Comparative Leaf Proteome Analysis of Maize (Zea mays L.) Exposed to Combined Drought and Heat Stress
Abstract
1. Introduction
2. Results
2.1. Effect of Combined Drought and Heat Stress on Physiological Traits of Maize and Biomass
2.1.1. Analysis of Variance
2.1.2. Sub-Stomatal CO2
2.1.3. Leaf Temperature
2.1.4. Photosynthetic Yield (PSII)
2.1.5. Shoot and Root Dry Weight
2.1.6. Stress Tolerance Indices
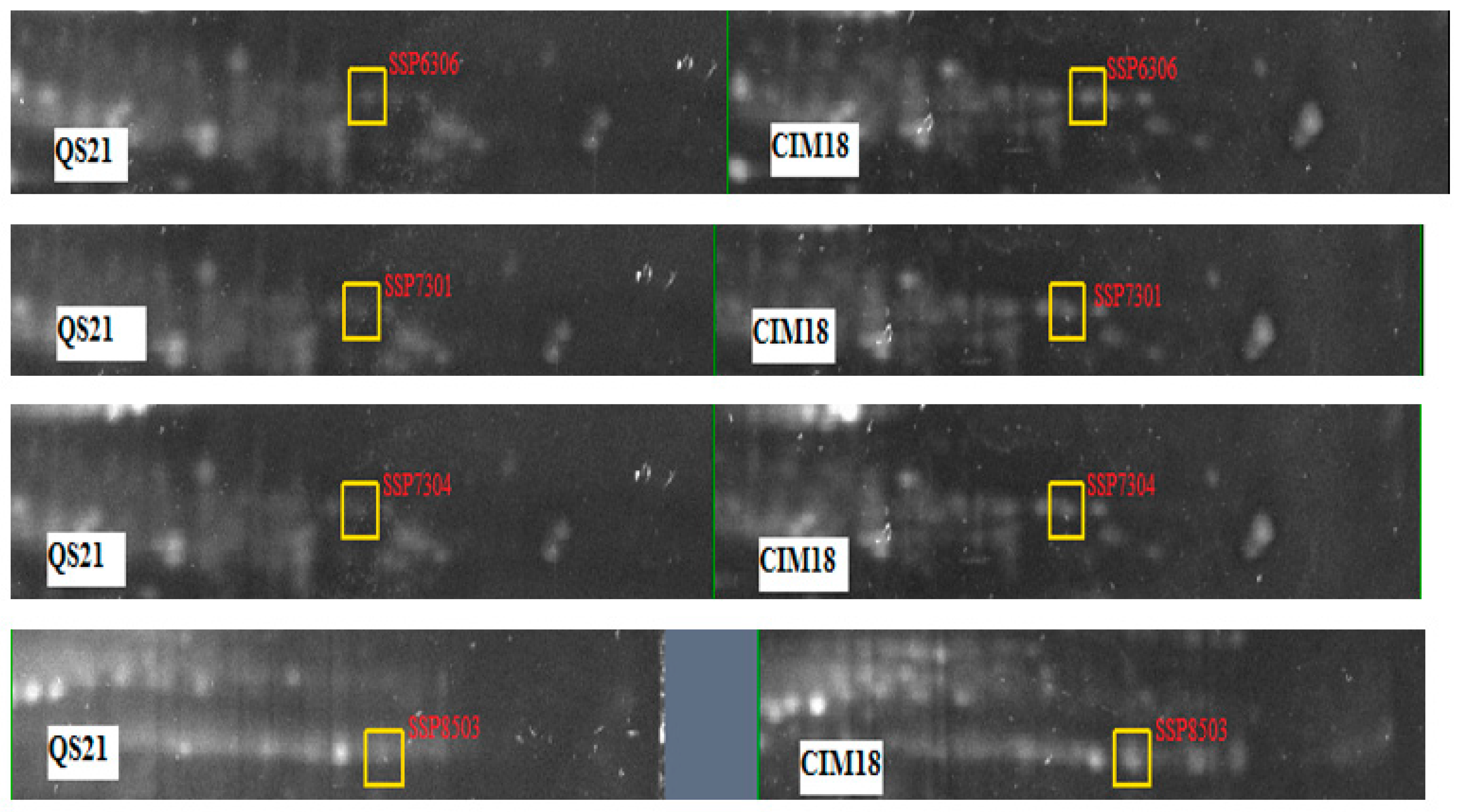
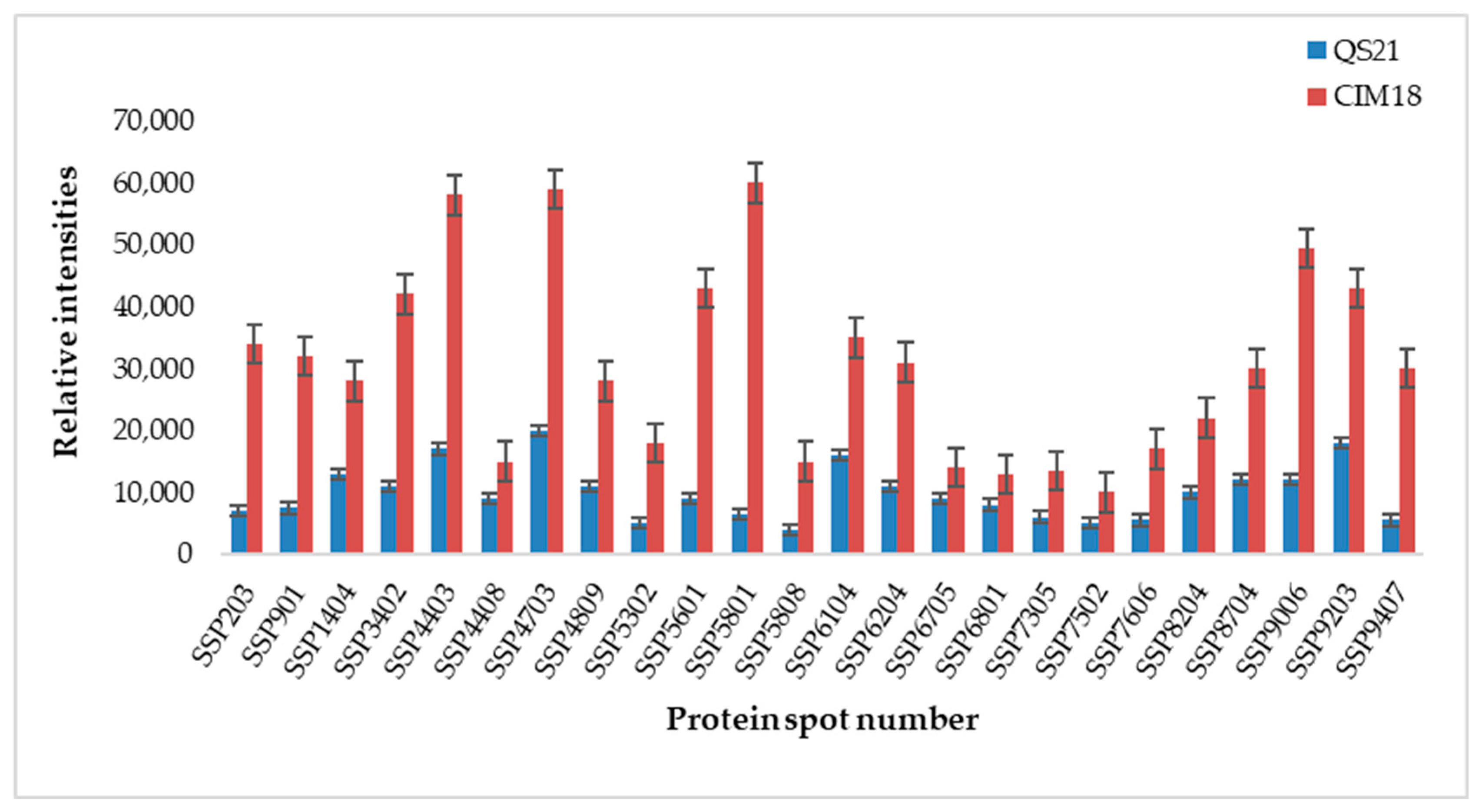
2.1.7. Quantitative Comparative Analysis of Protein Responses
3. Discussion
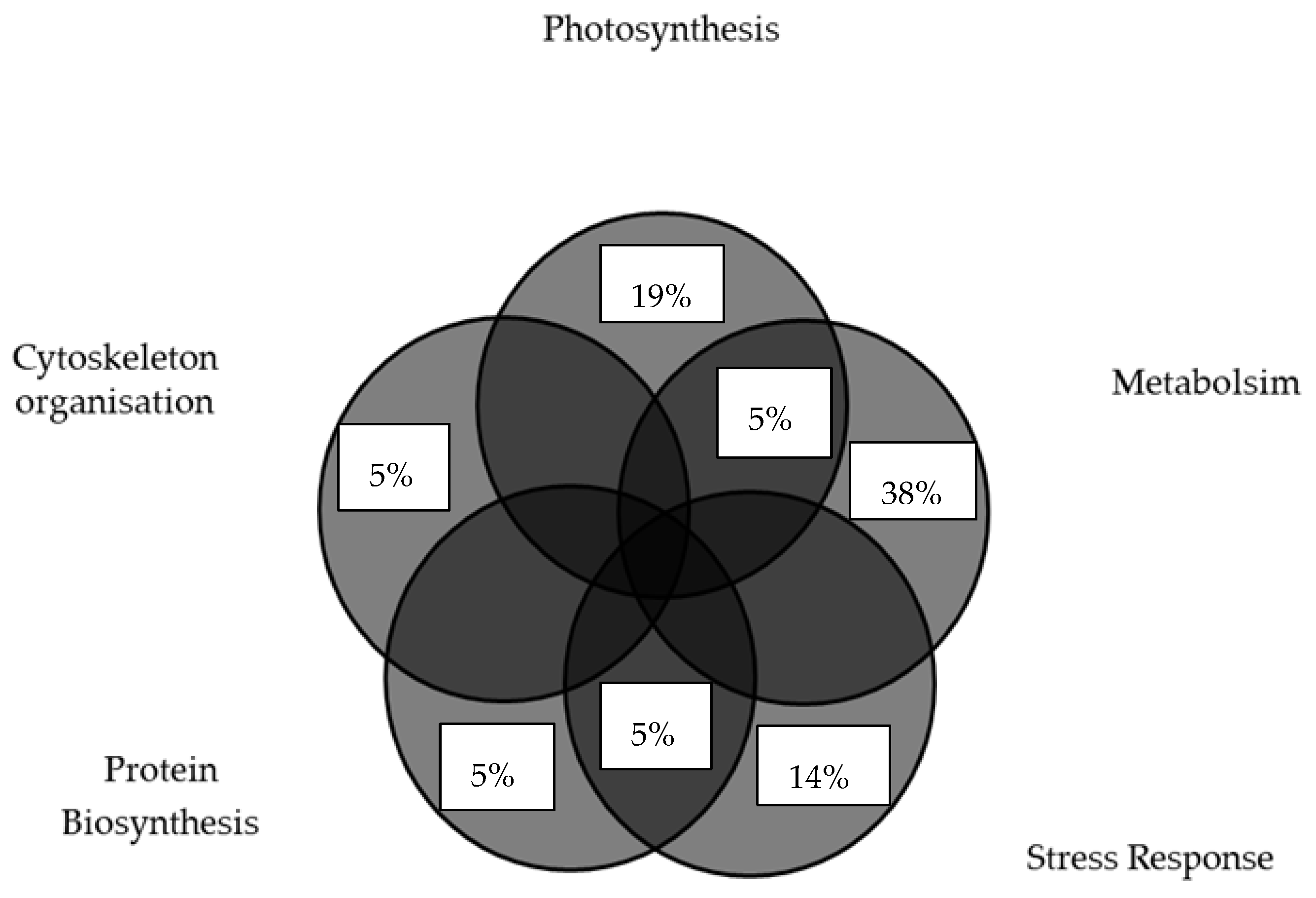
3.1. Photosynthesis- and Metabolism-Related Proteins
3.2. Heat-Stress-Responsive Proteins
3.3. Antioxidant Proteins
3.4. Structural Proteins
4. Materials and Methods
4.1. Plant Materials
4.2. Experimental Design
4.3. Drought and Heat Treatment
4.4. Data Collection
4.4.1. Physiological Traits
4.4.2. Plant Growth
4.5. Protein Extraction and Quantification
4.6. Protein Clean-Up and Two-Dimensional Electrophoresis
4.7. Gel Processing and Peptide Analysis
5. Data Analysis
6. Conclusions and Recommendations
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CDHS | Combined drought and heat stress |
| CIM | CIMMTY |
| QS | Quality Seed |
| CO2 | Carbon dioxide |
| ATP | Adenosine triphosphate |
| Rubisco | Ribulose- 1,5- bisphosphate carboxylase/oxygenase |
| HSP | Heat shock protein |
| GST | Glutathione S transferase |
| ROS | Reactive oxygen species |
| 2-D | Two-dimensional |
Appendix A

| Parameters | Inbred | Treatment | Inbred Line × Treatment |
|---|---|---|---|
| Sub-stomatal CO2 | <0.001 | NS | <0.001 |
| Leaf temperature | <0.001 | <0.001 | <0.001 |
| Photosynthetic yield | <0.001 | <0.001 | <0.001 |
| Shoot | <0.001 | <0.001 | <0.001 |
| Root | <0.001 | <0.001 | <0.001 |
| Line | Pedigree | Drought Tolerance | Heterotic Group |
|---|---|---|---|
| CIM1 | (CLQRCWQ50/CML312SR)-2-2-1-BB-1-B-B | - | A |
| CIM2 | [[CML202/CML144]F2-1-1-3-B-1-B*6/[GQL5/[GQL5/[MSRXPOOL9]C1F2-205-1(OSU23i)-5-3-X-X-1-BB]F2-4sx]-11-3-1-1-B*4]-B*5-1-B | drought-tolerant | B |
| CIM3 | [CML141/[CML141/CML395]F2-1sx]-4-2-1-B*4-1-BB-B | drought-tolerant | B |
| CIM4 | [CML144/[CML144/CML395]F2-5sx]-1-3-1-3-B*7-B | drought-tolerant | - |
| CIM5 | [CML144/SNSYNF2[N3/TUX-A-90]-102-1-2-2-BSR-B*4]-B-4-3-B*4-1-B-B | drought-tolerant | B |
| CIM6 | [CML150/CML373]-B-2-2-B*4-4-B-B | drought-tolerant | A |
| CIM7 | [CML159/[CML159/[MSRXPOOL9]C1F2-205-1(OSU23i)-5-3-X-X-1-BB]F2-3sx]-8-1-1-BBB-4-B-B | drought-tolerant | A |
| CIM8 | [CML182/TZMI703]-B-9-1-BB-#-BB-2-B-B | - | B |
| CIM9 | [CML202/CML144]F2-1-1-3-B-1-B*6-2-B | drought-tolerant | B |
| CIM10 | [CML205/CML176]-B-2-1-1-2-B*5-1-B-B | drought-tolerant | B |
| CIM11 | [CML389/CML176]-B-29-2-2-B*4-B | drought-tolerant | B |
| CIM12 | [GQL5/[GQL5/[MSRXPOOL9]C1F2-205-1(OSU23i)-5-3-X-X-1-BB]F2-4sx]-11-3-1-1-B*5-3-B-B | - | B |
| CIM13 | [GQL5/[GQL5/CML202]F2-3sx]-11-4-1-3-B*4-B | - | B |
| CIM14 | [TZMI703/CML176]-B-3-2-B*5-4-B-B | - | B |
| CIM15 | CLQRCWQ50-BB-1-2-B-B | - | B |
| CIM16 | CML176-#-B-2-B | drought-sensitive | B |
