Molecular Detection of the Seed-Borne Pathogen Colletotrichum lupini Targeting the Hyper-Variable IGS Region of the Ribosomal Cluster
Abstract
1. Introduction
2. Results
2.1. Evaluation of the Mycoflora Associated with Lupin Seeds
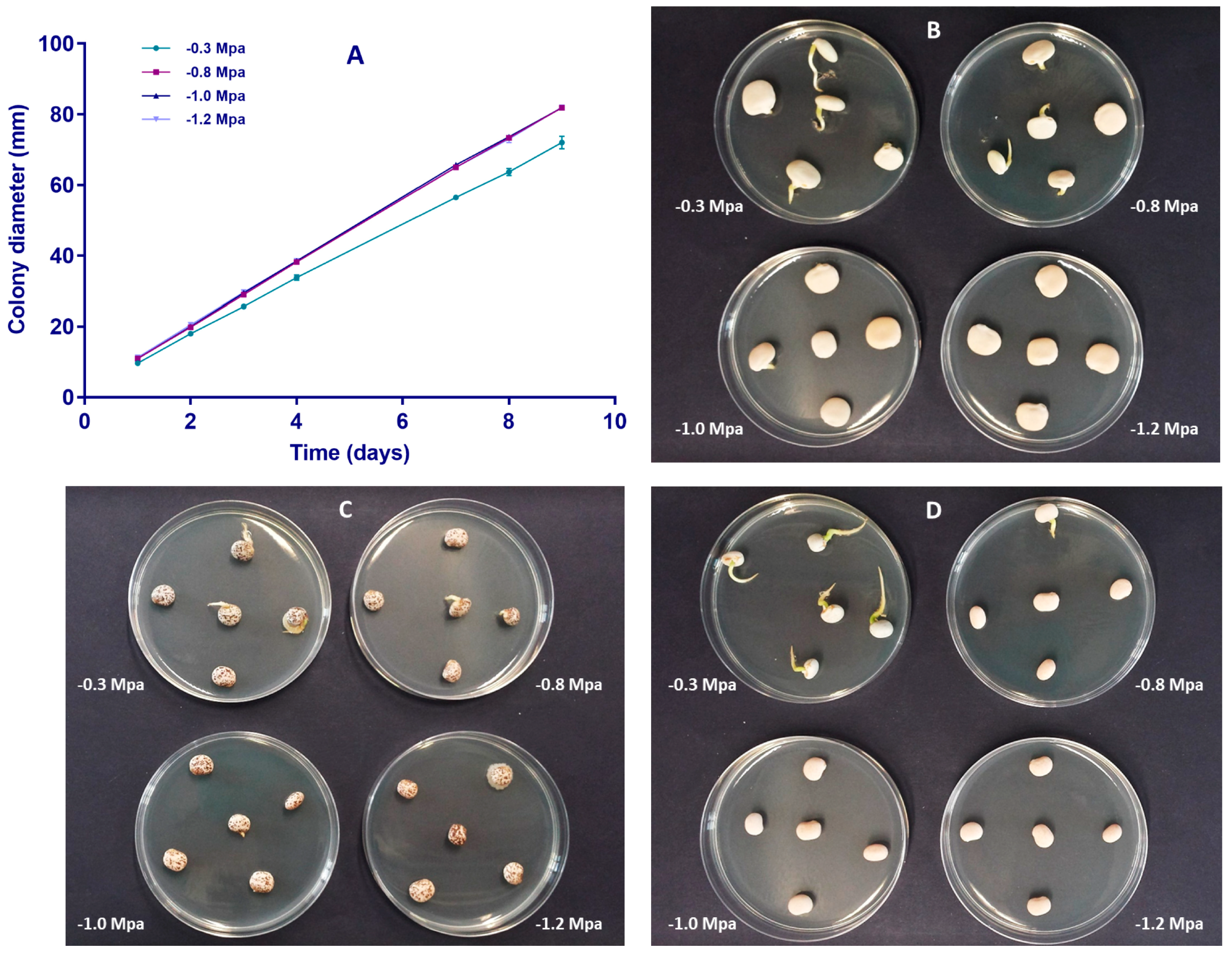
2.2. Artificial Inoculation of Lupin Seeds by the Water-Restriction Technique
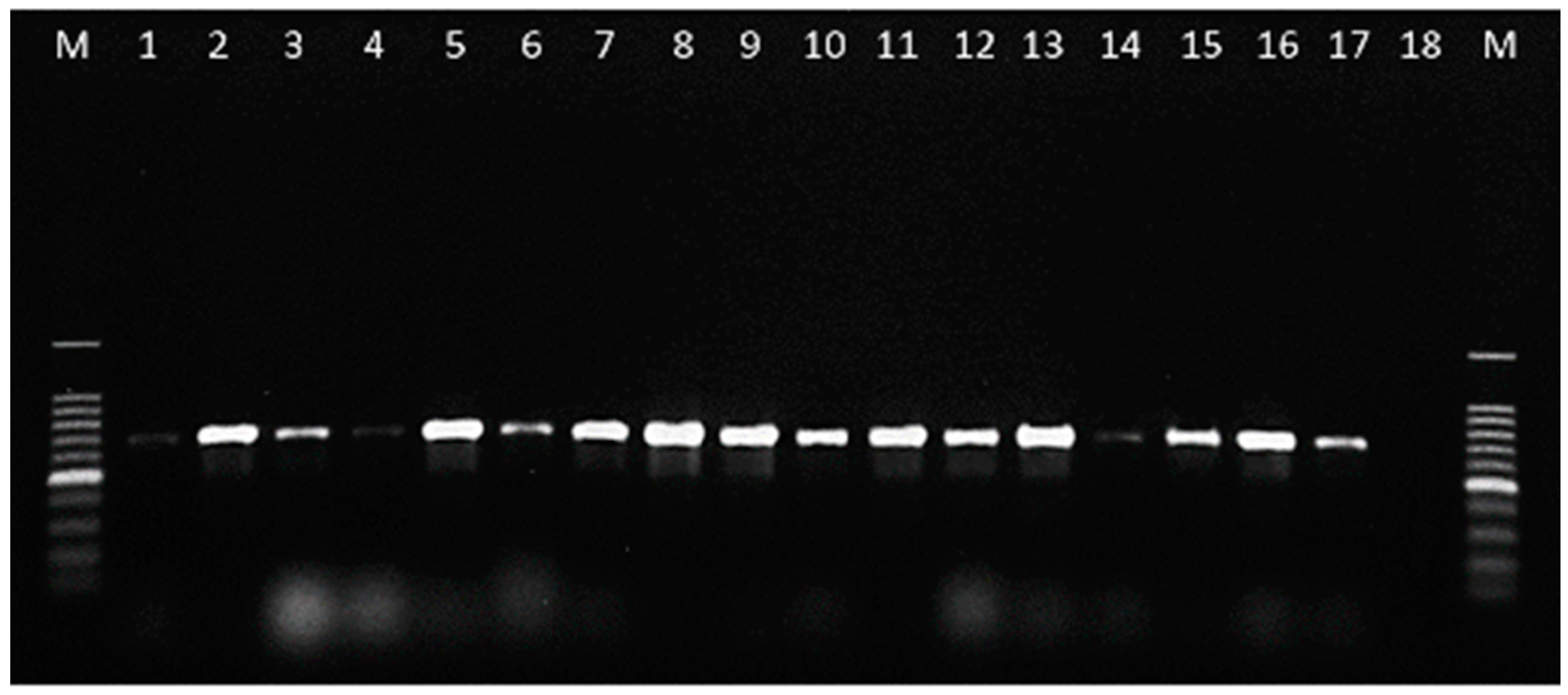
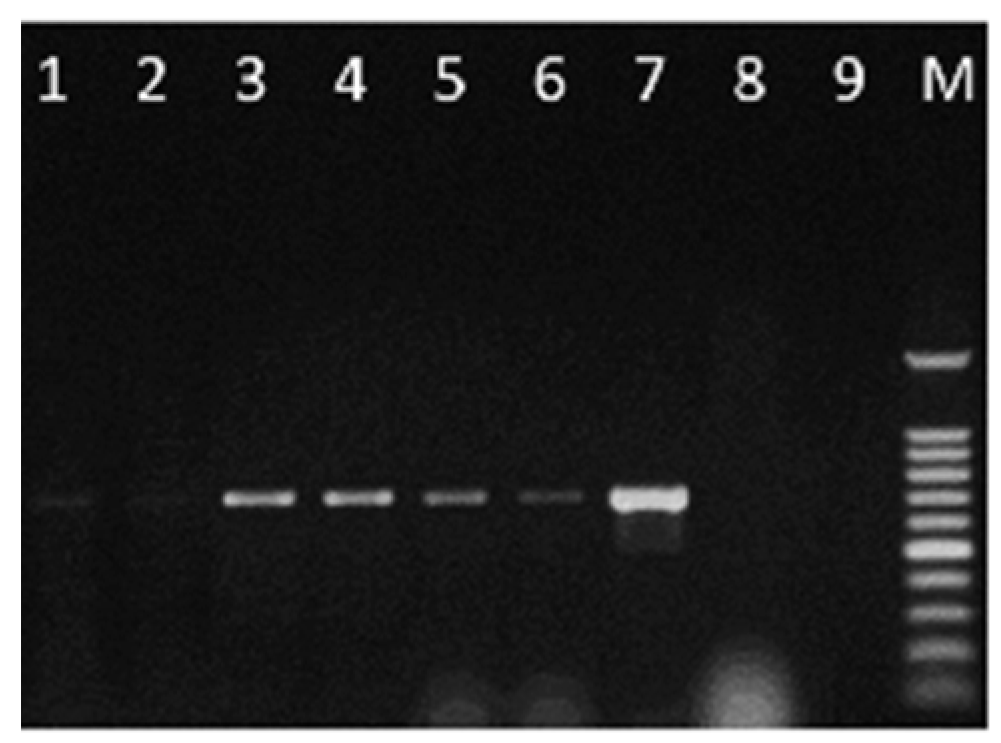
2.3. Development of a Specific PCR Assay for C. lupini
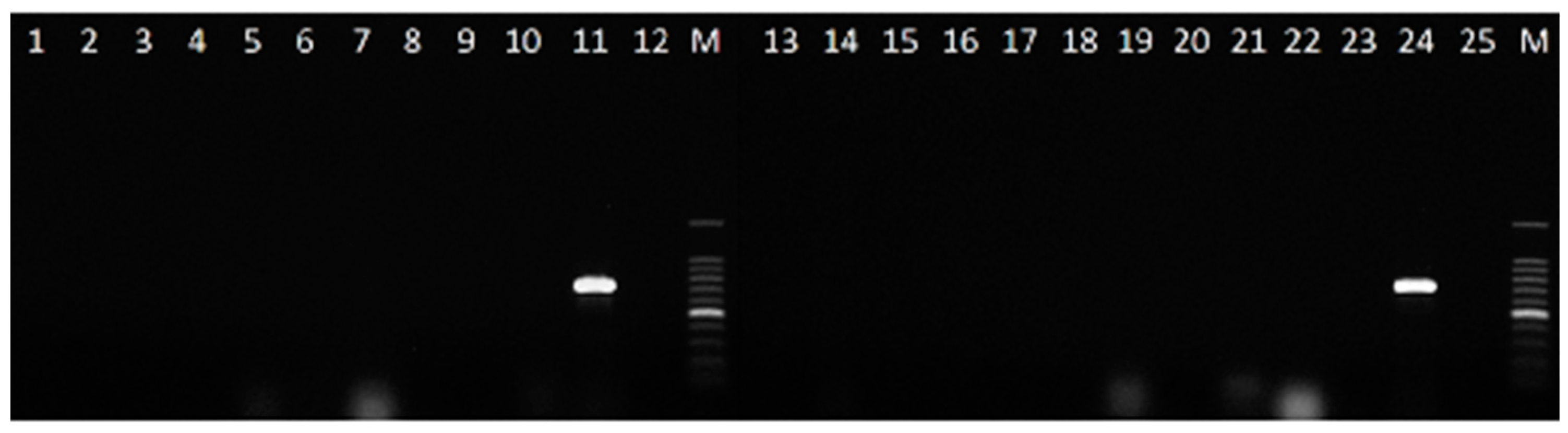
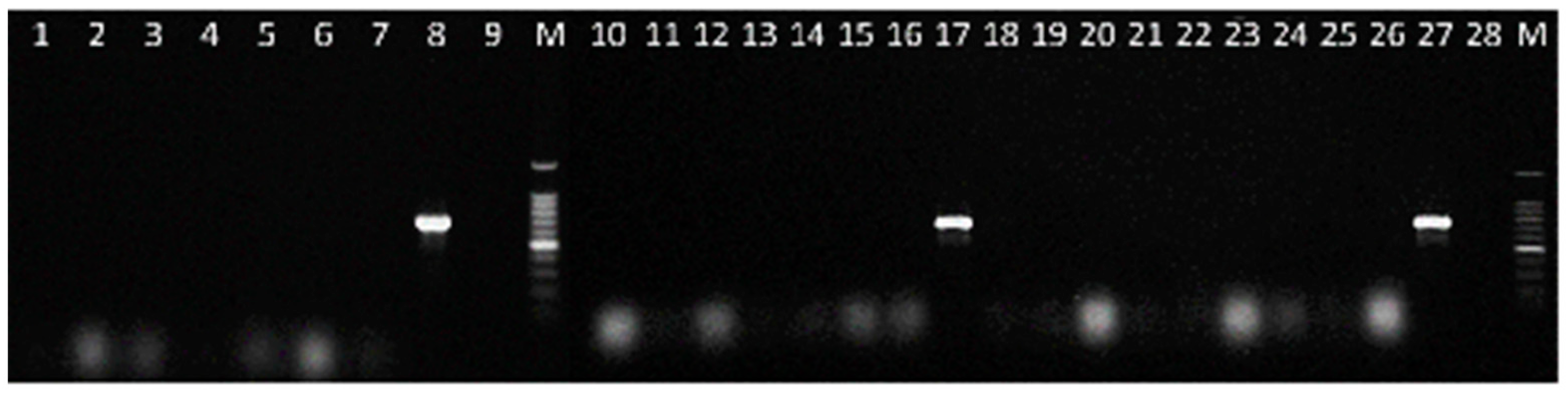
2.4. Detection of C. lupini in Artificially Infected Seed Samples
3. Discussion
4. Materials and Methods
4.1. Fungal Cultures
4.2. DNA Extraction
4.3. Evaluation of Mycoflora Associated with Lupin Seeds
4.4. Genome Data and Analysis of Ribosomal IGS Region
4.5. Development of Specific Oligonucleotide PCR Primers
4.6. Development of a Specific PCR Assay for C. lupini
4.7. Artificial Inoculation of Lupin Seeds by Water Restriction Technique
4.8. Detection of C. lupini in Seed Samples with Different Fungal Incidence
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gladstones, J.S. Lupins as crop plants. Field Crop Abstr. 1970, 23, 123–148. [Google Scholar]
- Herridge, D.F. The narrow-leafed lupin (Lupinus angustifolius L.) as a nitrogen-fixing rotation crop for cereal production. I. Indices of nitrogen fixation. Aust. J. Agric. Res. 1988, 39, 1003–1015. [Google Scholar] [CrossRef]
- Cowling, W.; Buirchell, B.; Tapia, M.E. Lupin. Lupinus spp. Promoting the Conservation and Use of Underutilized and Neglected Crops. 23; Institute of Plant Genetics and Crop Plant Research, Gatersleben, and International Plant Genetic Resources Institute: Rome, Italy, 1998. [Google Scholar]
