Abstract
Early leaf senescence negatively impacts the grain yield in wheat (Triticum aestivum L.). Induced mutants provide an important resource for mapping and cloning of genes for early leaf senescence. In our previous study, Els2, a single incomplete dominance gene, that caused early leaf senescence phenotype in the wheat mutant LF2099, had been mapped on the long arm of chromosome 2B. The objective of this study was to develop molecular markers tightly linked to the Els2 gene and construct a high-resolution map surrounding the Els2 gene. Three tightly linked single-nucleotide polymorphism (SNP) markers were obtained from the Illumina Wheat 90K iSelect SNP genotyping array and converted to Kompetitive allele-specific polymerase chain reaction (KASP) markers. To saturate the Els2 region, the Axiom® Wheat 660K SNP array was used to screen bulked extreme phenotype DNA pools, and 9 KASP markers were developed. For fine mapping of the Els2 gene, these KASP markers and previously identified polymorphic markers were analyzed in a large F2 population of the LF2099 × Chinese Spring cross. The Els2 gene was located in a 0.24-cM genetic region flanked by the KASP markers AX-111643885 and AX-111128667, which corresponded to a physical interval of 1.61 Mb in the Chinese Spring chromosome 2BL containing 27 predicted genes with high confidence. The study laid a foundation for a map-based clone of the Els2 gene controlling the mutation phenotype and revealing the molecular regulatory mechanism of wheat leaf senescence.
1. Introduction
Wheat is one of the most widely grown crops in the world, providing more than 20% of the calories and protein consumed by the human population [1]. Maintaining a high and stable yield of wheat cultivars is a huge challenge for both wheat breeders and researchers. Leaves are the main photosynthetic organ of wheat plants. A previous study showed that the photosynthetically active flag leaf can contribute 30%–50% of the assimilates for grain filling and yield [2]. The early senescence of functional leaves significantly reduces photosynthetic time and efficiency, seriously affecting grain yield and quality in wheat [3]. Therefore, discovering the genes responsible for early leaf senescence is important not only for the elucidation of the early leaf senescence mechanism, but also in terms of its applications in the development of wheat germplasms and cultivars.
Plant leaf senescence involves a complex and highly regulated process with the coordinated actions of multiple pathways [4,5]. In recent years, researchers have elucidated the molecular mechanisms of leaf senescence in model plants, and isolated and characterized several senescence-associated genes that are involved in chlorophyll (Chl) degradation [6,7,8,9], reactive oxygen species (ROS) scavenging [10,11,12], carbon and nitrogen imbalances [13,14,15], and hormone responses [16,17,18,19,20]. However, limited information is available about leaf senescence regulation in wheat. Several studies focused on the transcriptome of wheat leaf senescence [21,22,23,24]. These studies have provided global and potential gene dynamic expression changes and signalling pathways for leaf senescence regulation in wheat, but few genes involved in the regulation of wheat leaf senescence have been identified. Several early leaf senescence mutant genes in wheat were mapped using molecular markers [3,25,26]; however, our understanding of the molecular mechanism of the leaf senescence trait in wheat is limited because none of these genes have been fine mapped and cloned.
The induced mutants have also become important for forward genetic analysis, where genes underlying mutant traits can be mapped using molecular markers [25]. Various classes of molecular markers, particularly simple sequence repeat (SSR) markers, have been widely used to identify mutant genes in wheat [27,28,29,30,31]. However, it is difficult to develop a high-resolution genetic linkage map based only on SSR markers because of their limited number and scattered distribution on wheat chromosomes. Single-nucleotide polymorphisms (SNPs) are the most abundant type of molecular marker in plant, and a large number of SNPs have been discovered in wheat with the improvement of sequencing techniques [32,33,34,35,36]. At present, several kinds of wheat SNP arrays have been reported, such as the Illumina Wheat 9K iSelect SNP array [37], the Illumina Wheat 90K iSelect SNP genotyping array [38], the Axiom® HD Wheat genotyping (820K) array [39], the Wheat Breeders’ 35K Axiom array [40], 660K [34] and the Wheat 55K SNP array [41]. The 660K SNP array [42] has been effectively applied to develop Kompetitive allele-specific polymerase chain reaction (KASP) markers for saturation mapping of the target genes. Examples include the molecular mapping of the dwarfing genes Rht12 [43] and Rht24 [44], stripe rust resistances Yr26 [45], YrH62 [46] and Qyrnap.nwafu-2BS [47], and the chlorina mutant gene cn-A1 [48].
We previously reported the first case of the early leaf senescence mutant in wheat, which was controlled by the single incomplete dominant gene Els2 [49]. The Els2 gene was preliminarily mapped on chromosome arm 2BL in a 0.95 cM genetic interval between intron polymorphism (IP) markers 2BIP09 and 2BIP14 [49]. The objectives of this study were (1) to develop molecular markers closely linked to Els2 using the wheat 660K SNP array, and (2) to construct a high-resolution linkage map.