| CIM17 | CML181-B-1-5-B-B7 | drought-sensitive | - |
| CIM18 | CML182-BB-B | - | - |
| CIM19 | CML264Q-B-1-2-B-B | drought-sensitive | A |
| CIM20 | CML491-B-3-11-B-B | - | A |
| CIM21 | CML492-BB-2-1-B-B | drought-sensitive | B |
| QS1 | K054W | - | F |
| QS4 | V0548W | - | F |
| QS5 | V0298W | - | F |
| QS6 | B0388W | - | F |
| QS7 | EM362W | - | M |
| QS8 | EM583W | - | F |
| QS10 | E625W | - | F |
| QS14 | HM18W | - | O |
| QS15 | HM233W | - | T |
| QS16 | HM238W | - | M |
| QS17 | HM267W | - | F |
| QS18 | HM267W | - | F |
| QS19 | HM268W | - | F |
| QS20 | HM284W | - | H |
| QS21 | HM1472W | - | B |
| QS22 | JM226W | - | H |
| QS23 | JM2341W | - | H |
| QS25 | JM2561W | - | H |
| QS26 | JM2602W | - | H |
| QS27 | JM2621W | - | H |
| QS28 | JM2641W | - | H |
| QS29 | E5 | - | G |
| QS30 | E6 | - | G |
| QS32 | E27 | - | G |
References
- Cui, D.; Wu, D.; Liu, J.; Li, D.; Xu, C.; Li, S.; Li, P.; Zhang, H.; Liu, X.; Jiang, C.; et al. Proteomic analysis of seedling roots of two maize inbred lines that differ significantly in the salt stress response. PLoS ONE 2015, 10, e0116697. [Google Scholar] [CrossRef]
- Chávez-Arias, C.C.; Ramírez-Godoy, A.; Restrepo-Díaz, H. Influence of drought, high temperatures, and/or defense against arthropod herbivory on the production of secondary metabolites in maize plants. A review. Curr. Plant Biol. 2022, 32, 00268. [Google Scholar] [CrossRef]
- Grzesiak, M.T.; Marcinska, I.; Janowiak, F.; Rzepka, A.; Hura, T. The relationship between seedling growth and grain yield under drought conditions in maize and triticale genotypes. Acta Physiol. Plant 2012, 34, 1757–1764. [Google Scholar] [CrossRef]
- Millan, M.; Simonneau, T.; Coupe-Ledru, A.; Boulord, R.; Christophe, A.; Pallas, B. Relationships between leaf temperature, stomatal conductance and architecture: Potential impact on leaf burning among arange of genotypes in grapevine. OENO ONE 2023, 57, 345–359. [Google Scholar] [CrossRef]
- Mahmood, U.; Li, X.; Fan, Y.; Chang, W.; Niu, Y.; Li, J.; Qu, C.; Lu, K. Multi-omics revolution to promote plant breeding efficiency. Front. Plant Sci. 2022, 13, 1062952. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Ma, C.; Ma, H.; Qiu, Z.; Wen, X. Physiological and proteomic responses of pitaya to PEG-induced drought stress. Agriculture 2021, 11, 632. [Google Scholar] [CrossRef]
- Rollins, J.A.; Habte, E.; Templer, S.E.; Colby, T.; Schmidt, J.; von Korff, M. Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeumvulgare L.). J. Exp. Bot. 2013, 64, 3201–3212. [Google Scholar] [CrossRef]
- Dong, A.; Yang, Y.; Liu, S.; Zenda, T.; Liu, X.; Wang, Y.; Li, J.; Duan, H. Comparative proteomics analysis of two maize hybrids revealed drought-stress tolerance mechanisms. Biotechnol. Biotechnol. Equip. 2020, 34, 63–780. [Google Scholar] [CrossRef]
- Nemati, M.; Piro, A.; Norouzi, M.; Vahed, M.M.; Nisticò, D.M.; Mazzuca, S. Comparative physiological and leaf proteomic analyses revealed the tolerant and sensitive traits to drought stress in two wheat parental lines and their F6 progenies. EEB 2019, 158, 223–237. [Google Scholar] [CrossRef]