- Talhinhas, P.; Baroncelli, R.; Le Floch, G. Anthracnose of lupins caused by Colletotrichum lupini: A recent disease and a successful worldwide pathogen. J. Plant Pathol. 2016, 98, 5–14. [Google Scholar]
- Fernández-Pascual, M.; Pueyo, J.J.; Felipe, M.; Golvano, M.P.; Lucas, M.M. Singular features of the Bradyrhizobium-Lupinus symbiosis. Singul. Featur. Bradyrhizobium-Lupinus Symbiosis 2007, 1, 1–16. [Google Scholar]
- Carof, M.; Godinot, O.; Ridier, A. Diversity of protein-crop management in western France. Agron. Sustain. Dev. 2019, 39, 15. [Google Scholar] [CrossRef]
- Elmer, W.H.; Yang, H.A.; Sweetingham, M.W. Characterization of Colletotrichum gloeosporioides isolates from ornamental lupines in Connecticut. Plant Dis. 2001, 85, 216–219. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. FAOSTAT Statistics Database. Available online: http://www.fao.org/faostat/en/ (accessed on 1 January 2019).
- Gondran, J. Les maladies du lupin blanc doux en France. Perspect. Agric. 1984, 77, 31–41. [Google Scholar]
- Baier, A.C.; Linhares, A.G. Breeding for anthracnose tolerance in lupins. In Proceedings of the 6th International Lupin Conference, Temuco-Pucon, Chile, 25–30 November 1990; p. 127. [Google Scholar]
- Nirenberg, H.I.; Feiler, U.; Hagedorn, G. Description of Colletotrichum lupini comb. nov. in modern terms. Mycologia 2002, 94, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Lotter, H.C.; Berger, D.K. Anthracnose of lupins in South Africa is caused by Colletotrichum lupini var. setosum. Australas. Plant Pathol. 2005, 34, 385–392. [Google Scholar] [CrossRef]
- Thomas, G.J.; Sweetingham, M.W. Cultivar and environment influence the development of lupin anthracnose caused by Colletotrichum lupini. Australas. Plant Pathol. 2004, 33, 571–577. [Google Scholar] [CrossRef]