2. Results
2.1. Development of Kompetitive Allele-Specific Polymerase Chain Reaction (KASP) Markers Based on the 90K Single-Nucleotide Polymorphisms (SNP) Array
Previously, the early leaf senescence Els2 gene was mapped to chromosome 2BL using the Illumina wheat 90K SNP array [49]. Given that the SNP array has limitations in large scale population analysis, 15 chromosome-specific SNPs generated from 90K SNP were converted into 11 KASP primer pairs. They were subjected to a polymorphism analysis on the parental cultivars and 20 F2 plants (including six homozygous wide type, eight heterozygous, and six homozygous mutant plants). Three polymorphic KASP markers (BS00030364_51, BS00084417_51 and RFL_Contig4423_529) were identified (Figure 1A). By genotyping the previous mapping population consisting of 142 F2:3 lines from the LF2099 × Chinese Spring cross [49], all three KASP markers linked to the Els2 gene (Figure 1C, Table 1). The Els2 gene was then genetically restricted to a region of 1.1 cM, 0.54 cM from the KASP marker BS00084417_51 and 0.66 cM from the KASP marker RFL_Contig4423_529 (Figure 2A). However, the Els2 genetic region were not narrowed down by using KASP markers from the 90K SNP array.
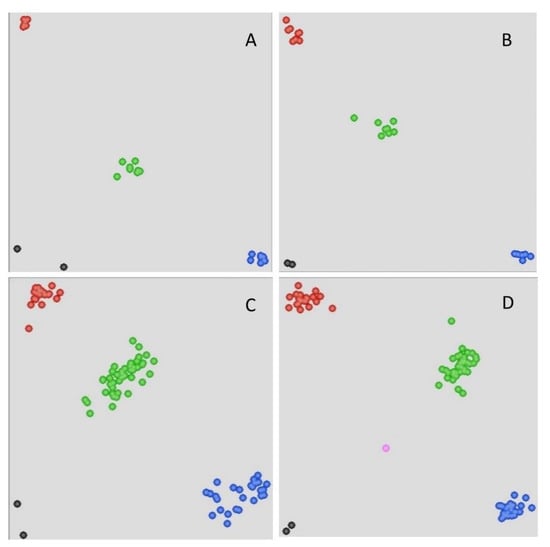
Figure 1.
Genotypic plots from selected Kompetitive allele-specific polymerase chain reaction (KASP) assays. The results of the parents and small population using markers (A) BS00084417_51 and (B) AX-111128667. Results of the F2 population using markers (C) RFL_Contig4423_529 and (D) AX-95682571. The central cluster (green) comprises heterozygous individuals, whereas the clusters near the axes are homozygous for either LF2099 (red) or Chinese Spring (blue). The black dots represent the nontemplate control, and the pink dots represent missing or failed data.

Table 1.
Primer sequences of Kompetitive allele-specific polymerase (KASP) markers developed from the 90K and 660K arrays.
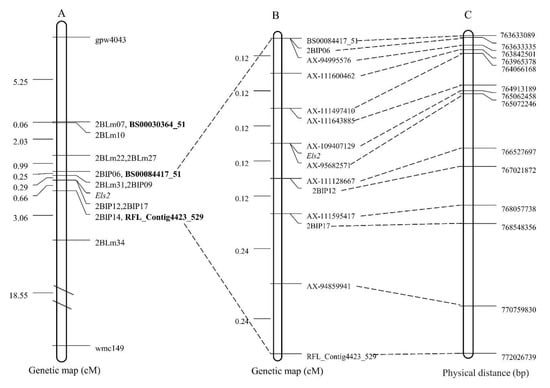
Figure 2.
The genetic linkage map and physical map of Chinese Spring genomic sequence fanking the Els2 gene. (A) The preliminary genetic linkage map of the Els2 gene [49]. (B) The high-resolution genetic map of the Els2 gene. (C) The corresponding physical location of the polymorphic linkage markers of the Els2 gene.
2.2. Development of KASP Markers Based on the 660K SNP Array
To saturate marker density in the Els2 region, we combined bulked segregant analysis (BSA) with 660K SNP arrays. A total of 10,097 SNP loci from the 660K SNP array were polymorphic between the two bulks and were located on 21 chromosomes, of which 3449 were located on chromosome 2B and the others were distributed on the other chromosomes (Figure 3A). These results were consistent with the previous 90K SNP genotyping result [49]. Of the 3449 SNP loci located on 2B, more than 1100 were positioned in the interval 750–775 Mb according to the wheat genome assembly (Figure 3B). Furthermore, two Els2-flanking KASP markers were located in the region 750–775 Mb (Figure 2A,C). To develop molecular markers more tightly linked to the Els2 gene, 25 SNPs located within the interval between BS00084417_51 and RFL_Contig4423_529 (763–772 Mb) were selected for conversion to KASP markers. The results indicated that nine KASP markers were tightly linked to the Els2 gene (Figure 1B,D, Table 1).
