Genome-Wide Characterization and Functional Analysis of CsDOF Transcription Factors in Camellia sinensis cv. Tieguanyin Under Combined Heat–Drought Stress
Abstract
1. Introduction
2. Results
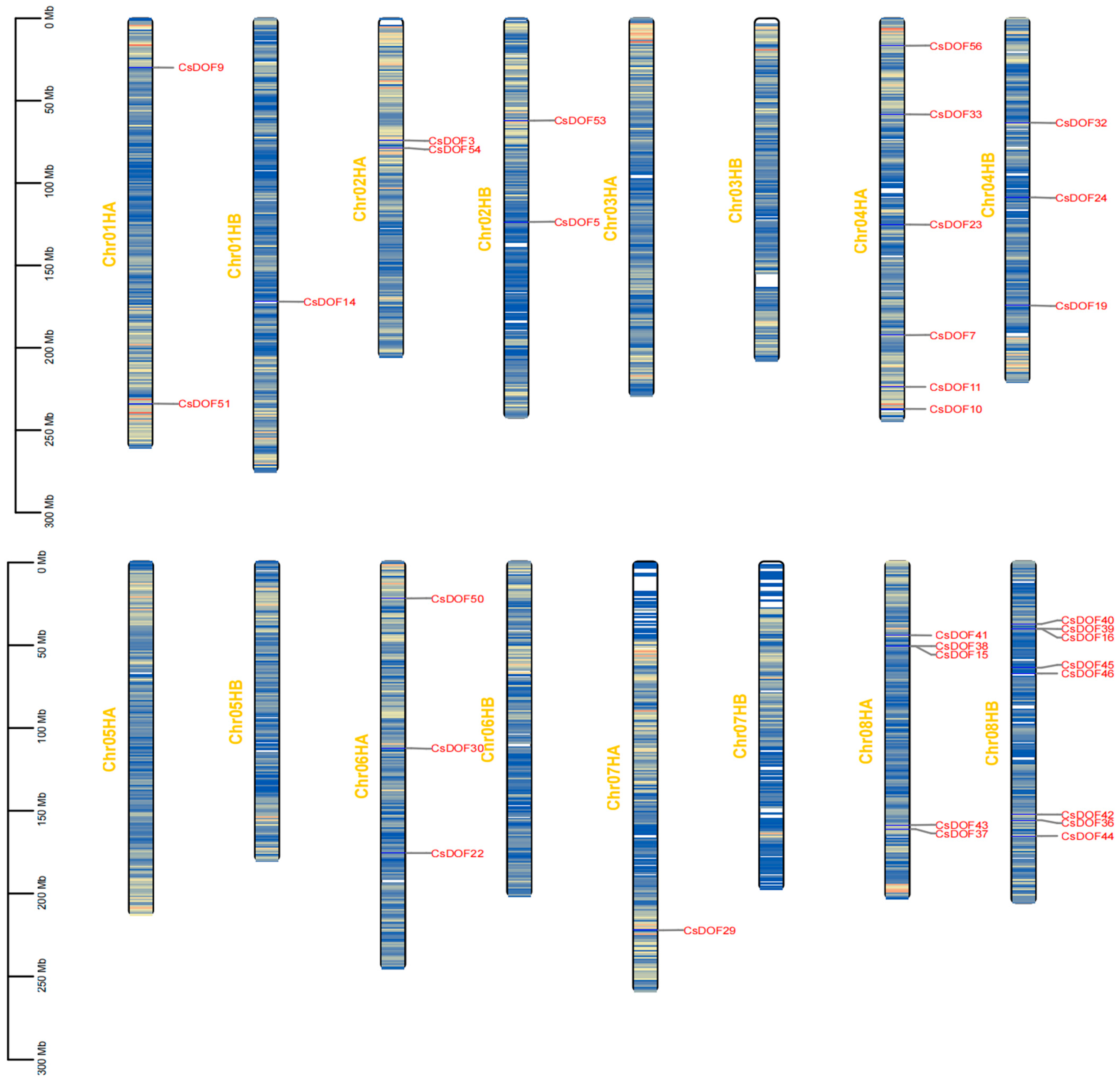
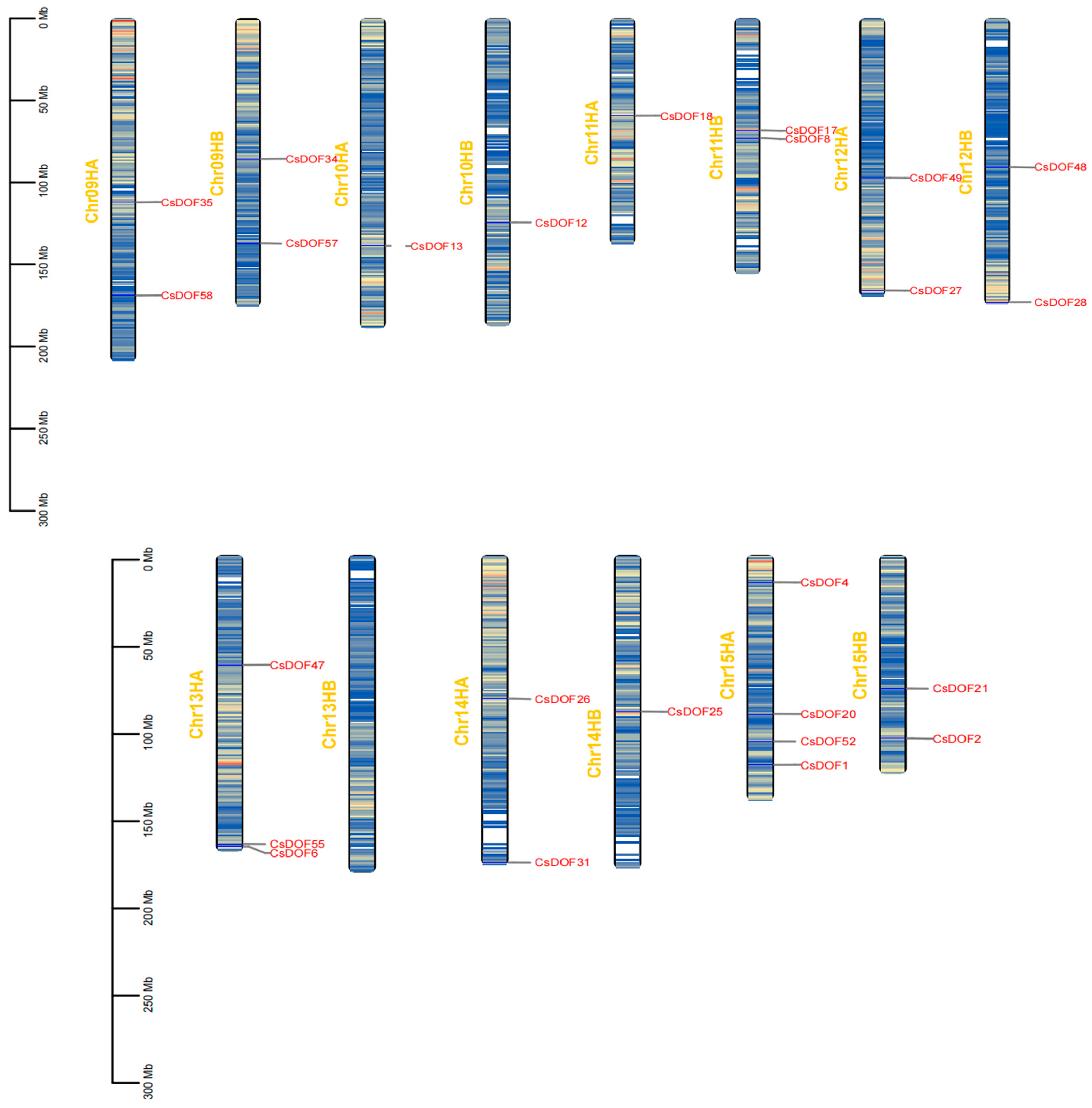
2.1. Comprehensive Analysis of Chromosomal Distribution and Protein