- Thomas, G.J.; Sweetingham, M.W.; Yang, H.A.; Speijers, J. Effect of temperature on growth of Colletotrichum lupini and on anthracnose infection and resistance in lupins. Australas. Plant Pathol. 2008, 37, 35–39. [Google Scholar] [CrossRef]
- Riegel, R.; Véliz, D.; von Baer, I.; Quitral, Y.; Muñoz, M. Genetic diversity and virulence of Colletotrichum lupini isolates collected in Chile. Trop. Plant Pathol. 2010, 35, 144–152. [Google Scholar] [CrossRef]
- Thomas, T.H. Lupins as crop plants: Biology, production and utilization. Edited by J. S. Gladstones, C. Atkins and J. Hamblin. 1998. Plant Growth Regul. 2001, 33, 246. [Google Scholar] [CrossRef]
- Gondran, J.; Pacault, D. L’anthracnose du lupin blanc. Phytoma-Déf. Végétaux 1997, 28–31. [Google Scholar]
- Decker, P. Anthracnose of blue lupin is seed-borne. Plant Dis. Rep. 1947, 31, 270–271. [Google Scholar]
- Sreenivasaprasad, S.; Talhinhas, P. Genotypic and phenotypic diversity in Colletotrichum acutatum, a cosmopolitan pathogen causing anthracnose on a wide range of hosts. Mol. Plant Pathol. 2005, 6, 361–378. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Marincowitz, S.; Barnes, I.; Bensch, K.; Braun, U.; Camporesi, E.; Damm, U.; et al. Genera of phytopathogenic fungi: GOPHY 1. Stud. Mycol. 2017, 86, 99–216. [Google Scholar] [CrossRef]
- Damm, U.; Sato, T.; Alizadeh, A.; Groenewald, J.Z.; Crous, P.W. The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Stud. Mycol. 2019, 92, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Baroncelli, R.; Talhinhas, P.; Pensec, F.; Sukno, S.A.; Le Floch, G.; Thon, M.R. The Colletotrichum acutatum species complex as a model system to study evolution and host specialization in plant pathogens. Front. Microbiol. 2017, 8, 2001. [Google Scholar] [CrossRef]