- Lima, L.L.; Balbi, B.P.; Mesquita, R.O.; da Silva, J.C.F.; Coutinho, F.S.; Carmo, F.M.S.; Vital, C.E.; Mehta, A.; Loureiro, M.E.; Fontes, E.P.B.; et al. Proteomic and metabolomic analysis of a drought tolerant soybean cultivar from Brazilian savanna. Crop Breed. Genet. Genom. 2019, 1, e190022. [Google Scholar]
- Boguszewska-Mankowska, D.; Gietler, M.; Nykiel, M. Comparative proteomic analysis of drought and high temperature response in roots of two potato cultivars. J. Plant Growth Regul. 2020, 92, 345–363. [Google Scholar] [CrossRef]
- Dong, S.; Zhou, Q.; Yan, C.; Song, S.; Wang, X.; Wu, Z.; Wang, X.; Ma, C. Comparative protein profiling of two soybean genotypes with different stress tolerance reveals major components in drought tolerance. Front. Sustain. Food Syst. 2023, 7, 1200608. [Google Scholar] [CrossRef]
- Younis, A.; Ramzan, F.; Ramzan, Y.; Zulfiqar, F.; Ahsan, M.; Lim, K.B. Molecular markers improve abiotic stress tolerance in crops: A review. Plants 2020, 9, 1374. [Google Scholar] [CrossRef]
- Dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological responses to drought, salinity, and heat stress in plants: A review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Mondal, S.; Karmakar, S.; Panda, D.; Pramanik, K.; Bose, B.; Singhal, R.K. Crucial plant processes under heat stress and tolerance through heat shock proteins. Plant Stress 2023, 10, 100227. [Google Scholar] [CrossRef]
- Asthir, B. Mechanisms of heat tolerance in crop plants. Biol. Plant 2015, 59, 620–628. [Google Scholar] [CrossRef]
- San-Eufrasio, B.; Bigatton, E.D.; Guerrero-Sánchez, V.M.; Chaturvedi, P.; Jorrín-Novo, J.V.; Rey, M.D.; Castillejo, M.A. Proteomics data analysis for the identification of proteins and derived proteotypic peptides of potential use as putative drought tolerance markers for Quercus ilex. Int. J. Mol. Sci. 2021, 22, 3191. [Google Scholar] [CrossRef]
- Xu, X.; Fonseca de Lima, C.F.; Vu, L.D.; De Smet, I. When drought meets heat–a plant omics perspective. Front. Plant Sci. 2023, 14, 1250878. [Google Scholar] [CrossRef] [PubMed]
- Halder, T.; Choudhary, M.; Liu, H.; Chen, Y.; Yan, G.; Siddique, K.H.M. Wheat Proteomics for Abiotic Stress Tolerance and Root System Architecture: Current Status and Future Prospects. Proteomes 2022, 10, 17. [Google Scholar] [CrossRef]
- Aina, O.; Bakare, O.O.; Fadaka, A.O.; Keyster, M.; Klein, A. Plant biomarkers as early detection tools in stress management in food crops: A review. Planta 2024, 259, 60. [Google Scholar] [CrossRef]
- Sehgal, A.; Sita, K.; Kumar, J.; Kumar, S.; Singh, S.; Siddique, K.H.; Nayyar, H. Effects of drought, heat and their interaction on the growth, yield and photosynthetic function of lentil (Lens culinaris Medikus) genotypes varying in heat and drought sensitivity. Front. Plant Sci. 2017, 8, 1776. [Google Scholar] [CrossRef]
- Marchin, R.M.; Backes, D.; Ossola, A.; Leishman, M.R.; Tjoelker, M.G.; Ellsworth, D.S. Extreme heat increases stomatal conductance and drought-induced mortality risk in vulnerable plant species. Glob. Change Biol. 2022, 3, 1133–1146. [Google Scholar] [CrossRef]