Physicochemical Properties of CsDOF Gene Family in Tieguanyin
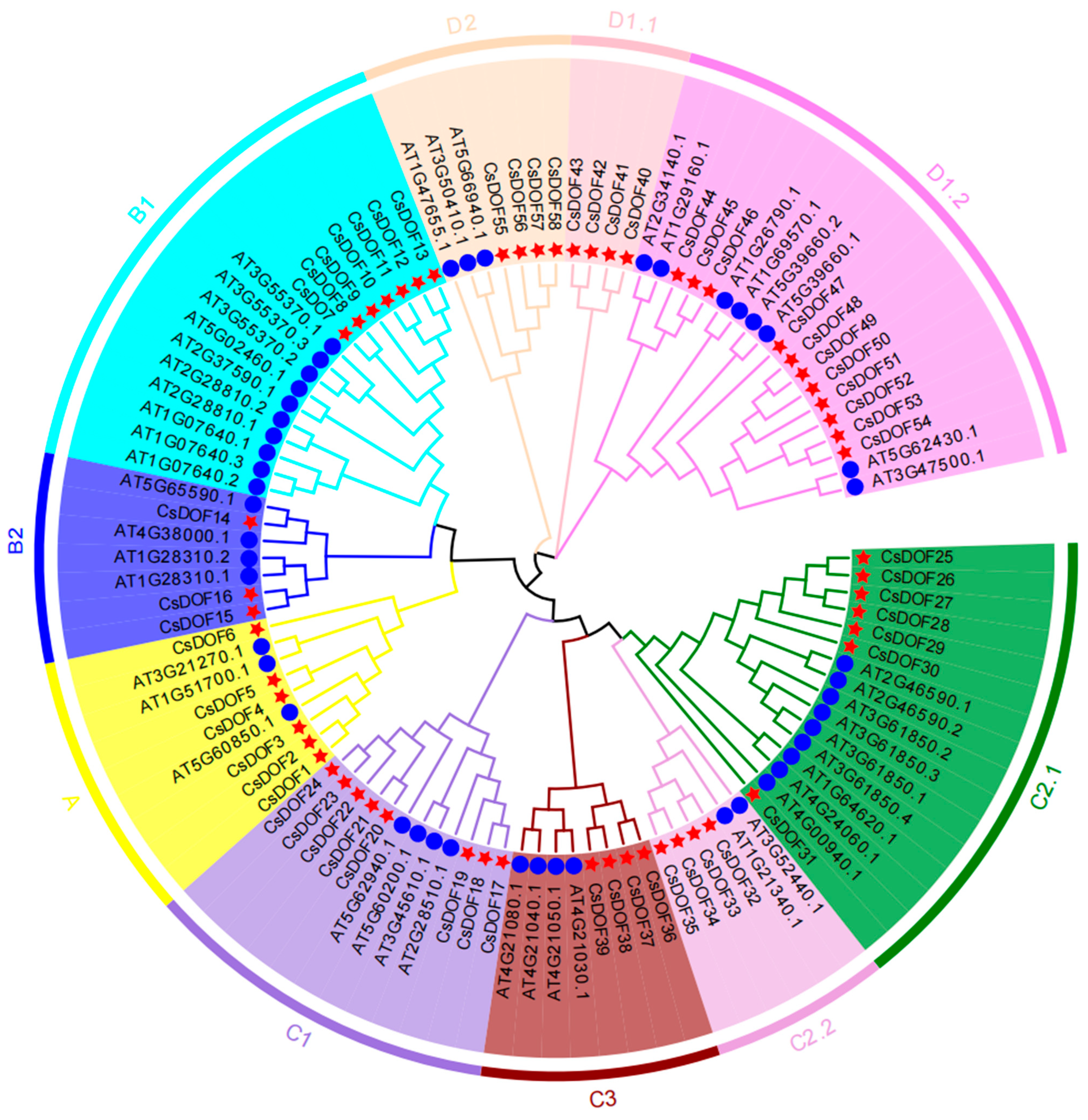
2.2. Phylogenetic Analysis and Functional Diversification of DOF Gene Family in