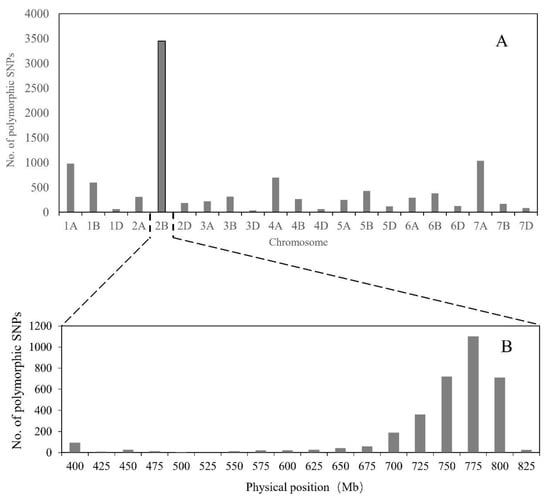
Figure 3.
Number of polymorphic single nucleotide polymorphisms (SNP) distributed on different wheat chromosomes (A) and distribution of SNP variants on chromosome 2B (B).
2.3. Construction of a High-Resolution Genetic Linkage Map
To construct a high-resolution genetic linkage map of Els2, we developed a large F2 population of 838 plants. The F2 population showed three classes of trait distribution, of which 198 were wide type plants, 419 were intermediate plants and 213 were mutant plants. The χ2 test shows a separation ratio of 1:2:1 (χ2 = 0.53 < χ20.05,2 = 5.99), which were consistent with our previous observations [49].
The Els2-flanking KASP markers BS00084417_51 and RFL_Contig4423_529 were used to identify recombinants in the F2 mapping population of the LF2099 × Chinese Spring cross. Only nine recombinant individuals were identified from 838 F2 individuals. Other molecular markers, including the previously reported IP markers 2BIP06, 2BIP12 and 2BIP17 [49], were then analyzed with these recombinants only. The F2:3 families produced by selfing the recombinant F2 plants were used to further estimate phenotypes. In combination with the marker genotypes and recombinant phenotype, a high-resolution genetic map around the Els2 locus was eventually constructed (Figure 2B). Among the markers flanking the Els2 gene, the KASP markers AX-111643885 and AX-111128667 were the most closely linked, at distances of 0.12 and 0.12 cM, respectively. Two KASP markers, AX-109407129 and AX-95682571, co-segregated with the Els2 gene.
2.4. Physical Mapping and Gene Annotation of the Els2 Goal Interval
To determine the physical locations of the Els2-linked markers, the sequences of all markers anchored in the high-resolution genetic map were used to blast against the Chinese Spring genomic sequence, and the relative physical positions of those markers were generally consistent with the genetic linkage map (Figure 2B,C). The genetic region of the markers AX-111643885 and AX-111128667 corresponded to a 1.61 Mb (764913189–766527697) genomic interval in the Chinese Spring reference genome (Figure 2C) and contained 27 annotated protein-coding genes (Table 2). Among them, one 50S ribosomal protein L14, five cytochrome P450 family proteins, three cysteine proteinases, two serine/threonine-protein kinase, and one RECEPTOR-like protein kinase were identified.

Table 2.
Gene models between the two flanking markers AX-111643885 and AX-111128667 based on the International Wheat Genome Consortium (IWGSC) RefSeq v1.1 annotation.
3. Discussion
Mapping functional genes using ethyl methanesulfonate (EMS) mutants combined with map-based cloning has been applied in many crops; however, the high-resolution mapping of a mutant gene in wheat may become problematic and tedious without high-density molecular markers. Several approaches have been used to develop high-density molecular markers around the target gene. Comparative genomics analyses are traditional and effective approaches for marker development in a wheat positional cloning project [50]. By applying comparative genomics analyses, we previously developed five new markers linked to the Els2 gene [49]. Although release of the Chinese Spring whole genomic assembly sequences [51] would reduce the importance of comparative genomics analyses in marker development, it is still an effective method for other species without a reference genome sequence.