- Avramova, V.; Nagel, K.A.; AbdElgawad, H.; Bustos, D.; DuPlessis, M.; Fiorani, F.; Beemster, G.T. Screening for drought tolerance of maize hybrids by multi-scale analysis of root and shoot traits at the seedling stage. J. Exp. Bot. 2016, 67, 2453–2466. [Google Scholar] [CrossRef] [PubMed]
- Pfunde, C.N. Genetics, Physiology, Proteomics of Quality Protein Maize Inbred Lines Under Drought and Heat Stress. Ph.D. Thesis, University of Fort Hare, Alice, South Africa, May 2017. [Google Scholar]
- Subramani, M.; Urrea, C.A.; Kalavacharla, V. Comparative Analysis of Untargeted Metabolomics in Tolerant and Sensitive Genotypes of Common Bean (Phaseolus vulgaris L.) Seeds Exposed to Terminal Drought Stress. Metabolites 2022, 12, 944. [Google Scholar] [CrossRef]
- Meng, X.; Liu, S.; Dong, T.; Xu, T.; Ma, D.; Pan, S.; Li, Z.; Zhu, M. Comparative Transcriptome and Proteome Analysis of Salt-Tolerant and Salt-Sensitive Sweet Potato and Overexpression of IbNAC7 Confers Salt Tolerance in Arabidopsis. Front. Plant Sci. 2020, 11, 572540. [Google Scholar] [CrossRef] [PubMed]
- Laurindo, B.S.; Laurindo, R.D.F.; Fontes, P.P.; Vital, C.E.; Delazari, F.T.; Baracat-Pereira, M.C.; da Silva, D.J.H. Comparative analysis of constitutive proteome between resistant and susceptible tomato genotypes regarding to late blight. Funct. Integr. Genom. 2018, 18, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Ghirardo, A.; Nosenko, T.; Kreuzwieser, J.; Winkler, J.B.; Kruse, J.; Albert, A.; Merl-Pham, J.; Lux, T.; Ache, P.; Zimmer, I.; et al. Protein expression plasticity contributes to heat and drought tolerance of date palm. Oecologia 2021, 197, 903–919. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Eldakak, M.; Paudel, B.; Kim, D.W.; Hemmati, H.; Basu, C.; Rohila, J.S. Leaf proteome analysis reveals prospective drought and heat stress response mechanisms in soybean. Biomed. Res. Int. 2016, 1, 6021047. [Google Scholar] [CrossRef]
- Abdulbaki, A.S.; Alsamadany, H.; Alzahrani, Y.; Olayinka, B.U. Rubisco and abiotic stresses in plants: Current assessment. Turk. J. Bot. 2022, 46, 541–552. [Google Scholar] [CrossRef]
- Loka, D.A.; Oosterhuis, D.M.; Baxevanos, D.; Noulas, C.; Hu, W. Single and combined effects of heat and water stress and recovery on cotton (Gossypium hirsutum L.) leaf physiology and sucrose metabolism. Plant Physiol. Biochem. 2020, 148, 166–179. [Google Scholar] [CrossRef]
- Vanlerberghe, G.C. Alternative oxidase: A mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 2013, 14, 6805–6847. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, H.; Guo, Z.; Meng, Y.; Yang, M.; Li, X.; Yang, Q. Adaptation responses in C4 photosynthesis of sweet maize (Zea mays L.) exposed to nicosulfuron. Ecotoxicol. Environ. Saf. 2021, 214, 112096. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Dan, X.; Liu, J.; Liv, Q.; Li, X. Translation elongation factor-1α is pivotal for plant heat tolerance despite its pronounced heat-induced aggregation. Plant Physiol. Biochem. 2024, 210, 108649. [Google Scholar] [CrossRef]
