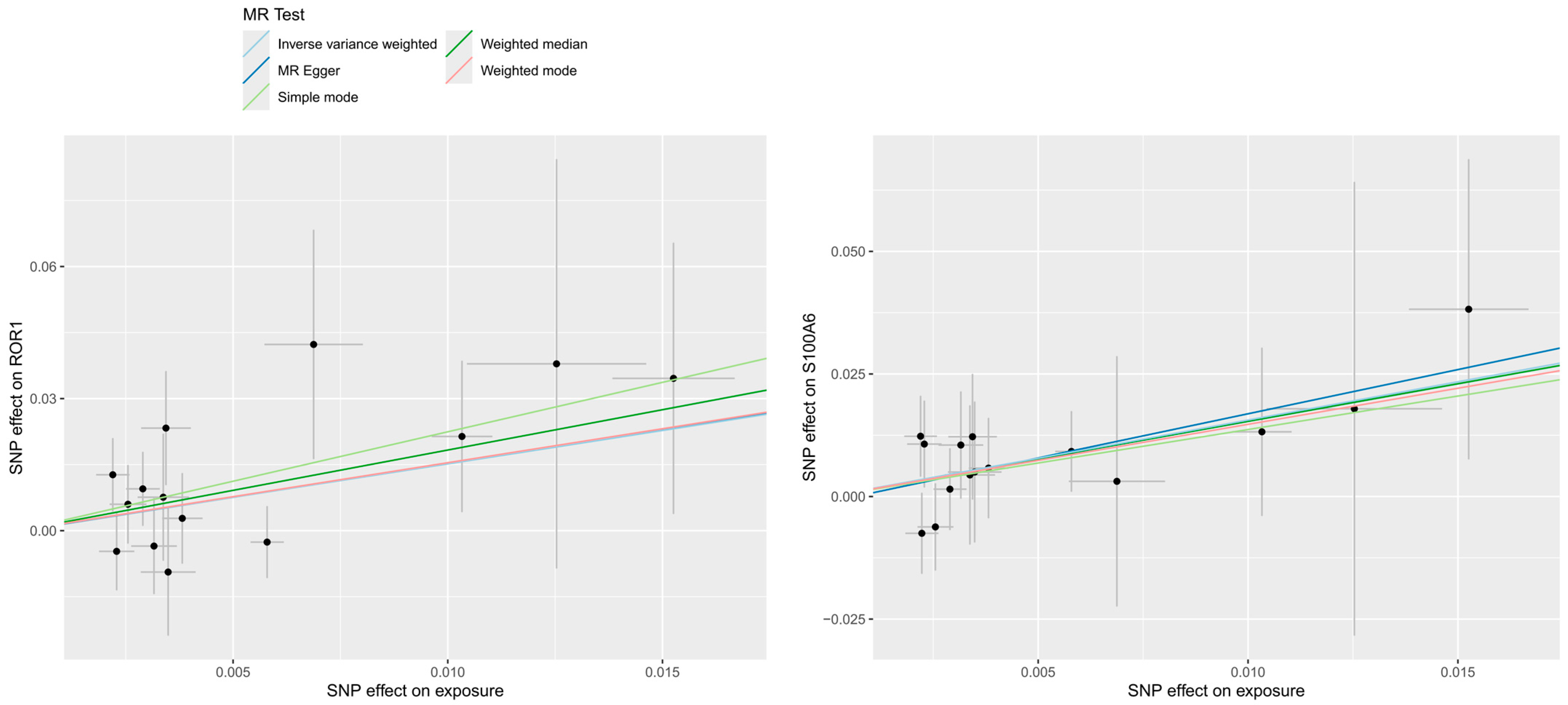
2.3.3. Sensitivity Analysis
2.4. KEGG Pathway Analysis
2.5. NGDC Search
2.6. Evaluation of Druggability
3. Results
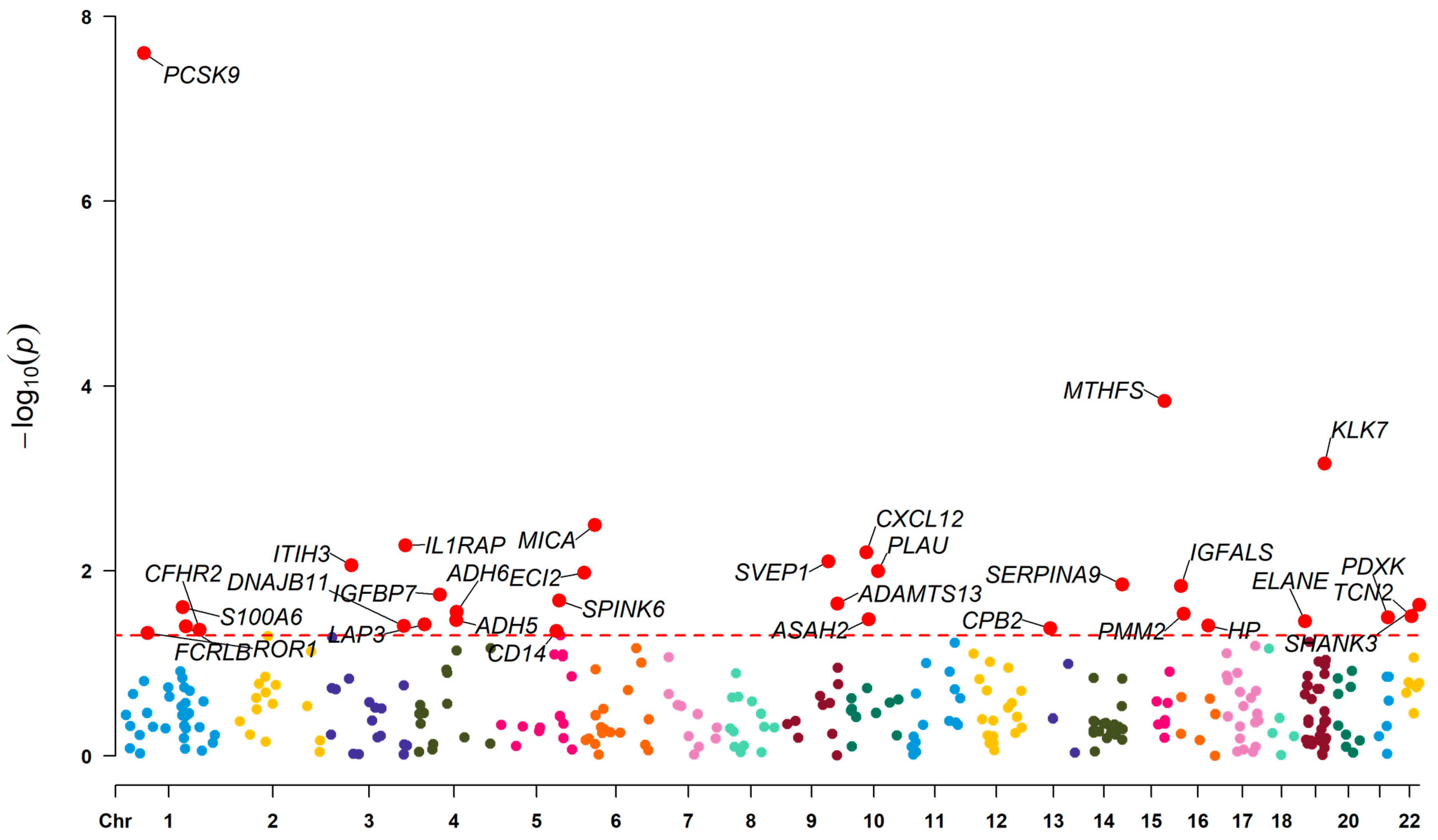
3.1. Effect of Plasma Proteins on CIHD
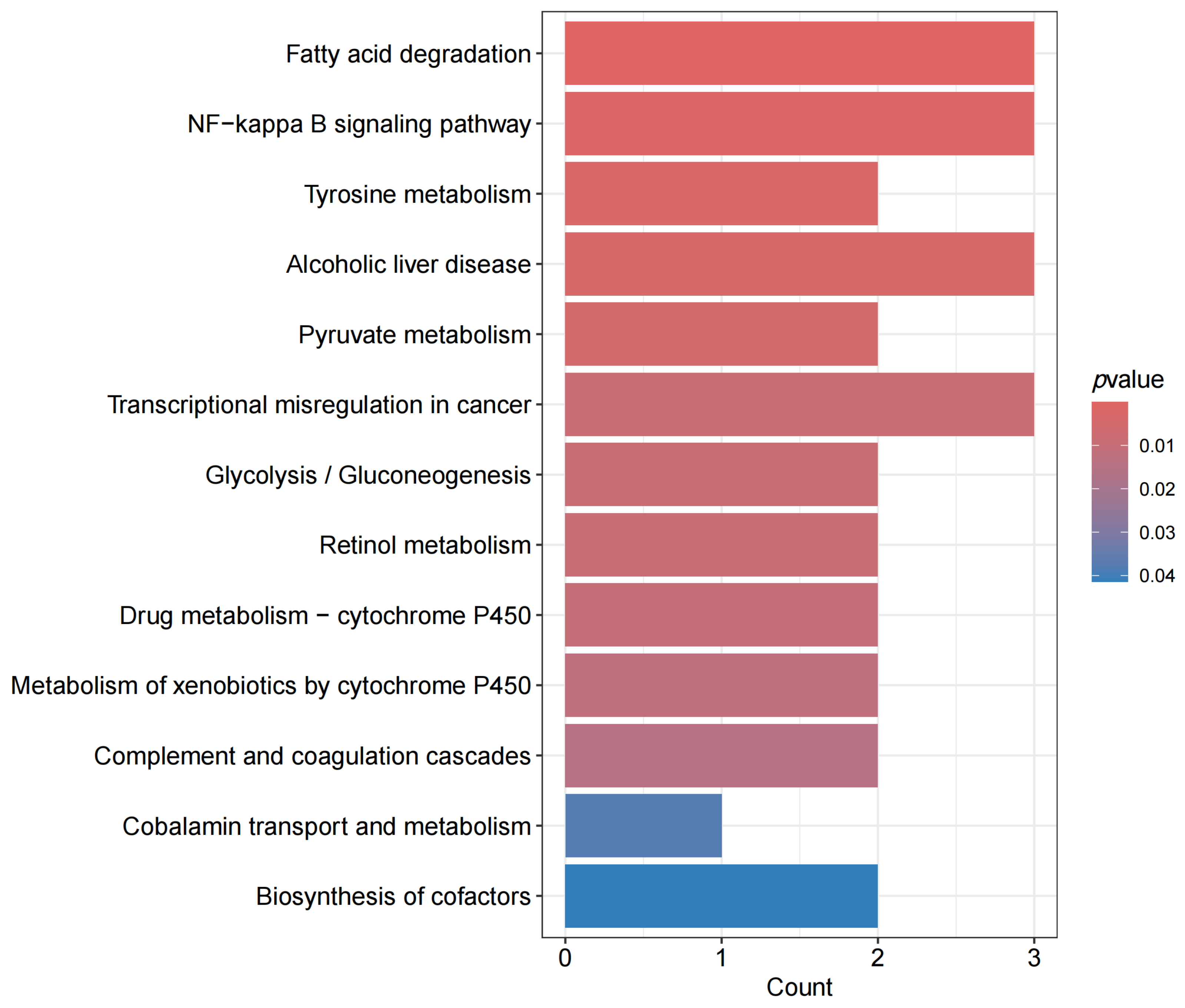
3.2. Pathway Analysis
3.3. Existing Evidence
3.4. Druggability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Full Name |
| CIHD | Chronic Ischemic Heart Disease |
| PCI | Percutaneous coronary intervention |
| CABG | Coronary artery bypass grafting |
| MR | Mendelian Randomization |
| IVs | Instrumental variables |
| RCTs | Randomised controlled trials |
| GWAS | Genome-wide association study |
| SNPs | Single-nucleotide polymorphisms |
| pQTLs | Protein Quantitative Trait Loci |
| LD | Linkage disequilibrium |
| FDR | False discovery rate |
| IVW | Inverse-variance weighted |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| NGDC | National Genomics Data Center |
| CNCB | China National Center for Bioinformation |
| CVD-A | Cardiovascular Disease Atlas |
| DSIGDB | Drug Signatures Database |
| PKC | Protein kinase C |
| TTP | Thrombotic thrombocytopenic purpura |
| ADH | Alcohol dehydrogenase |
| DME | Diabetic macular edema |
| ROS | Reactive oxygen species |
| TGF-β | Transforming growth factor-beta |
| CHF | Congestive heart failure |
| GPI | Glycosylphosphatidylinositol |
Appendix A
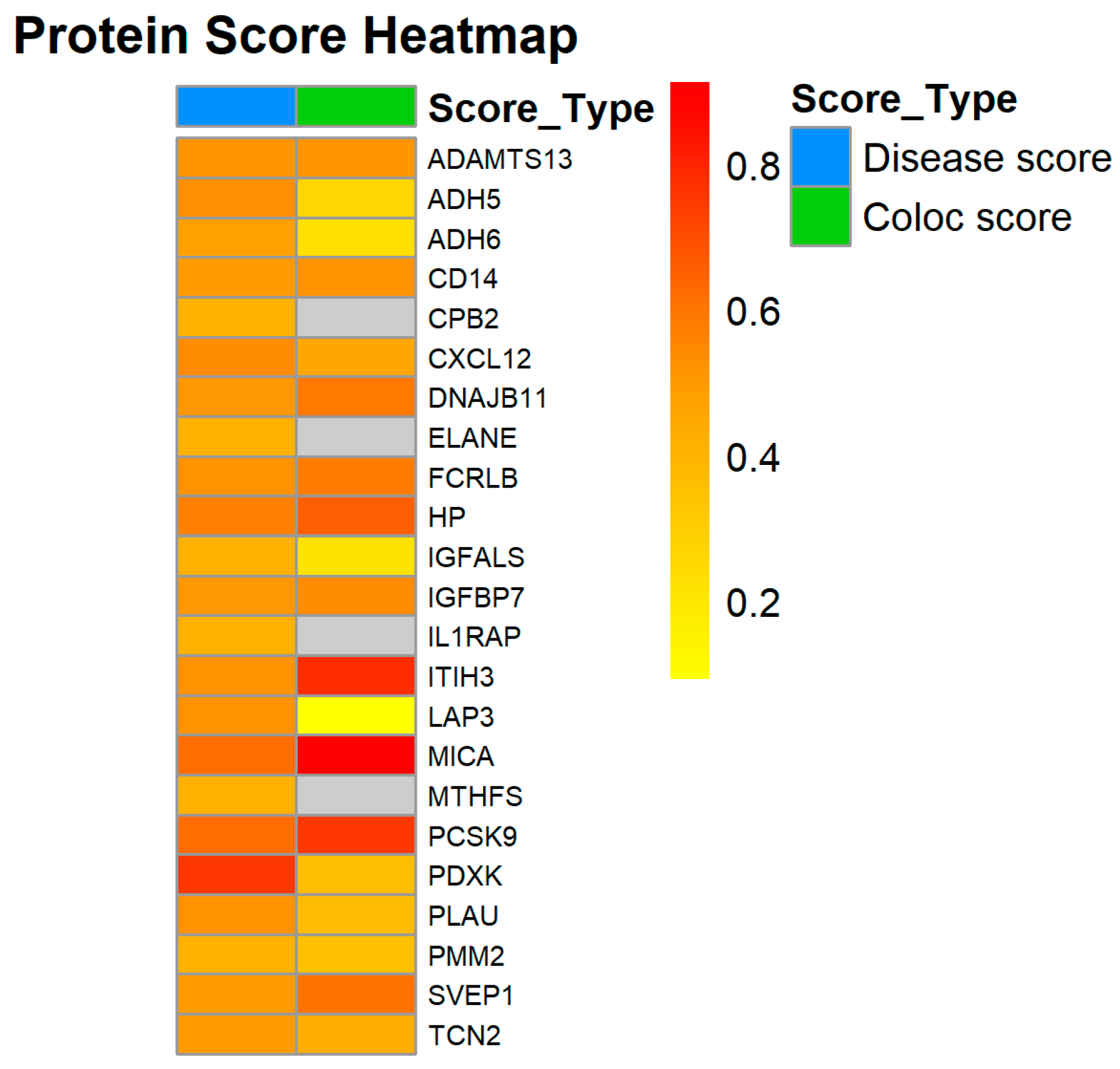
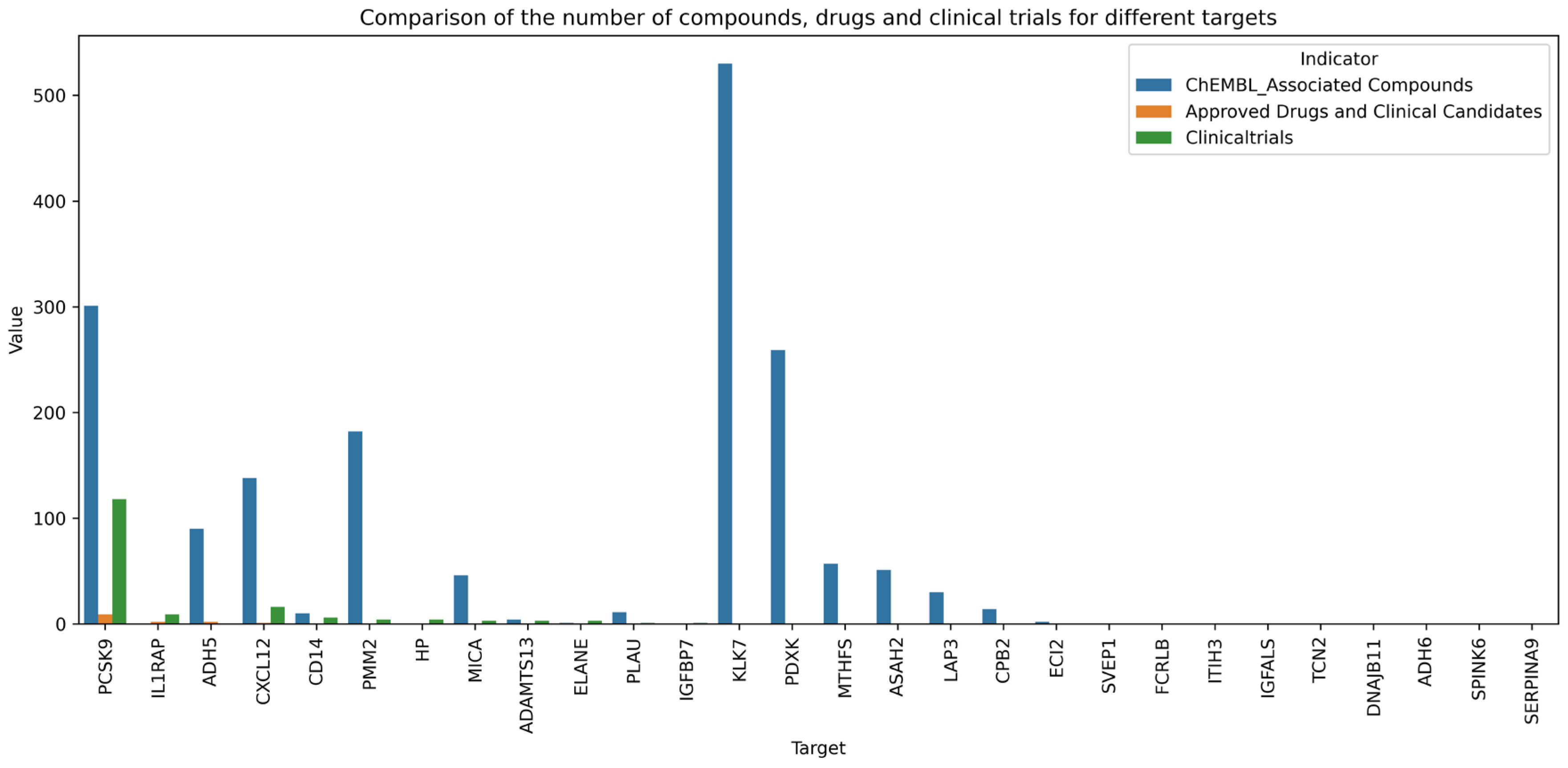
| Protein | OR | Disease Score | Coloc Score | Associated Compounds | Drugs and Candidates | ClinicalTrials |
|---|---|---|---|---|---|---|
| ADAMTS13 | 1.00188 | 0.51867 | 0.51364 | 4 | 0 | 3 |
| ADH5 | 0.99504 | 0.53561 | 0.26042 | 90 | 2 | 0 |
| ADH6 | 0.99511 | 0.48198 | 0.22744 | 0 | 0 | 0 |
| ASAH2 | 0.99741 | 0 | 0 | 51 | 0 | 0 |
| CD14 | 1.00343 | 0.49797 | 0.52008 | 10 | 0 | 6 |
| CPB2 | 1.00197 | 0.40938 | 0 | 14 | 0 | 0 |
| CXCL12 | 0.98999 | 0.5404 | 0.4607 | 138 | 1 | 16 |
| DNAJB11 | 1.00542 | 0.50799 | 0.59 | 0 | 0 | 0 |
| ECI2 | 1.00338 | 0 | 0 | 2 | 0 | 0 |
| ELANE | 1.00213 | 0.40938 | 0 | 1 | 0 | 3 |