The SNP genotyping assays, namely 90K [38] and 660K [42], have opened a new way to undertake a more efficient and faster marker saturation of target genes in wheat [47]. Compared to the 90K SNP chip, the 660K chip has several advantages, such as higher marker density and higher resolution, as well as a better distribution on the chromosomes [42,44]. In the present study, we used the 660K chip to screen candidate markers linked to Els2 and a significant number of polymorphic SNPs were identified (Figure 3A,B). To improve the efficiency of genotyping individuals, polymorphic SNP markers on 2BL were converted to KASP markers to genotype the F2 population. Using this strategy, the Els2 locus was further narrowed down to a 1.61 Mb region in the Chinese Spring chromosome 2BL containing 27 predicted genes with high confidence (Figure 2B,C, Table 2). It was a clearly effective approach to exploit constantly the updated genome information and high-throughput SNP platforms for high-resolution mapping of a mutant gene in wheat. In addition to the methods used in our study, the combination of the BSA strategy with next-generation sequencing technology (RNA-Seq or exome sequencing) has been used as a mapping strategy that offers the promise of a rapid discovery of genetic markers linked to target genes in wheat [3,52,53].
The Els2 genomic interval was narrowed down to the 1.61 Mb interval of the Chinese Spring 2BL containing 27 predicted protein-coding genes. Some of the annotated genes were associated with plant early leaf senescence, including the 50S ribosomal protein L14, cytochrome P450 family proteins, cysteine proteinase, serine/threonine-protein kinase, and RECEPTOR-like protein kinase (Table 2). In rice, the gene encoding the 50S ribosomal protein L21 in the chloroplastic precursor 50S ribosomal protein may be responsible for the early senescence mutant es-t [54]. Cytochrome P450 CYP89A9, which is in charge of non-fluorescent dioxobilin-type chlorophyll catabolite accumulation, is drawn into the formation of major chlorophyll catabolites during leaf senescence [55]. Cysteine proteases are the most abundant enzymes responsible for the proteolytic activity during leaf senescence [56]. A number of cDNA clones encoding cysteine proteinases in different plants have been previously reported to be up-regulated during leaf senescence [57]. Protein kinases and autophagy-related genes were also reported to play an important role in leaf senescence [58].
In the present study, we reported our work on the high-resolution genetic mapping of the novel early leaf senescence gene Els2. First, the development of 12 KASP markers from the high-density SNP arrays enabled us to construct high-resolution genetic maps around the target locus. Second, we physically delimited the Els2 gene to a 1.61-Mb genomic region including 27 putative genes. We believe our study has made a solid foundation for the future isolation of the Els2 locus.
4. Materials and Methods
4.1. Plant Materials and Population Construction
The novel early leaf senescence mutant LF2099 was identified from the chemical mutagen EMS-induced library derived from the common wheat accession H261. To develop a saturated genetic linkage map for Els2, the F2 and F2:3 mapping populations (consisting of 838 lines) produced by crossing LF2099 to the normal leaf senescence cultivar Chinese Spring were used throughout the study. All plants were grown on the experimental farm of Northwest A&F University, Yangling, China. Leaf senescence phenotypes were evaluated by visual observation. To ensure the accuracy of the results, the phenotypes of the recombinant F2 individuals were further validated by using the F3 lines. The F3 lines containing 30 seeds from every F2 plant were planted in rows to verify the phenotypes of the F2 plants in fields.
4.2. Combined Bulked Segregant Analysis Using the 660K SNP Array
The total genomic DNA from the parents and F2 plants were isolated from leaves by using a cetyltrimethylammonium bromide (CTAB) method [59]. The DNA quantity was measured spectrophotometrically, and the DNA integrity was confirmed on agarose gel. To obtain closer SNP markers and saturate the targeted gene, BSA was performed to identify polymorphic markers between the early and normal DNA bulks. The DNA of the 12 homozygous plants that exhibited early leaf senescence and normal leaf senescence, based on a validation of the phenotypes of the F3 family, was pooled to construct an early bulk sample and a normal bulk sample, separately. The F2 bulks were genotyped with the 660K SNP arrays from CapitalBio Corporation (Beijing, China) using the Affymetrix GeneTitan System based on the Axiom 2.0 Assay Manual Workflow protocol. Allele calling was performed using the Affymetrix proprietary software according to Axiom Best Practices Genotyping Workflow. The sequences of all SNPs were used to blast version1.0 of the assembled Chinese Spring survey sequence (International Wheat Genome Consortium (IWGSC) RefSeq v1.0) to determine their physical positions [51].