- Rahman, M.A.; Woo, J.H.; Song, Y.; Lee, S.H.; Hasan, M.M.; Azad, M.A.K.; Lee, K.W. Heat shock proteins and antioxidant genes involved in heat combined with drought stress responses in perennial rye grass. Life 2022, 12, 1426. [Google Scholar] [CrossRef]
- Klein, R.D.; Chidawanyika, T.; Tims, H.S.; Meulia, T.; Bouchard, R.A.; Pett, V.B. Chaperone function of two small heat shock proteins from maize. Plant Sci. 2014, 221, 48–58. [Google Scholar] [CrossRef]
- Dharan, S.S.; Sabu, K.K. Expression profiling of stress responsive genes in cell suspension of Elettaria cardamomum (L.) Maton under abiotic stress. J. Plant. Crops. 2023, 51, 23–30. [Google Scholar] [CrossRef]
- Yusof, N.A.; Masnoddin, M.; Charles, J.; Thien, Y.Q.; Nasib, F.N.; Michael, C.; Wong, V.L.; Murad, A.M.A.; Mahadi, N.M.; Bharudin, I. Can heat shock protein 70 (HSP70) serve as biomarkers in Antarctica for future ocean acidification, warming and salinity stress? Polar Biol. 2022, 45, 371–394. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, H.K.; Dong, Q.L.; Zhang, Y.Y.; Wang, Y.M.; Li, H.Y.; Xing, G.J.; Li, Q.Y.; Dong, Y.S. Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L). Front. Plant Sci. 2015, 6, 773. [Google Scholar] [CrossRef]
- Raja, V.; Qadir, S.U.; Alyemeni, M.N.; Ahmad, P. Impact of drought and heat stress individually and in combination on physio-biochemical parameters, antioxidant responses, and gene expression in Solanum lycopersicum. 3 Biotech 2020, 10, 208. [Google Scholar] [CrossRef]
- Yousaf, M.I.; Riaz, M.W.; Jiang, Y.; Yasir, M.; Aslam, M.Z.; Hussain, S.; Sajid Shah, S.A.; Shehzad, A.; Riasat, G.; Manzoor, M.A.; et al. Concurrent effects of drought and heat stresses on physio-chemical attributes, antioxidant status and kernel quality traits in maize (Zea mays L.) hybrids. Front. Plant Sci. 2022, 13, 898823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.W.; Feng, L.Y.; Cheng, J.; Tang, H.; Xu, F.; Zhu, F.; Zhao, Z.Y.; Yuan, M.; Chen, Y.E.; Wang, J.H.; et al. The roles of two transcription factors, ABI4 and CBFA, in ABA and plastid signalling and stress responses. Plant Mol. Biol. 2013, 83, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.S.; Singh, V.; Islam, S.; Islam, M.S.; Ahsan, R.; Kaundal, A.; Islam, T.; Ghosh, A. Genome-wide identification and expression profiling of glutathione S-transferase family under multiple abiotic and biotic stresses in Medicago truncatula L. PLoS ONE 2021, 16, 0247170. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Trivedi, P.K. Glutathione S-transferases role in combating abiotic stresses including arsenic detoxification in plants. Front. Plant Sci. 2018, 9, 751. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Yang, Z.; Wanga, W.; Guod, S.; Lic, Z.; Liuc, K.; Suia, N. Transcriptome analysis of maize inbred lines differing in drought tolerance provides novel insights into the molecular mechanisms of drought responses in roots. Plant Physiol. Biochem. 2020, 149, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Shi, M.; Wang, D.; Gong, Y.; Sha, Q.; Liv, P.; Yang, J.; Chu, P.; Guo, S. Research progress on the roles of actin-depolymerizing factor in plant stress responses. Front. Plant Sci. 2023, 14, 1278311. [Google Scholar] [CrossRef]