| FCRLB | 0.99773 | 0.52349 | 0.5825 | 0 | 0 | 0 |
| HP | 1.00192 | 0.57453 | 0.65644 | 0 | 0 | 4 |
| IGFALS | 1.00472 | 0.40938 | 0.2125 | 0 | 0 | 0 |
| IGFBP7 | 0.997 | 0.5061 | 0.54 | 0 | 0 | 1 |
| IL1RAP | 1.00169 | 0.40938 | 0 | 0 | 2 | 9 |
| ITIH3 | 0.99778 | 0.52057 | 0.78 | 0 | 0 | 0 |
| KLK7 | 1.0097 | 0 | 0 | 530 | 0 | 0 |
| LAP3 | 1.00725 | 0.52349 | 0.09353 | 30 | 0 | 0 |
| MICA | 1.00195 | 0.62257 | 0.91274 | 46 | 0 | 3 |
| MTHFS | 0.99508 | 0.40938 | 0 | 57 | 0 | 0 |
| PCSK9 | 1.00719 | 0.62208 | 0.75407 | 301 | 9 | 118 |
| PDXK | 1.00503 | 0.75629 | 0.35876 | 259 | 0 | 0 |
| PLAU | 1.00333 | 0.52756 | 0.36407 | 11 | 0 | 1 |
| PMM2 | 0.99531 | 0.40938 | 0.35 | 182 | 0 | 4 |
| SERPINA9 | 0.99464 | 0 | 0 | 0 | 0 | 0 |
| SPINK6 | 0.9972 | 0 | 0 | 0 | 0 | 0 |
| SVEP1 | 0.99752 | 0.49549 | 0.60818 | 0 | 0 | 0 |
| TCN2 | 1.0018 | 0.496 | 0.42125 | 0 | 0 | 0 |
| Limitation Category | Specific Description | Potential Impact | Improvement Strategies |
|---|---|---|---|
| Confounding by comorbidities | The study did not fully investigate comorbidities (e.g., diabetes, hypertension) and medication use of the research subjects. These factors are known to directly alter plasma protein abundance. | May lead to overestimation or underestimation of the causal association between candidate proteins and CIHD; reduces the accuracy of druggability evaluation for targets (e.g., proteins affected by antidiabetic drugs). | 1. In subsequent studies, collect detailed clinical data (comorbidity diagnosis, medication records) and adjust for confounding factors using multivariate MR analysis (e.g., including comorbidity-related SNPs as covariates). |
| 2. Stratify analyses by comorbidity status (e.g., diabetic vs. non-diabetic CIHD patients) to verify the consistency of protein marker effects across subgroups. | |||