- Dubrulle, G.; Pensec, F.; Picot, A.; Rigalma, K.; Pawtowski, A.; Gironde, S.; Harzic, N.; Nodet, P.; Baroncelli, R.; Le Floch, G. Phylogenetic diversity and temperature effect on growth and pathogenicity of Colletotrichum lupini. Plant Dis. 2019, in press. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D., Shinsky, J., White, T., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Lio, D.D.; Cobo-Díaz, J.F.; Masson, C.; Chalopin, M.; Kebe, D.; Giraud, M.; Verhaeghe, A.; Nodet, P.; Sarrocco, S.; Floch, G.L.; et al. Combined Metabarcoding and Multi-locus approach for Genetic characterization of Colletotrichum species associated with common walnut (Juglans regia) anthracnose in France. Sci. Rep. 2018, 8, 10765. [Google Scholar] [CrossRef] [PubMed]
- Schena, L.; Abdelfattah, A.; Mosca, S.; Li Destri Nicosia, M.G.; Agosteo, G.E.; Cacciola, S.O. Quantitative detection of Colletotrichum godetiae and C. acutatum sensu stricto in the phyllosphere and carposphere of olive during four phenological phases. Eur. J. Plant Pathol. 2017, 149, 337–347. [Google Scholar] [CrossRef]
- Suarez, M.B.; Walsh, K.; Boonham, N.; O’Neill, T.; Pearson, S.; Barker, I. Development of real-time PCR (TaqMan®) assays for the detection and quantification of Botrytis cinerea in planta. Plant Physiol. Biochem. 2005, 43, 890–899. [Google Scholar] [CrossRef]
- Sanzani, S.M.; Schena, L.; De Cicco, V.; Ippolito, A. Early detection of Botrytis cinerea latent infections as a tool to improve postharvest quality of table grapes. Postharvest Biol. Technol. 2012, 68, 64–71. [Google Scholar] [CrossRef]