Arabidopsis and Camellia sinensis (CsDOF)
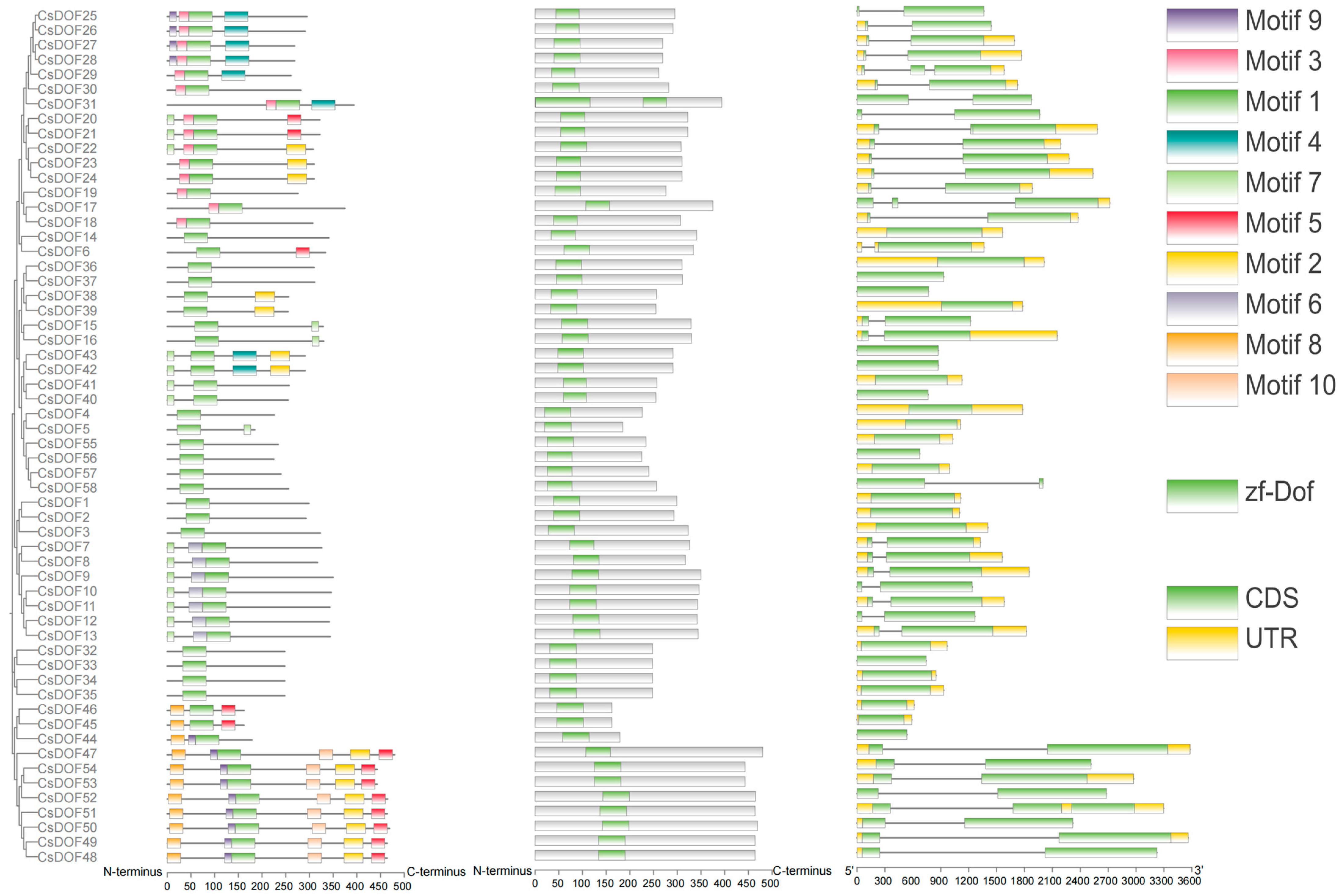
2.3. Characterization of the CsDOF Gene Family: Conserved Motifs, Exon–Intron Organization, and