| Single ancestry limitation | All protein markers were identified based on European population data (Icelandic plasma proteome and European CIHD GWAS), lacking validation in non- European populations (e.g., Asian, African). | Ethnic differences in genetic backgrounds (e.g., linkage disequilibrium patterns) and environmental factors (e.g., diet) may limit the generalizability of results. For example, HP protein-related inflammatory complex concentrations differ significantly between South Asian and white males. | 1. Validate the “pQTLs-protein-CIHD” causal chain in large-scale plasma proteome and GWAS datasets of Asian (e.g., Chinese Biobank) and African populations. |
| 2. Integrate ethnicity-specific genetic variations to optimize biomarker adaptability (e.g., adjusting SNP selection criteria based on population-specific LD structures). | |||
| Potential pQTL detection bias | MR analysis relies on pQTL- protein abundance associations, with low-abundance proteins easily overlooked due to limited pQTL data, insufficient SNPs making leave-one-out MR reliability assessment impossible, and the used SomaScan platform (average duplicate correlation = 0.85) having affinity-based method limitations. | May miss critical low- abundance protein markers for CIHD, weaken MR causal inference robustness, and lead to inaccurate protein quantification due to platform- specific biases, affecting the reliability of identified protein markers and therapeutic targets. | 1. Integrate multi-platform proteomic data (e.g., SomaScan + Olink) to improve the detection rate of low-abundance proteins and expand pQTL coverage. |
| 2. Increase sample size in pQTL studies to enhance the number and strength of instrumental variables; use MR-PRESSO (MR Pleiotropy RESidual Sum and Outlier) method to further validate result stability when SNPs are limited. | |||
| 3. Develop filters for SomaScan data to correct aptamer binding artifacts caused by PAVs or protein modifications, improving quantification accuracy. | |||
| Lack of experimental validation | The study only identified protein markers and their associated pathways through bioinformatics analyses (MR, KEGG, database mining); no in vitro (e.g., cell models) or in vivo (e.g., animal models) experiments were conducted to verify their biological functions in CIHD. | Cannot confirm the direct regulatory role of candidate