- Schweigkofler, W.; O’Donnell, K.; Garbelotto, M. Detection and quantification of airborne conidia of Fusarium circinatum, the causal agent of pine pitch canker, from two California sites by using a real-time PCR approach combined with a simple spore trapping method. Appl Env. Microbiol 2004, 70, 3512–3520. [Google Scholar] [CrossRef] [PubMed]
- Zambounis, A.G.; Paplomatas, E.; Tsaftaris, A.S. Intergenic spacer–RFLP analysis and direct quantification of australian Fusarium oxysporum f. sp. vasinfectum isolates from soil and infected cotton tissues. Plant Dis. 2007, 91, 1564–1573. [Google Scholar]
- Dita, M.A.; Waalwijk, C.; Buddenhagen, I.W.; Souza, M.T., Jr.; Kema, G.H.J. A molecular diagnostic for tropical race 4 of the banana fusarium wilt pathogen. Plant Pathol. 2010, 59, 348–357. [Google Scholar] [CrossRef]
- Liew, E.C.Y.; Maclean, D.J.; Irwin, J.A.G. Specific PCR based detection of Phytophthora medicaginis using the intergenic spacer region of the ribosomal DNA. Mycol. Res. 1998, 102, 73–80. [Google Scholar] [CrossRef]
- Schena, L.; Nigro, F.; Ippolito, A. Real-Time PCR detection and quantification of soilborne fungal pathogens: The case of Rosellinia necatrix, Phytophthora nicotianae, P. citrophthora and Verticillium dahliae. Phytopathol. Mediterr. 2004, 43, 273–280. [Google Scholar]
- Bilodeau, G.J.; Koike, S.T.; Uribe, P.; Martin, F.N. Development of an assay for rapid detection and quantification of Verticillium dahliae in soil. Phytopathology 2011, 102, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Pecchia, S.; Da Lio, D. Development of a rapid PCR-Nucleic Acid Lateral Flow Immunoassay (PCR-NALFIA) based on rDNA IGS sequence analysis for the detection of Macrophomina phaseolina in soil. J. Microbiol. Methods 2018, 151, 118–128. [Google Scholar] [CrossRef] [PubMed]
- ISTA International Seed Testing Association. Available online: https://www.seedtest.org/ (accessed on 15 January 2019).