- Sniegowska-Swierk, K.; Dubas, E.; Rapacz, M. Drought-induced changes in the actin cytoskeleton of barley (Hordeum vulgare L.) leaves. Acta Physiol. Plant 2015, 37, 73. [Google Scholar] [CrossRef]
- Bawa, G.; Liu, Z.; Zhou, Y.; Fan, S.; Ma, Q.; Tissue, D.T.; Sun, X. Cotton proteomics: Dissecting the stress response mechanisms in cotton. Front. Plant Sci. 2022, 13, 1035801. [Google Scholar] [CrossRef]
- Hu, X.; Wu, L.; Zhao, F.; Zhang, D.; Li, N.; Zhu, G.; Li, C.; Wang, W. Phosphoproteomic analysis of the response of maize leaves to drought, heat and their combination stress. Front. Plant Sci. 2015, 6, 298. [Google Scholar] [CrossRef]
- Nieto-Sotelo, J.; Martinez, L.; Ponce, G.; Cassab, G.I.; Alagon, A.; Meely, R.B.; Ribaut, J.; Yang, R. Maize HSP101 plays important roles in both induced and basal thermotolerance and primary radicle growth. Plant Cell 2002, 14, 1621–1633. [Google Scholar] [CrossRef]
- Farshadfar, E.; Poursiahbidi, M.M.; Safavi, S.M. Assessment of drought tolerance in land races of bread wheat based on resistance/tolerance indices. Int. J. Adv. Biol. Biomed. Res 2013, 1, 14–158. [Google Scholar]

| Rank | PSII | Ci (mol mol−1) | T Leaf (°C) | SDW (g) | RDW (g) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | QS17 | 0.69 a | CIM20 | 9012.00 a | CIM15 | 35.10 a | QS22 | 0.17 a | CIM12 | 0.09 a |
| 2 | QS1 | 0.69 ab | QS23 | 9012.00 a | QS25 | 34.60 a | CIM12 | 0.14 ab | QS22 | 0.08 ab |
| 3 | QS30 | 0.68 a–c | CIM9 | 5506.00 b | QS1 | 34.35 ab | CIM18 | 0.14 a–c | CIM18 | 0.08 a–c |
| 4 | QS16 | 0.67 a–c | CIM13 b | 5506.00 | QS32 | 34.30 ab | CIM20 | 0.14 a–c | CIM10 | 0.08 a–c |
| 5 | CIM18 | 0.67 a–c | CIM21 | 3935.00 c | QS18 | 33.95 ab | QS8 | 0.14 a–c | QS8 | 0.07 a–d |
| 6 | CIM14 | 0.67 a–c | CIM5 | 3935.00 c | CIM10 | 33.80 ab | QS26 | 0.14 a–d | QS26 | 0.07 a–d |
| 7 | CIM11 | 0.67 a–c | CIM6 | 3935.00 c | QS29 | 33.80 ab | QS18 | 0.13 a–e | QS18 | 0.07 b–e |
| 8 | CIM12 | 0.67 a–c | QS3 | 3935.00 c | CIM16 | 33.55 a–c | CIM10 | 0.13 b–f | CIM7 | 0.06 b–f |
| 9 | CIM1 | 0.66 a–c | QS5 | 3789.00 d | CIM6 | 33.50 a–c | QS6 | 0.12 b–f | QS5 | 0.06 c–g |
| 10 | QS7 | 0.66 a–c | QS20 | 3591.00 e | CIM21 | 33.40 a–d | QS7 | 0.12 b–f | QS6 | 0.06 c–g |
| Bottom | ||||||||||
| 1 | CIM2 | 0.60 a–e | CIM16 | 827.00 w | CIM19 | 31.10 b–j | QS16 | 0.09 e–i | QS28 | 0.04 f–h |
| 2 | QS19 | 0.60 a–e | QS15 | 827.000 w | QS23 | 31.05 b–j | QS15 | 0.09 e–i | CIM2 | 0.04 f–h |
| 3 | QS28 | 0.57 a–e | QS16 | 772.000 x | QS21 | 30.30 c–j | CIM15 | 0.09 e–i | CIM11 | 0.04 f–h |
| 4 | CIM16 | 0.53 a–e | CIM17 | 544.000 y | QS8 | 30.10 d–j | CIM17 | 0.09 f–i | CIM13 | 0.04 f–h |
| 5 | CIM21 | 0.52 a–e | QS21 | 531.000 Z | CIM7 | 29.80 e–j | CIM21 | 0.09 f–i | CIM16 | 0.04 f–h |
| 6 | QS4 | 0.49 b–e | QS27 | 531.000 z | QS27 | 29.35 f–j | QS10 | 0.09 f–i | QS1 | 0.04 f–h |
| 7 | CIM13 | 0.49 c–e | CIM1 | 438.00 za | CIM20 | 29.00 g–j | QS23 | 0.09 f–i | QS 10 | 0.04 f–h |
| 8 | CIM19 | 0.43 de | CIM2 | 361.00 zb | QS17 | 28.70 h-j | CIM19 | 0.08 g–i | QS15 | 0.04 gh |