proteins in CIHD pathogenesis (e.g., whether CXCL12 truly protects cardiomyocytes by inhibiting inflammation); limits the translational value of therapeutic targets. | 1. Establish in vitro models (e.g., hypoxia-induced cardiomyocyte injury models, endothelial cell inflammation models) to detect changes in candidate protein expression and assess the effects of protein overexpression/knockdown on cell viability and inflammatory factor release. |
| 2. Construct animal models (e.g., CIHD mouse models with CXCL12/PLAU/CD14 gene knockout) to evaluate the impact of key targets on myocardial ischemia–reperfusion injury and atherosclerotic plaque stability. | |||
| 3. Conduct preliminary clinical validation (e.g., detect plasma levels of candidate proteins in CIHD patients vs. healthy controls) to verify their diagnostic and prognostic |
References
- Stone, P.H.; Libby, P.; Boden, W.E. Fundamental Pathobiology of Coronary Atherosclerosis and Clinical Implications for Chronic Ischemic Heart Disease Management—The Plaque Hypothesis: A Narrative Review. JAMA Cardiol. 2023, 8, 192–201. [Google Scholar] [CrossRef]
- World Health Organization. ICD-11 Cause of Death and Disease Statistics. Version 2025-01, ICD-11 Mortality and Morbidity Statistics, 2025. Available online: https://icd11.pumch.cn/ (accessed on 29 May 2025).
- Global Burden of Disease Study 2021 Collaborators. Global Burden of Disease Study 2021 (GBD 2021); Institute for Health Metrics and Evaluation (IHME): Seattle, WA, USA, 2024. [Google Scholar]
- GBD 2021 Risk Factors Collaborators. Global burden and strength of evidence for 88 risk factors in 204 countries and 811 subnational locations, 1990–2021: A systematic analysis for the Global Burden of Disease Study 2021. Lancet 2024, 403, 2162–2203. [Google Scholar] [CrossRef]
- Shi, H.; Xia, Y.; Cheng, Y.; Liang, P.; Cheng, M.; Zhang, B.; Liang, Z.; Wang, Y.; Xie, W. Global burden of ischaemic heart disease from 2022 to 2050: Projections of incidence, prevalence, deaths, and disability-adjusted life years. Eur. Heart J.-Qual. Care Clin. Outcomes 2025, 11, 355–366. [Google Scholar] [CrossRef]