- Walcott, R.R. Detection of seedborne pathogens. HortTechnology 2003, 13, 40–47. [Google Scholar] [CrossRef]
- Mancini, V.; Murolo, S.; Romanazzi, G. Diagnostic methods for detecting fungal pathogens on vegetable seeds. Plant Pathol. 2016, 65, 691–703. [Google Scholar] [CrossRef]
- Marcinkowska, J.Z. Methods of finding and identification of pathogens in seeds. Plant Breed. Seed Sci. 2002, 46, 31–48. [Google Scholar]
- Ioos, R.; Fourrier, C.; Iancu, G.; Gordon, T.R. Sensitive detection of Fusarium circinatum in pine seed by combining an enrichment procedure with a real-time polymerase chain reaction using dual-labeled probe chemistry. Phytopathology 2009, 99, 582–590. [Google Scholar] [CrossRef]
- Guillemette, T.; Iacomi-Vasilescu, B.; Simoneau, P. Conventional and real-time PCR-based assay for detecting pathogenic Alternaria brassicae in cruciferous seed. Plant Dis. 2004, 88, 490–496. [Google Scholar] [CrossRef]
- Phan, H.T.T.; Ford, R.; Bretag, T.; Taylor, P.W.J. A rapid and sensitive polymerase chain reaction (PCR) assay for detection of Ascochyta rabiei, the cause of ascochyta blight of chickpea. Australas. Plant Pathol. 2002, 31, 31–39. [Google Scholar] [CrossRef]
- Chadha, S.; Gopalakrishna, T. Detection of Magnaporthe grisea in infested rice seeds using polymerase chain reaction. J. Appl. Microbiol. 2006, 100, 1147–1153. [Google Scholar] [CrossRef]
- Taylor, J.L. A simple, sensitive, and rapid method for detecting seed contaminated with highly virulent Leptosphaeria maculans. Appl Env. Microbiol 1993, 59, 3681–3685. [Google Scholar]
- Kumar, P.; Akhtar, J.; Kandan, A.; Kumar, S.; Batra, R.; Dubey, S.C. Advance Detection Techniques of Phytopathogenic Fungi: Current Trends and Future Perspectives. In Current Trends in Plant Disease Diagnostics and Management Practices; Springer: Berlin, Germany, 2016; pp. 265–298. [Google Scholar]
- Schena, L.; Nigro, F.; Ippolito, A.; Gallitelli, D. Real-time quantitative PCR: A new technology to detect and study phytopathogenic and antagonistic fungi. Eur. J. Plant Pathol. 2004, 110, 893–908. [Google Scholar] [CrossRef]
- Abdel-Hafez, S.I.I. Mycoflora of bean, broad bean, lentil, lupine and pea seeds in Saudi Arabia. Mycopathologia 1984, 88, 45–49. [Google Scholar] [CrossRef]
- El-Nagerabi, S.A.F.; Elshafie, A.E. Composition of mycoflora and aflatoxins in lupine seeds from the Sudan. Phytopathol. Mediterr. 2000, 39, 257–262. [Google Scholar]
- Nedzinskiene, T.L.; Asakaviciute, R. Development of fungi on Lupinus angustifolius L. and Lupinus luteus L. Res. Plant Biol. 2011, 1, 20–29. [Google Scholar]
- Alomran, M.M.; Lupien, S.L.; Dugan, F.M. Mycobiota of Lupinus albus seeds from a public germplasm collection. North Am. Fungi 2013, 8, 1–15. [Google Scholar] [CrossRef]
- Pszczolkowska, A.; Okorski, A.; Kotecki, A.; Gas, M.; Kulik, T.; Reczek, A. Incidence of seed-borne fungi on Lupinus mutabilis depending on a plant morphotype, sowing date and plant density. J. Elem. 2016, 21, 501–512. [Google Scholar] [CrossRef][Green Version]
- Machado, J.C.; Coutinho, W.M.; Guimarães, R.M.; Vieira, M.; Ferreira, D.F. Use of osmotic solutes to control seed germination of rice and common bean in seed health blotter tests. Seed Sci. Technol. 2008, 36, 66–75. [Google Scholar] [CrossRef]
- de Carvalho, J.C.B. Uso da restrição hídrica na inoculação de Colletotrichum lindemuthianum em sementes de feijoeiro (Phaseolus vulgaris L.); Universidade Federal de Lavras: Lavras, Brazil, 1999. [Google Scholar]
- Machado, A.Q. Uso da restrição hídrica em testes de sanidade de sementes de algodoeiro; Universidade Federal de Lavras: Lavras, Brazil, 2002. [Google Scholar]
- Machado, J.C.; Guimaraes, R.M.; Vieira, M.; Souza, R.M.; Pozza, E.A. Use of water restriction technique in seed pathology. Seed Test. Int. 2004, 128, 14–18. [Google Scholar]
- de Falleiro, B.A.S.; de Almeida, P.B.A.; Coutinho, W.M.; Suassuna, N.D.; Kobayasti, L. Use of osmotic solutions for inhibition of sunflower seed germination in blotter test. Trop. Plant Pathol. 2010, 35, 343–350. [Google Scholar] [CrossRef][Green Version]
- Farias, C.R.J.; Del Ponte, E.M.; Corrêa, C.L.; Afonso, A.P.S.; Pierobom, C.R. Infecção de sementes de trigo com Bipolaris sorokiniana pela técnica de restrição hídrica. Trop. Plant Pathol. 2010, 35, 253–257. [Google Scholar] [CrossRef]