4.3. Conversion of SNP Markers to KASP Markers
The polymorphic SNP markers from 90K and 660K were converted to KASP markers using the PolyMarker software (available online: http://polymarker.tgac.ac.uk) [60]. Chromosome-specific KASP markers were used to screen the parents and small population to confirm the polymorphisms before genotyping the entire mapping population. The KASP reactions were performed in a 384-well plate format following the protocol of LGC Genomics. Each KASP reaction mixtures consisted of final volumes of 5 μL containing 50–100 ng of genomic DNA, 2.5 µL of 2 × KASP Master mix (V4.0, LGC Genomics, Hoddesdon, UK), 0.056 µL of assay primer mix (12 µM of each allele-specific primer and 30 µM of common primer) and 2.444 µL of water. The assay reaction was performed in a 384 well Thermal Cycler (Bio-Rad) in the following cycling conditions: denaturation at 95 °C for 15 min, 9 cycles of 95 °C for 20 s and touchdown starting at 65 °C for 60 s (decreased by 0.8 °C per cycle), 30–40 cycles of amplification (95 °C for 20 s; 57 °C for 60 s). The amplified products were visualized by a microplate reader (FLUOstar Omega, BMG LABTECH, Germany) and analyzed by the software Klustering Caller (LGC, Middlesex, UK) [61].
4.4. Construction of High-Density Genetic Linkage Map
The linkage analysis of the polymorphic molecular markers and the Els2gene was conducted using JoinMap4.0 software [62] with default parameters and a LOD score of 3.0 as a threshold. The genetic distance was calculated and presented in Kosambi centiMorgans (cM). The genetic map was drawn with the software Mapdraw V2.1 (Huazhong Agricultural Sciences, Wuhan, China) [63].
4.5. Physical Mapping and Gene Annotation of the Els2 Region
All 14 markers used for the fine mapping of the Els2 were anchored to the wheat reference genome sequence [51]. All sequences including forward and reverse primers were searched against the Chinese Spring whole genome assembly sequences (Reference Sequence v1.0, IWGSC, http://www.wheatgenome.org/) using the BLASTN algorithm applying default parameters. Obtained physical positions of mapped markers were visualized using the software Mapdraw V2.1 (Huazhong Agricultural Sciences, Wuhan, China) [63]. The information on the Chinese Spring genomic sequence was used to identify the genes that were included in the interval between the two Els2-flanking markers. IWGSC RefSeq v1.1 with gene annotations became available on the website https://wheat-urgi.versailles.inra.fr/Seq-Repository/Annotation. Annotated genes in the target region were extracted for analyzing genes involved in plant leaf senescence.
Author Contributions
Conceptualization, Y.X. and C.W.; methodology, N.W., Y.X., S.L. and Y.G.; software, N.W.; validation, N.W. and Y.X.; formal analysis, N.W. and Y.X.; investigation, Y.L. and S.W.; resources, C.W.; data curation, S.W., Y.L., and S.L.; writing—original draft, Y.X., N.W., S.L. and S.W.; writing—review and editing, Y.L. and Y.G.; supervision, and funding acquisition, C.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Key Research and Development Program of China (2016YFD0102101) and National Natural Science Foundation of China (31101139).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fu, D.L.; Uauy, C.; Distelfeld, A.; Blechl, A.; Epstein, L.; Chen, X.M.; Sela, H.A.; Fahima, T.; Dubcovsky, J. A Kinase-START Gene Confers Temperature-Dependent Resistance to Wheat Stripe Rust. Science 2009, 323, 1357–1360. [Google Scholar] [CrossRef]
- Sylvester-Bradley, R.; Scott, R.; Wright, C. Physiology in the production and improvement of cereals. In Physiology in the Production and Improvement of Cereals; HGCA: London, UK, 1990. [Google Scholar]
- Li, M.M.; Li, B.B.; Guo, G.H.; Chen, Y.X.; Xie, J.Z.; Lu, P.; Wu, Q.H.; Zhang, D.Y.; Zhang, H.Z.; Yang, J.; et al. Mapping a leaf senescence gene els1 by BSR-Seq in common wheat. Crop J. 2018, 6, 236–243. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Woo, H.R.; Nam, H.G. Toward Systems Understanding of Leaf Senescence: An Integrated Multi-Omics Perspective on Leaf Senescence Research. Mol. Plant 2016, 9, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, G.K.; Tripathy, B.C. Modulation of biosynthesis of photosynthetic pigments and light-harvesting complex in wild-type and gun5 mutant of Arabidopsis thaliana during impaired chloroplast development. Protoplasma 2016, 253, 747–752. [Google Scholar] [CrossRef]