- Chaitman, B.R.; Sano, J. Novel therapeutic approaches to treating chronic angina in the setting of chronic ischemic heart disease. Clin. Cardiol. 2007, 30, I25–I30. [Google Scholar] [CrossRef]
- Tori, R. Insight into the Updated AHA/ACC Guidelines on Treating Chronic Heart Disease. In The Cardiology Advisor; Haymarket Media Inc.: New York, NY, USA, 2023. [Google Scholar]
- Virani, S.S.; Newby, L.K.; Arnold, S.V.; Bittner, V.; Brewer, L.C.; Demeter, S.H.; Dixon, D.L.; Fearon, W.F.; Hess, B.; Johnson, H.M.; et al. 2023 AHA/ACC/ACCP/ASPC/NLA/PCNA Guideline for the Management of Patients with Chronic Coronary Disease: A Report of the American Heart Association/American College of Cardiology Joint Committee on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2023, 82, 833–955. [Google Scholar] [CrossRef]
- Sun, L.Y.; Gaudino, M.; Chen, R.J.; Eddeen, A.B.; Ruel, M. Long-term Outcomes in Patients With Severely Reduced Left Ventricular Ejection Fraction Undergoing Percutaneous Coronary Intervention vs. Coronary Artery Bypass Grafting. JAMA Cardiol. 2020, 5, 631–641. [Google Scholar] [CrossRef]
- Doenst, T.; Haverich, A.; Serruys, P.; Bonow, R.O.; Kappetein, P.; Falk, V.; Velazquez, E.; Diegeler, A.; Sigusch, H. PCI and CABG for Treating Stable Coronary Artery Disease: JACC Review Topic of the Week. J. Am. Coll. Cardiol. 2019, 73, 964–976. [Google Scholar] [CrossRef]
- AACVPR. Guidelines for Cardiac Rehabilitation Programs, 6th ed.; Human Kinetics: Champaign, IL, USA, 2021. [Google Scholar]
- Karakasis, P.; Patoulias, D.; Theofilis, P.; Pamporis, K.; Sagris, M.; Vlachakis, P.K.; Koufakis, T.; Antoniadis, A.P.; Fragakis, N. GLP-1 Receptor Agonists and Myocardial Perfusion: Bridging Mechanisms to Clinical Outcomes. Int. J. Mol. Sci. 2025, 26, 3050. [Google Scholar] [CrossRef] [PubMed]
- Ischemic Heart Disease Clinical Trials. Krannert Cardiovascular Research Center, IU School of Medicine. Available online: https://www.mayo.edu/research/centers-programs/cardiovascular-research-center/research/ischemic-heart-disease-program (accessed on 29 May 2025).