| 9 | CIM5 | 0.40 e | QS4 | 361.00 zb | CIM18 | 28.60 ij | CIM1 | 0.08 hi | CIM19 | 0.04 gh |
| 10 | QS5 | 0.08 f | QS6 | −793.00 zc | CIM12 | 28.30 ij | QS21 | 0.07 i | CIM1 | 0.03 h |
| INBRED LINE | STI | DRI | K2STI | Inbred Line | STI | DRI | K2STI |
|---|---|---|---|---|---|---|---|
| CIM1 | 0.16 | 0.15 | 0.09 | QS14 | 0.26 | 0.17 | 0.21 |
| CIM18 | 0.32 | 0.42 | 0.58 | QS15 | 0.23 | 0.11 | 0.13 |
| CIM11 | 0.25 | 0.44 | 0.40 | QS16 | 0.18 | 0.18 | 0.13 |
| CIM12 | 0.22 | 0.61 | 0.40 | QS17 | 0.26 | 0.28 | 0.31 |
| CIM13 | 0.16 | 0.27 | 0.14 | QS18 | 0.30 | 0.30 | 0.41 |
| CIM14 | 0.20 | 0.22 | 0.17 | QS19 | 0.28 | 0.26 | 0.33 |
| CIM15 | 0.16 | 0.15 | 0.09 | QS20 | 0.25 | 0.18 | 0.21 |
| CIM16 | 0.19 | 0.17 | 0.13 | QS21 | 0.21 | 0.28 | 0.21 |
| CIM17 | 0.21 | 0.12 | 0.12 | QS22 | 0.43 | 0.54 | 1.07 |
| CIM10 | 0.16 | 0.15 | 0.09 | QS23 | 0.17 | 0.14 | 0.10 |
| CIM2 | 0.12 | 0.14 | 0.05 | QS25 | 0.25 | 0.23 | 0.25 |
| CIM20 | 0.18 | 0.25 | 0.15 | QS26 | 0.29 | 0.32 | 0.39 |
| CIM21 | 0.21 | 0.54 | 0.33 | QS27 | 0.22 | 0.21 | 0.18 |
| CIM19 | 0.14 | 0.17 | 0.08 | QS28 | 0.20 | 0.16 | 0.14 |
| CIM3 | 0.21 | 0.28 | 0.21 | QS29 | 0.22 | 0.20 | 0.19 |
| CIM4 | 0.22 | 0.21 | 0.18 | QS30 | 0.31 | 0.19 | 0.31 |
| CIM5 | 0.28 | 0.26 | 0.33 | QS32 | 0.21 | 0.16 | 0.15 |
| CIM6 | 0.26 | 0.22 | 0.26 | QS4 | 0.22 | 0.15 | 0.16 |
| CIM7 | 0.30 | 0.24 | 0.35 | QS5 | 0.32 | 0.23 | 0.38 |
| CIM8 | 0.27 | 0.21 | 0.27 | QS6 | 0.35 | 0.26 | 0.47 |
| CIM9 | 0.24 | 0.19 | 0.20 | QS7 | 0.25 | 0.29 | 0.29 |
| QS10 | 0.20 | 0.16 | 0.14 | QS8 | 0.26 | 0.43 | 0.41 |
| QS1 | 0.26 | 0.22 | 0.26 |
| Spot # | Protein Names | Accession | % Coverage | Subcellular Location | Function | GO Annotation |
|---|---|---|---|---|---|---|
| 4606 | Actin-1 | sp|P02582|ACT1_MAIZE | 9.33 | Nucleus (GO:0005634) Cytoskeleton (GO:0005856) | Cytoskeleton organization | GO:0007010 |
| 8406 | Adenylate kinase, chloroplastic | sp|P43188|KADC_MAIZE | 58.11 | Plastid; chloroplast (GO:0009507) | Metabolism signalling | GO:0006139 GO:0046940 |
| 7403 | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic | sp|P09315|G3PA_MAIZE | 8.933 | Plastid; chloroplast (GO:0009507) | Metabolism, photosynthesis | GO:0006006, GO:0006006 GO:0009416, GO:0019253 |
| 4607 | DIMBOA UDP-glucosyltransferase BX9 | sp|B4G072|BX9_MAIZE | 36.80 | Cytoplasm (GO:0005737) | Metabolism | GO:0008194, GO:0016740 GO:0016757,GO:0047254 GO:0080043, GO:0080044 |
| 8509 | Glyceraldehyde-3-phosphate dehydrogenase 2, cytosolic | sp|Q09054|G3PC2_MAIZE | 48.66 | Cytoplasm (GO:0005737) | Metabolism | GO:0006006, GO:0006096 |
| 4605 | Glutamine synthetase root isozyme 3 | sp|P38561|GLNA3_MAIZE | 22.47 | Cytoplasm (GO:0005737) | Metabolism | GO:0006542 |
| 5605 | Malate dehydrogenase [NADP], chloroplastic | sp|P15719|MDHP_MAIZE | 18.98 | Plastid; chloroplast (GO:0009507) | Metabolism | GO:0006099, GO:0006107 GO:0006108, GO:0019674 |
| 0405 | Glutamine synthetase, chloroplastic | sp|P25462|GLNAC_MAIZE | 35.70 | Plastid; chloroplast (GO:0009507) | Metabolism | GO:0006542 |