- Morita, R.; Sato, Y.; Masuda, Y.; Nishimura, M.; Kusaba, M. Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef]
- Rong, H.; Tang, Y.; Zhang, H.; Wu, P.; Chen, Y.; Li, M.; Wu, G.; Jiang, H. The Stay-Green Rice like (SGRL) gene regulates chlorophyll degradation in rice. J. Plant Physiol. 2013, 170, 1367–1373. [Google Scholar] [CrossRef]
- Yamatani, H.; Sato, Y.; Masuda, Y.; Kato, Y.; Morita, R.; Fukunaga, K.; Nagamura, Y.; Nishimura, M.; Sakamoto, W.; Tanaka, A.; et al. NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll-protein complexes during leaf senescence. Plant J. 2013, 74, 652–662. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, T.; Peng, H.; Luo, S.; Tan, J.; Jiang, K.; Heng, Y.; Zhang, X.; Guo, X.; Zheng, J.; et al. Rice premature leaf senescence 2, encoding a glycosyltransferase (GT), is involved in leaf senescence. Front. Plant Sci. 2018, 9, 560. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Hong, X.; Hu, D.; Liu, C.; Yang, J.; Li, Y.; Huang, Y.; Feng, Y.; Gong, H.; et al. Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J. Exp. Bot. 2015, 66, 973–987. [Google Scholar] [CrossRef]
- Chen, L.J.; Wuriyanghan, H.; Zhang, Y.Q.; Duan, K.X.; Chen, H.W.; Li, Q.T.; Lu, X.; He, S.J.; Ma, B.; Zhang, W.K.; et al. An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 2013, 163, 1752–1765. [Google Scholar] [CrossRef] [PubMed]
- Bi, Z.; Zhang, Y.; Wu, W.; Zhan, X.; Yu, N.; Xu, T.; Liu, Q.; Li, Z.; Shen, X.; Chen, D.; et al. ES7, encoding a ferredoxin-dependent glutamate synthase, functions in nitrogen metabolism and impacts leaf senescence in rice. Plant Sci. 2017, 259, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Ueda, N.; Aoki, K.; Kuromori, T.; Hirayama, T.; Shinozaki, K.; Yamaya, T.; Sakakibara, H. AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 2004, 45, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Turano, F.J. The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6872–6877. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, Y.X.; Di, C.; Zhang, Q.L.; Zhang, K.; Wang, C.C.; You, Q.; Yan, H.; Dai, S.Y.; Yuan, J.S.; et al. JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. J. Exp. Bot. 2016, 67, 751–762. [Google Scholar] [CrossRef]
- Lee, I.C.; Hong, S.W.; Whang, S.S.; Lim, P.O.; Nam, H.G.; Koo, J.C. Age-Dependent Action of an ABA-Inducible Receptor Kinase, RPK1, as a Positive Regulator of Senescence in Arabidopsis Leaves. Plant Cell Physiol. 2011, 52, 651–662. [Google Scholar] [CrossRef]
- Zhang, Q.; Xia, C.; Zhang, L.; Dong, C.; Liu, X.; Kong, X. Transcriptome Analysis of a Premature Leaf Senescence Mutant of Common Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2018, 19, 782. [Google Scholar] [CrossRef]
- Borrill, P.; Harrington, S.A.; Simmonds, J.; Uauy, C. Identification of Transcription Factors Regulating Senescence in Wheat through Gene Regulatory Network Modelling. Plant Physiol. 2019, 180, 1740–1755. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.L.; Holm, P.B. Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol. J. 2007, 5, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.L.; Holm, P.B.; Krupinska, K. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol. 2008, 10, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Jaiswal, V.; Kumar, S.; Balyan, H.S.; Gupta, P.K. Chromosome mapping of four novel mutants in bread wheat (Triticum aestivum L.). Acta Physiol. Plant 2015, 37, 66. [Google Scholar] [CrossRef]
- Harrington, S.A.; Cobo, N.; Karafiatova, M.; Dolezel, J.; Borrill, P.; Uauy, C. Identification of a Dominant Chlorosis Phenotype Through a Forward Screen of the Triticum turgidum cv. Kronos TILLING Population. Front. Plant Sci. 2019, 10, 963. [Google Scholar] [CrossRef]