- Yan, T.-C.; Yue, Z.-X.; Xu, H.-Q.; Liu, Y.-H.; Hong, Y.-F.; Chen, G.-X.; Tao, L.; Xie, T. A systematic review of state-of-the-art strategies for machine learning-based protein function prediction. Comput. Biol. Med. 2023, 154, 106446. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.A.; Varma, V.R.; An, Y.; Varma, S.; Candia, J.; Fantoni, G.; Tiwari, V.; Anerillas, C.; Williamson, A.; Saito, A.; et al. A brain proteomic signature of incipient Alzheimer’s disease in young APOE ε4 carriers identifies novel drug targets. Sci. Adv. 2021, 7, eabi8178. [Google Scholar] [CrossRef] [PubMed]
- Savage, S.R.; Yi, X.; Lei, J.T.; Wen, B.; Zhao, H.; Liao, Y.; Jaehnig, E.J.; Somes, L.K.; Shafer, P.W.; Lee, T.D.; et al. Pan-cancer proteogenomics expands the landscape of therapeutic targets. Cell 2024, 187, 4389–4407.e15. [Google Scholar] [CrossRef]
- Naryzhny, S. Puzzle of Proteoform Variety—Where Is a Key? Proteomes 2024, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Small, D.S.; Thompson, S.G. A review of instrumental variable estimators for Mendelian randomization. Stat. Methods Med. Res. 2017, 26, 2333–2355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Mu, Y.; Jiang, X.; Zhao, Y.; Wang, Q.; Shen, Z. Causal relationship between thyroid dysfunction and gastric cancer: A two-sample Mendelian randomization study. Front. Endocrinol. 2024, 15, 1335149. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Lan, L.; Zheng, Z.; Zhang, H.; Wen, Y. Association between genetically predicted rheumatoid arthritis and alopecia areata: A two-sample Mendelian randomization study. Front. Immunol. 2023, 14, 1269640. [Google Scholar] [CrossRef]
- Ferkingstad, E.; Sulem, P.; Atlason, B.A.; Sveinbjornsson, G.; Magnusson, M.I.; Styrmisdottir, E.L.; Gunnarsdottir, K.; Helgason, A.; Oddsson, A.; Halldorsson, B.V.; et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021, 53, 1712–1721. [Google Scholar] [CrossRef]
- Elsworth, B.; Lyon, M.; Alexander, T.; Liu, Y.; Matthews, P.; Hallett, J.; Bates, P.; Palmer, T.; Haberland, V.; Smith, G.D.; et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv 2020. bioRxiv 2020.08.10.244293v1. [Google Scholar] [CrossRef]
- Burgess, S.; Smith, G.D.; Davies, N.M.; Dudbridge, F.; Gill, D.; Glymour, M.M.; Hartwig, F.P.; Kutalik, Z.; Holmes, M.V.; Minelli, C.; et al. Guidelines for performing Mendelian randomization investigations: Update for summer 2023. Wellcome Open Res. 2023, 4, 186. [Google Scholar] [CrossRef]
- Zhou, S.; Tao, B.; Guo, Y.; Gu, J.; Li, H.; Zou, C.; Tang, S.; Jiang, S.; Fu, D.; Li, J. Integrating plasma protein-centric multi-omics to identify potential therapeutic targets for pancreatic cancer. J. Transl. Med. 2024, 22, 557. [Google Scholar] [CrossRef]
- Yavorska, O.O.; Burgess, S. Mendelian Randomization: An R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017, 46, 1734–1739. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef]
- Cao, J.; Wang, N.; Luo, Y.; Ma, C.; Chen, Z.; Chenzhao, C.; Zhang, F.; Qi, X.; Xiong, W. A cause–effect relationship between Graves’ disease and the gut microbiome contributes to the thyroid–gut axis: A bidirectional two-sample Mendelian randomization study. Front. Immunol. 2023, 14, 977587. [Google Scholar] [CrossRef]
- Slob, E.A.W.; Burgess, S. A comparison of robust Mendelian randomization methods using summary data. Genet. Epidemiol. 2020, 44, 313–329. [Google Scholar] [CrossRef]
- Pan, T.; Bai, L.; Zhu, D.; Wei, Y.; Zhao, Q.; Feng, F.; Wang, Z.; Xu, Y.; Zhou, X. The causal relationship between genetically predicted blood metabolites and idiopathic pulmonary fibrosis: A bidirectional two-sample Mendelian randomization study. PLoS ONE 2024, 19, e0300423. [Google Scholar] [CrossRef]
- Wu, H.; Yang, J.; Wang, H.; Li, L. Mendelian randomization to explore the direct or mediating associations between socioeconomic status and lung cancer. Front. Oncol. 2023, 13, 1143059. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Qian, Q.; Xue, R.; Xu, C.; Wang, F.; Zeng, J.; Xiao, J. CVD Atlas: A multi-omics database of cardiovascular disease. Nucleic Acids Res. 2025, 53, D1348–D1355. [Google Scholar] [CrossRef] [PubMed]