- da Machado, J.C.; Machado, A.Q.; Pozza, E.A.; Machado, C.F.; Zancan, W.L.A. Inoculum potential of Fusarium verticillioides and performance of maize seeds. Trop. Plant Pathol. 2013, 38, 213–217. [Google Scholar] [CrossRef]
- da Botelho, L.S.; Barrocas, E.N.; da Machado, J.C.; de Martins, R.S. Detection of Sclerotinia sclerotiorum in soybean seeds by conventional and quantitative PCR techniques. J. Seed Sci. 2015, 37, 55–62. [Google Scholar] [CrossRef][Green Version]
- da Siqueira, C.S.; Barrocas, E.N.; da Machado, J.C.; Corrêa, C.L. Transmission of Stenocarpella maydis by maize seeds. Rev. Ciênc. Agronômica 2016, 47, 393–400. [Google Scholar]
- Cullen, D.W.; Lees, A.K.; Toth, I.K.; Duncan, J.M. Conventional PCR and real-time quantitative PCR detection of Helminthosporium solani in soil and on potato tubers. Eur. J. Plant Pathol. 2001, 107, 387–398. [Google Scholar] [CrossRef]
- Shea, G.; Thomas, G.; Buirchell, B.; Salam, M.; McKirdy, S.; Sweetingham, M. Case study: Industry response to the lupin anthracnose incursion in Western Australia. In Lupins for Health and Wealth (Fremantle, Western Australia ed.); Palta, J.A., Berger, J.B., Eds.; International Lupin Association: Canterbury, New Zealand, 2008; Volume 1, pp. 546–551. [Google Scholar]
- Kim, W.K.; Mauthe, W.; Hausner, G.; Klassen, G.R. Isolation of high molecular weight DNA and double-stranded RNAs from fungi. Can. J. Bot. 1990, 68, 1898–1902. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A Method for Designing Primer Sets for Speciation Studies in Filamentous Ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Baroncelli, R.; Amby, D.B.; Zapparata, A.; Sarrocco, S.; Vannacci, G.; Le Floch, G.; Harrison, R.J.; Holub, E.; Sukno, S.A.; Sreenivasaprasad, S.; et al. Gene family expansions and contractions are associated with host range in plant pathogens of the genus Colletotrichum. BMC Genomics 2016, 17, 555. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Anderson, J.B.; Stasovski, E. Molecular phylogeny of northern hemisphere species of Armillaria. Mycologia 1992, 84, 505–516. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Proceedings of the Nucleic Acids Symposium Series; Oxford University Press: London, UK, 1999; pp. 95–98. [Google Scholar]
- Heydecker, W.; Higgins, J.; Turner, Y.J. Invigoration of seeds. Seed Sci. Technol. 1975, 3, 881–888. [Google Scholar]
| Lupinus spp. | albus | luteus | angustifolius | Method 1 | Loci Used for Molecular Characterization | ||
|---|---|---|---|---|---|---|---|
| cultivar | Multitalia | Mister | Tango | ||||
| Contamination (%) | 84 | 82 | 60 | Accession of Reference Sequences | |||
| Germination (%) | 85.5 | 70.5 | 93.5 | ITS | TUB | TEF | |
| Alternaria alternata | 42 | 4 | 2 | M&G | MK560162 | MK567923 | - |
| Alternaria infectoria | 2 | - | - | M&G | MK560163 | MK567924 | - |
| Alternaria tenuissima | 4 | - | - | M&G | MK560164 | MK567925 | - |
| Arthrinium phaeospermum | 2 | - | - | M&G | MK560165 | - | - |
| Aspergillus spp. | 6 | 40 | 10 | M | - | - | - |
| Botrytis cinerea | 2 | - | - | M&G | MK560166 | - | - |
| Chaetomium sp. | - | - | 2 | M | - | - | - |
| Cladosporium oxysporum | 14 | 28 | 22 | M&G | MK560167 | MK567926 | - |
| Curvularia hawaiiensis | 2 | - | - | M&G | MK560168 | MK567927 | - |
| Diaporthe sp. | 2 | - | - | M | - | - | - |
| Eurotium sp. | - | - | 2 | M&G | MK560169 | - | - |
| Fusarium incarnatum | 2 | - | - | M | - | - | MK567921 |
| Fusarium tricinctum | 2 | - | - | M | - | - | MK567922 |
| Lecythophora sp. | - | - | 2 | M&G | MK560170 | MK567928 | - |
| Mucor sp. | 8 | 8 | - | M | - | - | - |
| Penicillium spp. | 12 | 22 | 22 | M | - | - | - |
| Sordaria fimicola | - | 2 | - | M&G | MK560171 | MK567929 | - |
| Trichoderma spp. | 2 | - | 2 | M | - | - | - |
| Meyerozima caribbica | 2 | - | - | M&G | MK560172 | - | - |
| Sterile mycelium | - | - | 2 | M | - | - | - |
| Colletotrichum spp. | Isolate 1 | Host | Country 2 | GenBank Accession Numbers 3 | ||
|---|---|---|---|---|---|---|
| ITS | rDNA cluster | IGS | ||||
| C. lupini | PT30, RB020 | Lupinus albus | PT | MK463722 | - | MK567906 |