- Ansari, M.J.; Al-Ghamdi, A.; Usmani, S.; Kumar, R.; Nuru, A.; Singh, K.; Dhaliwal, H.S. Characterization and gene mapping of a brittle culm mutant of diploid wheat (Triticum monococcum L.) with irregular xylem vessels development. Acta Physiol. Plant 2013, 35, 2407–2419. [Google Scholar] [CrossRef]
- Li, N.; Jia, J.; Xia, C.; Liu, X.; Kong, X. Characterization and mapping of novel chlorophyll deficient mutant genes in durum wheat. Breed Sci. 2013, 63, 169–175. [Google Scholar] [CrossRef]
- Xu, T.; Bian, N.; Wen, M.; Xiao, J.; Yuan, C.; Cao, A.; Zhang, S.; Wang, X.; Wang, H. Characterization of a common wheat (Triticum aestivum L.) high-tillering dwarf mutant. Theor. Appl. Genet. 2017, 130, 483–494. [Google Scholar] [CrossRef]
- Ansari, M.J.; Al-Ghamdi, A.; Kumar, R.; Usmani, S.; Al-Attal, Y.; Nuru, A.; Mohamed, A.A.; Singh, K.; Dhaliwal, H.S. Characterization and gene mapping of a chlorophyll-deficient mutant clm1 of Triticum monococcum L. Biol. Plant. 2013, 57, 442–448. [Google Scholar] [CrossRef]
- Zhang, L.L.; Liu, C.; An, X.Y.; Wu, H.Y.; Feng, Y.; Wang, H.; Sun, D.J. Identification and genetic mapping of a novel incompletely dominant yellow leaf color gene, Y1718, on chromosome 2BS in wheat. Euphytica 2017, 213, 141. [Google Scholar] [CrossRef]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Zhang, N.; Fan, X.L.; Zhang, W.; Zhao, C.H.; Yang, L.J.; Pan, R.Q.; Chen, M.; Han, J.; Zhao, X.Q.; et al. Utilization of a Wheat660K SNP array-derived high-density genetic map for high-resolution mapping of a major QTL for kernel number. Sci. Rep. 2017, 7, 3788. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhao, S.; Kong, X.; Li, Y.; Zhao, G.; He, W.; Appels, R.; Pfeifer, M.; Tao, Y.; Zhang, X.; et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 2013, 496, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.; Duran, C.; Berkman, P.J.; Lorenc, M.T.; Stiller, J.; Manoli, S.; Hayden, M.J.; Forrest, K.L.; Fleury, D.; Baumann, U.; et al. Single nucleotide polymorphism discovery from wheat next-generation sequence data. Plant Biotechnol. J. 2012, 10, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, C.R.; Chao, S.; Wang, S.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenac, C.; Brown-Guedira, G.L.; Akhunova, A.; et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 8057–8062. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Winfield, M.O.; Allen, A.M.; Burridge, A.J.; Barker, G.L.; Benbow, H.R.; Wilkinson, P.A.; Coghill, J.; Waterfall, C.; Davassi, A.; Scopes, G.; et al. High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 2016, 14, 1195–1206. [Google Scholar] [CrossRef]
- Klindworth, D.L.; Saini, J.; Long, Y.; Rouse, M.N.; Faris, J.D.; Jin, Y.; Xu, S.S. Physical mapping of DNA markers linked to stem rust resistance gene Sr47 in durum wheat. Theor. Appl. Genet. 2017, 130, 1135–1154. [Google Scholar] [CrossRef]
- Ren, T.; Hu, Y.; Tang, Y.; Li, C.; Yan, B.; Ren, Z.; Tan, F.; Tang, Z.; Fu, S.; Li, Z. Utilization of a Wheat55K SNP Array for Mapping of Major QTL for Temporal Expression of the Tiller Number. Front. Plant Sci. 2018, 9, 333. [Google Scholar] [CrossRef]
- Sun, C.; Dong, Z.; Zhao, L.; Ren, Y.; Zhang, N.; Chen, F. The Wheat 660K SNP array demonstrates great potential for marker-assisted selection in polyploid wheat. Plant Biotechnol. J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, W.; Li, Y.; Shan, Q.; Ye, X.; Wang, D.; Yu, K.; Lu, W.; Xin, P.; Pei, Z.; et al. A wheat dominant dwarfing line with Rht12, which reduces stem cell length and affects gibberellic acid synthesis, is a 5AL terminal deletion line. Plant J. 2019, 97, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wen, W.; Xie, L.; Fu, L.; Xu, D.; Fu, C.; Wang, D.; Chen, X.; Xia, X.; Chen, Q.; et al. Molecular Mapping of Reduced Plant Height Gene Rht24 in Bread Wheat. Front. Plant Sci. 2017, 8, 1379. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zeng, Q.; Wang, Q.; Liu, S.; Yu, S.; Mu, J.; Huang, S.; Sela, H.; Distelfeld, A.; Huang, L.; et al. SNP-based pool genotyping and haplotype analysis accelerate fine-mapping of the wheat genomic region containing stripe rust resistance gene Yr26. Theor. Appl. Genet. 2018, 131, 1481–1496. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Q.; Xu, L.; Chen, X.; Li, B.; Mu, J.; Zeng, Q.; Huang, L.; Han, D.; Kang, Z. Combining Single Nucleotide Polymorphism Genotyping Array with Bulked Segregant Analysis to Map a Gene Controlling Adult Plant Resistance to Stripe Rust in Wheat Line 03031-1-5 H62. Phytopathology 2017, 108, 103–113. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Q.; Liu, S.; Huang, S.; Mu, J.; Zeng, Q.; Huang, L.; Han, D.; Kang, Z. Saturation Mapping of a Major Effect QTL for Stripe Rust Resistance on Wheat Chromosome 2B in Cultivar Napo 63 Using SNP Genotyping Arrays. Front. Plant Sci. 2017, 8, 653. [Google Scholar] [CrossRef]