- Yoo, M.; Shin, J.; Kim, J.; Ryall, K.A.; Lee, K.; Lee, S.; Jeon, M.; Kang, J.; Tan, A.C. DSigDB: Drug signatures database for gene set analysis. Bioinformatics 2015, 31, 3069–3071. [Google Scholar] [CrossRef]
- Zdrazil, B.; Felix, E.; Hunter, F.; Manners, E.J.; Blackshaw, J.; Corbett, S.; de Veij, M.; Ioannidis, H.; Lopez, D.M.; Mosquera, J.F.; et al. The ChEMBL Database in 2023: A drug discovery platform spanning multiple bioactivity data types and time periods. Nucleic Acids Res. 2024, 52, D1180–D1192. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Huang, J.; Ding, H.; Tao, Y.; Nan, J.; Xiao, C.; Wang, Y.; Wu, R.; Ni, C.; Zhong, Z.; et al. Flavin-containing monooxygenase 2 confers cardioprotection in ischemia models through its disulfide bond catalytic activity. J. Clin. Investig. 2024, 134, e177077. [Google Scholar] [CrossRef]
- Huss, J.M.; Kelly, D.P. Mitochondrial energy metabolism in heart failure: A question of balance. J. Clin. Investig. 2005, 115, 547–555. [Google Scholar] [CrossRef]
- Gao, J.-H.; Yu, X.-H.; Tang, C.-K. CXC chemokine ligand 12 (CXCL12) in atherosclerosis: An underlying therapeutic target. Clin. Chim. Acta 2019, 495, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Diamandis, M.; Paterson, A.D.; Rommens, J.M.; Veljkovic, D.K.; Blavignac, J.; Bulman, D.E.; Waye, J.S.; Derome, F.; Rivard, G.E.; Hayward, C.P.M. Quebec platelet disorder is linked to the urokinase plasminogen activator gene (PLAU) and increases expression of the linked allele in megakaryocytes. Blood 2009, 113, 1543–1546. [Google Scholar] [CrossRef] [PubMed]
- Pepine, C.J.; Nichols, W.W. The pathophysiology of chronic ischemic heart disease. Clin. Cardiol. 2007, 30, I4–I9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yan, F.; Luan, F.; Chai, Y.; Li, N.; Wang, Y.-W.; Chen, Z.-L.; Xu, D.-Q.; Tang, Y.-P. The pathological mechanisms and potential therapeutic drugs for myocardial ischemia reperfusion injury. Phytomed. Int. J. Phytother. Phytopharm. 2024, 129, 155649. [Google Scholar] [CrossRef]
- Reiner, A.P.; Lange, E.M.; Jenny, N.S.; Chaves, P.H.; Ellis, J.; Li, J.; Walston, J.; Lange, L.A.; Cushman, M.; Tracy, R.P. Soluble CD14: Genomewide association analysis and relationship to cardiovascular risk and mortality in older adults. Arter. Thromb. Vasc. Biol. 2013, 33, 158–164. [Google Scholar] [CrossRef]
- Fernández-Real, J.M.; del Pulgar, S.P.; Luche, E.; Moreno-Navarrete, J.M.; Waget, A.; Serino, M.; Sorianello, E.; Sánchez-Pla, A.; Pontaque, F.C.; Vendrell, J.; et al. CD14 modulates inflammation-driven insulin resistance. Diabetes 2011, 60, 2179–2186. [Google Scholar] [CrossRef]
- Zhang, S.; Jin, K.; Li, T.; Zhou, M.; Yang, W. Comprehensive analysis of INHBA: A biomarker for anti-TGFβ treatment in head and neck cancer. Exp. Biol. Med. 2022, 247, 1317–1329. [Google Scholar] [CrossRef]
- Sun, Y.; Cai, H.; Ge, J.; Shao, F.; Huang, Z.; Ding, Z.; Dong, L.; Chen, J.; Zhang, J.; Zang, Y. Tubule--derived INHBB promotes interstitial fibroblast activation and renal fibrosis. J. Pathol. 2022, 256, 25–37. [Google Scholar] [CrossRef]
- Massagué, J.; Sheppard, D. TGF-β signaling in health and disease. Cell 2023, 186, 4007–4037. [Google Scholar] [CrossRef] [PubMed]
- Reiss, A.B.; Shah, N.; Muhieddine, D.; Zhen, J.; Yudkevich, J.; Kasselman, L.J.; DeLeon, J. PCSK9 in cholesterol metabolism: From bench to bedside. Clin. Sci. 2018, 132, 1135–1153. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Li, J.; Jiang, N.; Chen, L.; Tang, L.; Mao, Y.; Li, W.; Zhou, G.; Li, Y.; et al. The immune modulation effects of gemcitabine plus cisplatin induction chemotherapy in nasopharyngeal carcinoma. Cancer Med. 2022, 11, 3437–3444. [Google Scholar] [CrossRef] [PubMed]
- Kalaria, T.R.; Harris, N.; Sensi, H.; Valentine, R.; Asif, U.; Sharrod-Cole, H.; Coley-Grant, D.; Min, S.S.; Ford, C.; Gama, R. High-sensitivity cardiac troponin I: Is ethnicity relevant? J. Clin. Pathol. 2021, 74, 709–711. [Google Scholar] [CrossRef]
- Bailey, D.; Colantonio, D.; Kyriakopoulou, L.; Cohen, A.H.; Chan, M.K.; Armbruster, D.; Adeli, K. Marked biological variance in endocrine and biochemical markers in childhood: Establishment of pediatric reference intervals using healthy community children from the CALIPER cohort. Clin. Chem. 2013, 59, 1393–1405. [Google Scholar] [CrossRef]