| 1606 | ATP synthase subunit beta, mitochondrial | sp|P19023|ATPBM_MAIZE | 1.989 | Mitochondrion; mitochondrion inner membrane (GO:0005743) | Metabolism; ATP metabolism | GO:0046034, GO:0006754 GO:0015986, GO:0042776 |
| 5404 | Triosephosphate isomerase, cytosolic | sp|P12863|TPIS_MAIZE | 22.13 | Cytoplasm (GO:0005737) | Metabolism; carbohydrate metabolism | GO:0006094, GO:0006096 GO:0016052 |
| 8611 | Ribulose bisphosphate carboxylase large chain | sp|P00874|RBL_MAIZE | 49.16 | Plastid; chloroplast (GO:0009507) | Photosynthesis | GO:0009853, GO:0015977 GO:0015979, GO:0019253 |
| 5609 | Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic | sp|Q9ZT00|RCA_MAIZE | 19.63 | Chloroplast stroma (GO:0009570) | Photosynthesis | GO:0046863, GO:0016887 GO:0005524 |
| 4807 | Pyruvate, phosphate dikinase 1, chloroplastic | sp|P11155|PPDK1_MAIZE | 28.83 | Plastid; chloroplast (GO:0009507) Cytoplasm (GO:0005737) | Photosynthesis | GO:0015979 |
| 8503 | ATP synthase subunit gamma, chloroplastic | sp|P0C1M0|ATPG_MAIZE | 42.34 | Plastid; chloroplast thylakoid membrane (GO:0009535) | Photosynthesis Proton transmembrane transport | GO:0006754, GO:1902600 |
| 3503 | 60S acidic ribosomal protein P0 | sp|O24573|RLA0_MAIZE | 19.12 | Ribosome (GO:0005840) Cytosolic large ribosomal subunit (GO:0022625) | Protein biosynthesis Stress response to anoxia | GO:0002181 GO:0034059 |
| 0406 | Elongation factor 1-alpha | sp|Q41803|EF1A_MAIZE | 6.711 | Cytoplasm (GO:0005737) | Protein biosynthesis | GO:0006412 GO:0006414 |
| 5205 | 17.8 kDa class II heat shock protein | sp|P24632|HSP22_MAIZE | 37.20 | Cytoplasm (GO:0005737) | Stress response, protein folding Heat response, salt stress response Hydrogen peroxide response Protein oligomerization | GO:0006950, GO:0006457 GO:0009408, GO:0009651 GO:0042542, GO:0051259 |
| 4804 | Heat shock 70 kDa protein | sp|P11143|HSP70_MAIZE | 19.22 | Cytoplasm (GO:0005737) | Stress response to heat Protein folding | GO:0009408 GO:0042026 |
| 7304 | Glutathione S-transferase 3 | sp|P04907|GSTF3_MAIZE | 16.67 | Cytoplasm (GO:0005737) | Stress response to herbicide | GO:0009635 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pfunde, C.; Mutengwa, C.S.; Bradley, G.; Chiuta, N.E. Comparative Leaf Proteome Analysis of Maize (Zea mays L.) Exposed to Combined Drought and Heat Stress. Plants 2025, 14, 3419. https://doi.org/10.3390/plants14223419
Pfunde C, Mutengwa CS, Bradley G, Chiuta NE. Comparative Leaf Proteome Analysis of Maize (Zea mays L.) Exposed to Combined Drought and Heat Stress. Plants. 2025; 14(22):3419. https://doi.org/10.3390/plants14223419
Chicago/Turabian StylePfunde, Cleopatra, Charles Shelton Mutengwa, Graeme Bradley, and Nyasha Esnath Chiuta. 2025. "Comparative Leaf Proteome Analysis of Maize (Zea mays L.) Exposed to Combined Drought and Heat Stress" Plants 14, no. 22: 3419. https://doi.org/10.3390/plants14223419
APA StylePfunde, C., Mutengwa, C. S., Bradley, G., & Chiuta, N. E. (2025). Comparative Leaf Proteome Analysis of Maize (Zea mays L.) Exposed to Combined Drought and Heat Stress. Plants, 14(22), 3419. https://doi.org/10.3390/plants14223419