- Jiang, H.B.; Wang, N.; Jian, J.T.; Wang, C.S.; Xie, Y.Z. Rapid mapping of a chlorina mutant gene cn-A1 in hexaploid wheat by bulked segregant analysis and single nucleotide polymorphism genotyping arrays. Crop Pasture Sci. 2019, 70, 827–836. [Google Scholar] [CrossRef]
- Wang, N.; Xie, Y.Z.; Li, Y.Z.; Wu, S.N.; Wei, H.H.; Wang, C.S. Molecular mapping a novel early leaf-senescence gene Els2 in common wheat by SNP genotyping arrays. Crop Pasture Sci. 2020, 71, 356–367. [Google Scholar] [CrossRef]
- Wang, M.; Wang, S.; Xia, G. From genome to gene: A new epoch for wheat research? Trends Plant Sci. 2015, 20, 380–387. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Martinez, S.A.; Shorinola, O.; Conselman, S.; See, D.; Skinner, D.Z.; Uauy, C.; Steber, C.M. Exome sequencing of bulked segregants identified a novel TaMKK3-A allele linked to the wheat ERA8 ABA-hypersensitive germination phenotype. Theor. Appl. Genet. 2020, 133, 719–736. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.H.; Li, J.T.; Wu, P.P.; Li, Y.H.; Qiu, D.; Qu, Y.F.; Xie, J.Z.; Zhang, H.J.; Yang, L.; Fu, T.T.; et al. Development of SNP, KASP, and SSR Markers by BSR-Seq Technology for Saturation of Genetic Linkage Map and Efficient Detection of Wheat Powdery Mildew Resistance Gene Pm61. Int. J. Mol. Sci. 2019, 20, 750. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Rao, Y.; Liu, H.; Fang, Y.; Dong, G.; Huang, L.; Leng, Y.; Guo, L.; Zhang, G.; Hu, J.; et al. Characterization and fine mapping of an early senescence mutant (es-t) in Oryza sativa L. Chin. Sci. Bull. 2011, 56, 2437–2443. [Google Scholar] [CrossRef][Green Version]
- Christ, B.; Sussenbacher, I.; Moser, S.; Bichsel, N.; Egert, A.; Muller, T.; Krautler, B.; Hortensteiner, S. Cytochrome P450 CYP89A9 Is Involved in the Formation of Major Chlorophyll Catabolites during Leaf Senescence in Arabidopsis. Plant Cell 2013, 25, 1868–1880. [Google Scholar] [CrossRef]
- Diaz-Mendoza, M.; Velasco-Arroyo, B.; Gonzalez-Melendi, P.; Martinez, M.; Diaz, I. C1A cysteine protease-cystatin interactions in leaf senescence. J. Exp. Bot. 2014, 65, 3825–3833. [Google Scholar] [CrossRef]
- Chen, G.H.; Huang, L.T.; Yap, M.N.; Lee, R.H.; Huang, Y.J.; Cheng, M.C.; Chen, S.C. Molecular characterization of a senescence-associated gene encoding cysteine proteinase and its gene expression during leaf senescence in sweet potato. Plant Cell Physiol. 2002, 43, 984–991. [Google Scholar] [CrossRef]
- Zhou, C.; Cai, Z.; Guo, Y.; Gan, S. An arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 2009, 150, 167–177. [Google Scholar] [CrossRef]
- Doyle, J.J.T.; Doyle, J.L. Isolation of Plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Ramirez-Gonzalez, R.H.; Uauy, C.; Caccamo, M. PolyMarker: A fast polyploid primer design pipeline. Bioinformatics 2015, 31, 2038–2039. [Google Scholar] [CrossRef]
- Wu, J.; Liu, S.; Wang, Q.; Zeng, Q.; Mu, J.; Huang, S.; Yu, S.; Han, D.; Kang, Z. Rapid identification of an adult plant stripe rust resistance gene in hexaploid wheat by high-throughput SNP array genotyping of pooled extremes. Theor. Appl. Genet. 2018, 131, 43–58. [Google Scholar] [CrossRef]
- Van Ooijen, J.W. JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Available online: https://www.scienceopen.com/document?vid=baa76c8c-fb55-4c13-a6ca-24c71002ab5a (accessed on 15 December 2006).
- Liu, R.H.; Meng, J.L. MapDraw:A Microsoft Excel Macro for Drawing Genetic Linkage Maps Based on Given Genetic Linkage Data. Hereditas 2003, 25, 317–321. [Google Scholar] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).