- Burgess, M.W.; Keshishian, H.; Mani, D.; Gillette, M.A.; Carr, S.A. Simplified and efficient quantification of low-abundance proteins at very high multiplex via targeted mass spectrometry. Mol. Cell. Proteom. 2014, 13, 1137–1149. [Google Scholar] [CrossRef]
- Rooney, M.R.; Chen, J.; Ballantyne, C.M.; Hoogeveen, R.C.; Boerwinkle, E.; Yu, B.; Walker, K.A.; Schlosser, P.; Selvin, E.; Chatterjee, N.; et al. Correlations Within and Between Highly Multiplexed Proteomic Assays of Human Plasma. Clin. Chem. 2025, 71, 677–687. [Google Scholar] [CrossRef]
- Gazzaz, Z.J.; Iftikhar, R.; Jameel, T.; Baig, M.; Murad, M.A. Association of Dyslipidemia and Comorbidities with Risk Factors Among Diabetic Patients: A Retrospective Analysis. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 935–941. [Google Scholar] [CrossRef] [PubMed]


| Protein Markers | Effect | Strength of Evidence | Pathway Involvement | Drugability | Reasons |
|---|---|---|---|---|---|
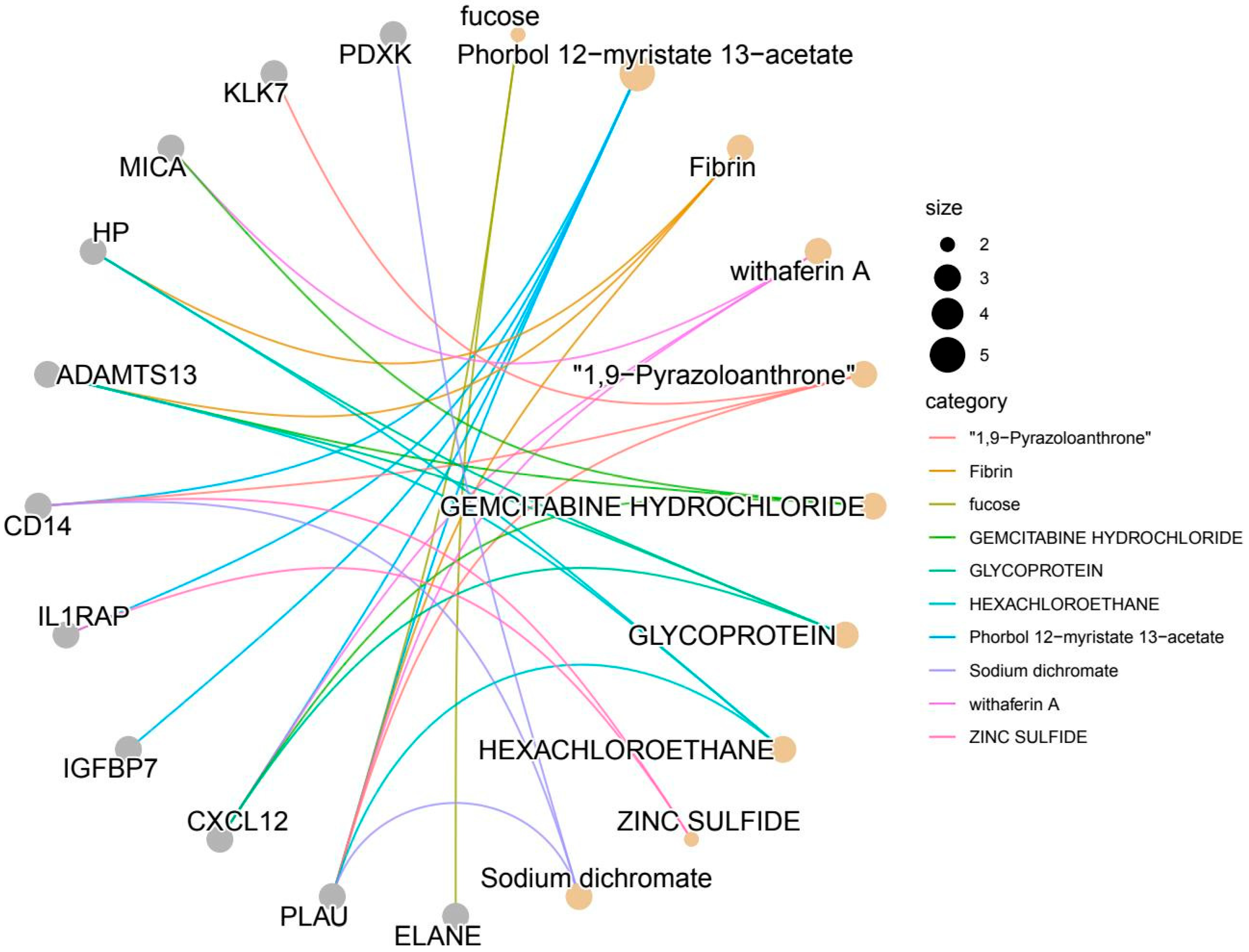
| CXCL12 | Protective factor | Genetic association: IVW p = 0.006, OR = 0.990 (significantly protective) | Core pathway: NF-κB signaling (regulating inflammation), CXCR4/CXCL12 axis (atherosclerosis) | Drug enrichment: 4 compounds (such as gemcitabine) are enriched (DSIGDB) | It exhibits the strongest genetic association (p = 0.006), the most comprehensive clinical evidence (16 trials), remarkable druggability (138 compounds + candidate drugs), and directly regulates the core inflammatory axis of CIHD. |
| Database support: NGDC disease score = 0.540, coloc = 0.461 (both high) | Number of pathways: 2 core pathways | Number of compounds: 138 (ChEMBL) | |||
| Clinical evidence: 16 related trials (clinicaltrials.gov) | Function: Inhibiting monocyte infiltration (key anti-inflammatory step) | Clinical progress: OLAPTESED PEGOL (CXCR4 antagonist, early trial) | |||
| Overall rating: High | Overall rating: Medium | Overall rating: High | |||
| PLAU | Risk factor | Genetic association: IVW p = 0.010, OR = 1.003 (significant risk) | Core pathway: NF-κB signaling, complement—coagulation cascade, transcriptional dysregulation (cancer/cardiovascular crossover) | Drug enrichment: 6 compounds (such as phorbol ester) are enriched (DSIGDB, up to) | This pathway exhibits the highest level of participation (covering inflammation, coagulation, and transcriptional regulation) and the greatest degree of drug enrichment (6 types of drugs). However, there is limited clinical evidence (only 1 case). Therefore, it is necessary to promote its clinical translation. |
| Database support: NGDC disease score = 0.528, coloc = 0.364 (single high) | Number of pathways: 3 core pathways | Number of compounds: 11 (ChEMBL) | |||
| Clinical evidence: 1 relevant trial | Function: Regulates thrombosis + plaque stability (dual core step) | Clinical progress: No marketed drug, but multi-drug regulatory potential (such as fuprostone) | |||
| Overall rating: Medium | Overall rating: High | Overall rating: Medium |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, C.; Qiao, G.; Wu, J.; Lu, Y.; Liu, M.; Zhang, C. Identification of Protein Markers for Chronic Ischemic Heart Disease Through Integrated Analysis of the Human Plasma Proteome and Genome-Wide Association Data. Proteomes 2025, 13, 55. https://doi.org/10.3390/proteomes13040055
Ren C, Qiao G, Wu J, Lu Y, Liu M, Zhang C. Identification of Protein Markers for Chronic Ischemic Heart Disease Through Integrated Analysis of the Human Plasma Proteome and Genome-Wide Association Data. Proteomes. 2025; 13(4):55. https://doi.org/10.3390/proteomes13040055
Chicago/Turabian StyleRen, Chunyang, Gan Qiao, Jianping Wu, Yongxiang Lu, Minghua Liu, and Chunxiang Zhang. 2025. "Identification of Protein Markers for Chronic Ischemic Heart Disease Through Integrated Analysis of the Human Plasma Proteome and Genome-Wide Association Data" Proteomes 13, no. 4: 55. https://doi.org/10.3390/proteomes13040055
APA StyleRen, C., Qiao, G., Wu, J., Lu, Y., Liu, M., & Zhang, C. (2025). Identification of Protein Markers for Chronic Ischemic Heart Disease Through Integrated Analysis of the Human Plasma Proteome and Genome-Wide Association Data. Proteomes, 13(4), 55. https://doi.org/10.3390/proteomes13040055