| C. lupini | CBS129944, RB042 | Cinnamomum sp. | PT | MH865693 | - | MK567907 |
| C. lupini | CSL1294, RB116 | Lupinus polyphyllus | GB | MK463723 | - | MK567908 |
| C. lupini | G52, RB119 | Lupinus albus | DE | MK463724 | - | MK567909 |
| C. lupini | 96A649, RB120 | Lupinus polyphyllus | AU | MK463725 | - | MK567910 |
| C. lupini | IMI504884, RB121 | Lupinus albus | CA | KJ018635 | - | MK567911 |
| C. lupini | C3, RB122 | Lupinus luteus | PL | MK463726 | - | MK567912 |
| C. lupini | IMI504885, RB123 | Lupinus albus | ZA | MK463727 | - | MK567913 |
| C. lupini | 70555, RB124 | Lupinus albus | CL | MK463728 | - | MK567914 |
| C. lupini | CBS109224, RB125 | Lupinus albus | AT | JQ948172 | - | MK567915 |
| C. lupini | PT702, RB127 | Olea europea | ES | MK463729 | - | MK567916 |
| C. lupini | IMI350308, RB147 | Lupinus sp. | GB | MK463730 | - | MK567917 |
| C. lupini | CBS109221, RB172 | Lupinus albus | DE | JQ948169 | - | MK567918 |
| C. lupini | CBS109225, RB173 | Lupinus albus | US | JQ948155 | MK541036 | - |
| C. lupini | IMI375715, RB217 | Lupinus albus | AU | JQ948161 | - | MK567919 |
| C. lupini | IMI504893, RB221 | Lupinus sp. | FR | MK463733 | MK541037 | - |
| C. lupini | CBS509.97, RB235 | Lupinus albus | FR | JQ948159 | - | MK567920 |
| C. abscissum | IMI504790, RB197 | Citrus x sinensis | US | KT153558 | MK541030 | - |
| C. costaricense | CBS211.78, RB184 | Coffea sp. | CR | JQ948181 | MK541033 | - |
| C. cuscutae | IMI304802, RB216 | Cuscuta sp. | DM | JQ948195 | - | - |
| C. melonis | CBS134730, RB237 | Malus domestica | BR | KC204997 | - | - |
| C. paranaense | IMI384185, RB218 | Caryocar brasiliense | BR | JQ948191 | - | - |
| C. tamarilloi | CBS129955, RB018 | Solanum betaceum | CO | JQ948189 | MK541029 | - |
| Colletotrichum sp. | CBS101611, RB170 | Fern | CR | JQ948196 | - | - |
| C. nymphaeae | IMI 504889, RB190 | Fragaria x ananassa | DK | KT153561 | MK541035 | - |
| C. simmondsii | CBS 122122, RB179 | Carica papaya | AU | JQ948276 | MK541034 | - |
| C. fioriniae | IMI 504882, RB111 | Fragaria x ananassa | NZ | KT153562 | MK541031 | - |
| C. acutatum | CBS 112980, RB175 | Pinus radiata | ZA | JQ948356 | - | - |
| C. godetiae | CBS 193.32, RB019 | Olea europaea | IT | JQ948415 | - | - |
| C. phormii | CBS 102054, RB171 | Phormium sp. | NZ | JQ948448 | - | - |
| C. salicis | CBS 607.94, RB157 | Salix sp. | NL | JQ948460 | MK541032 | - |
| C. coccodes | RB302 | Solanum lycopersicum | IT | MK531998 | - | - |
| C. spinaciae | RB305 | Spinacia oleracea | IT | MK531997 | - | - |
| C. higginsianum | IMI 349063, RB300 | Brassica chinensis | TT | JQ005760 | - | - |
| C. graminicola | CBS130836, RB301 | Zea mays | US | JQ005767 | - | - |
| C. fructicola | CSL386, RB003 | Fragaria x ananassa | US | KM246513 | - | - |
| C. orchidophilum | IMI309357, RB209 | Phalaenopsis sp. | GB | JQ948153 | - | - |
| C. tofieldiae | IMI288810, RB164 | Dianthus sp. | GB | GU227803 | - | - |
| C. trichellum | RB306 | Hedera sp. | IT | MK532000 | - | - |
| C. truncatum | RB308 | Glycine max | AR | MK531999 | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pecchia, S.; Caggiano, B.; Da Lio, D.; Cafà, G.; Le Floch, G.; Baroncelli, R. Molecular Detection of the Seed-Borne Pathogen Colletotrichum lupini Targeting the Hyper-Variable IGS Region of the Ribosomal Cluster. Plants 2019, 8, 222. https://doi.org/10.3390/plants8070222
Pecchia S, Caggiano B, Da Lio D, Cafà G, Le Floch G, Baroncelli R. Molecular Detection of the Seed-Borne Pathogen Colletotrichum lupini Targeting the Hyper-Variable IGS Region of the Ribosomal Cluster. Plants. 2019; 8(7):222. https://doi.org/10.3390/plants8070222
Chicago/Turabian StylePecchia, Susanna, Benedetta Caggiano, Daniele Da Lio, Giovanni Cafà, Gaetan Le Floch, and Riccardo Baroncelli. 2019. "Molecular Detection of the Seed-Borne Pathogen Colletotrichum lupini Targeting the Hyper-Variable IGS Region of the Ribosomal Cluster" Plants 8, no. 7: 222. https://doi.org/10.3390/plants8070222
APA StylePecchia, S., Caggiano, B., Da Lio, D., Cafà, G., Le Floch, G., & Baroncelli, R. (2019). Molecular Detection of the Seed-Borne Pathogen Colletotrichum lupini Targeting the Hyper-Variable IGS Region of the Ribosomal Cluster. Plants, 8(7), 222. https://doi.org/10.3390/plants8070222