Phylogenetic Relationships
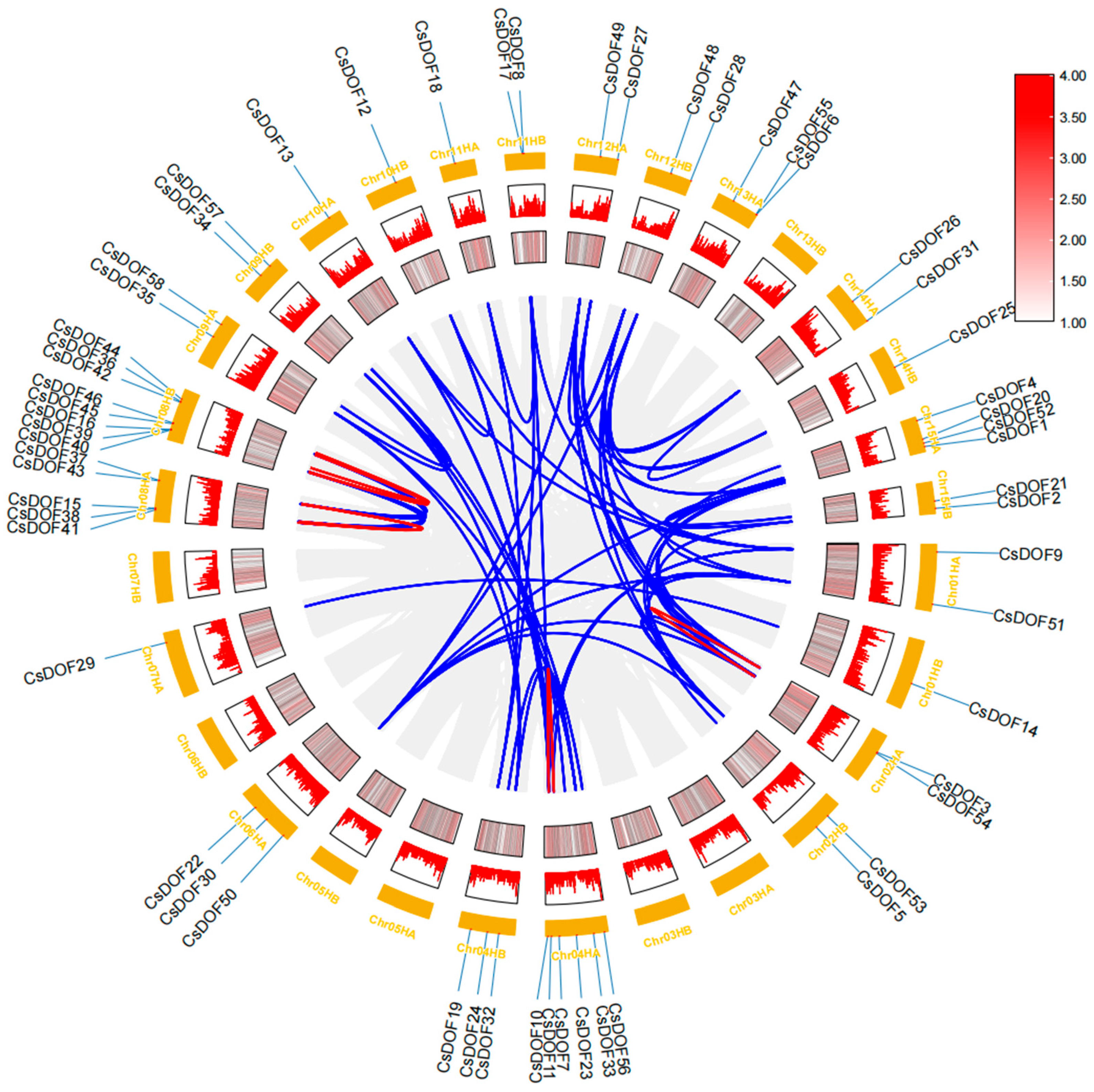
2.4. Genome-Wide Analysis of CsDOF Genes in Tieguanyin: Uneven Chromosomal Distribution and Duplication-Driven Expansion
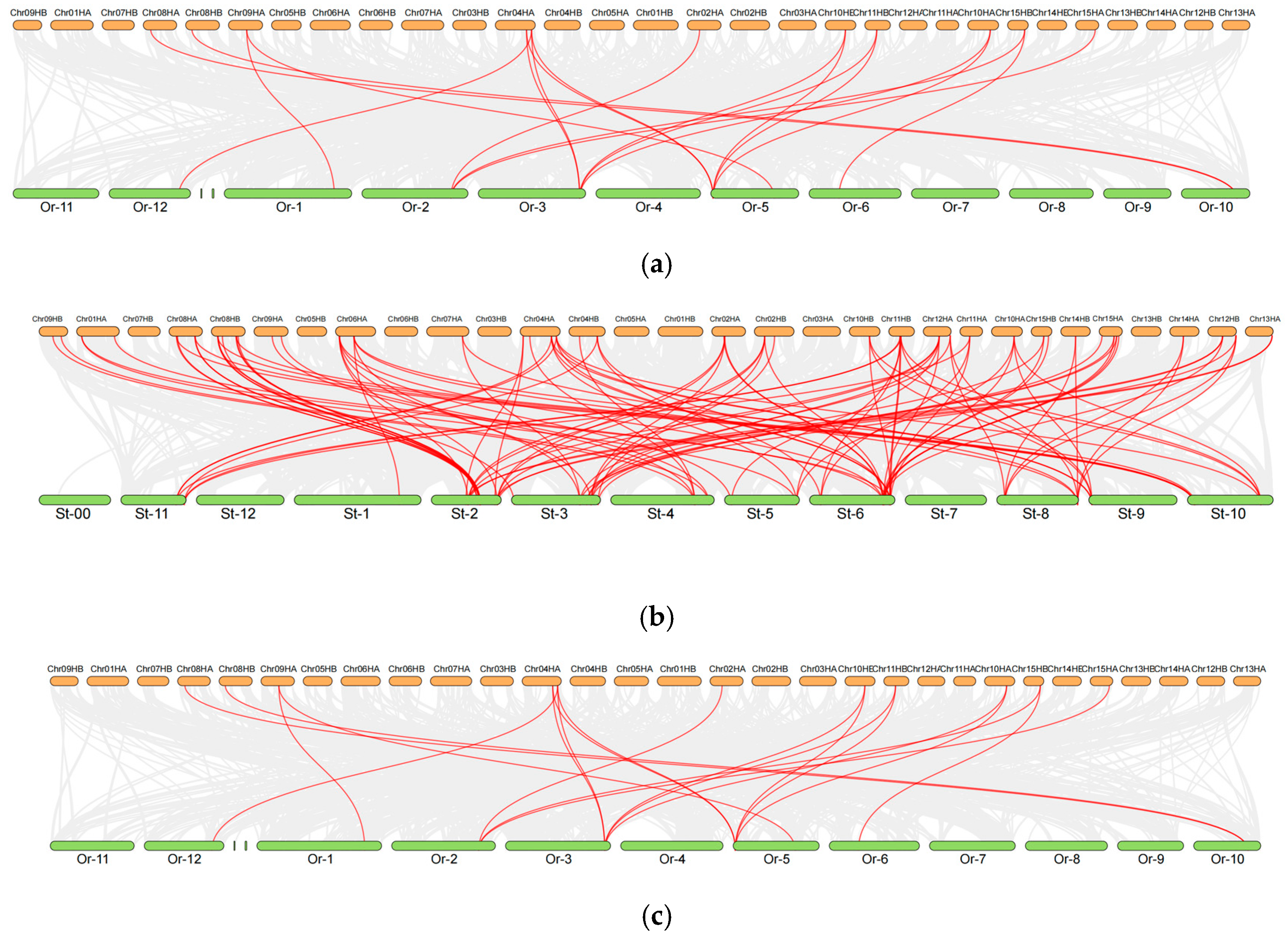
2.5. Cross-Species Collinearity Analysis of DOF Genes in Camellia sinensis and Model Plants Reveals Evolutionary Trajectories and Breeding Implications
2.6. Prediction Analysis of Cis-Acting Elements for CsDOF Gene Families
2.7. Expression Profile of CsDOF Gene in Roots, Stems, and Leaves of Tieguanyin
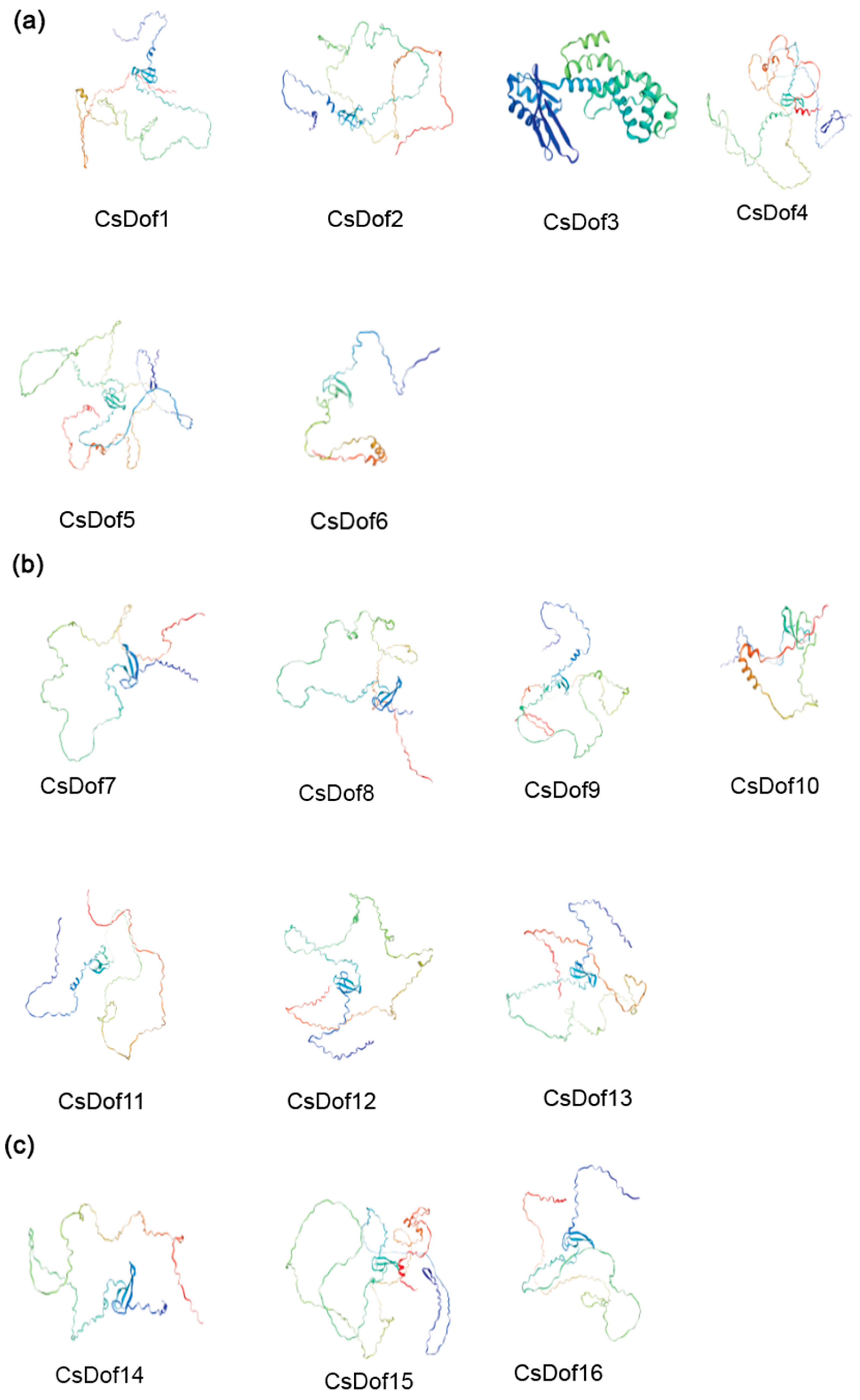
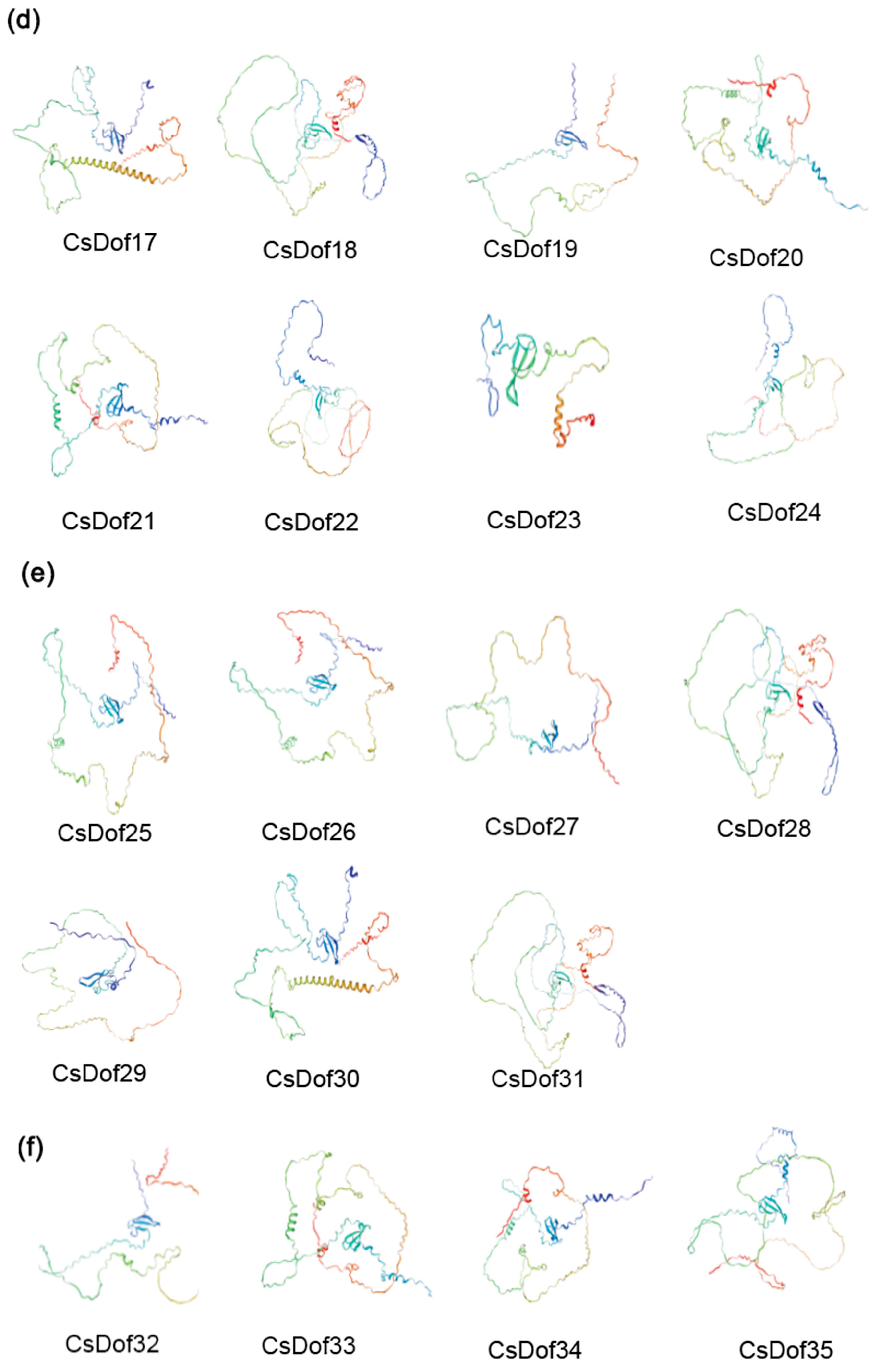
2.8. Predicted Protein Secondary and Tertiary Structures of the CsDOF Genes
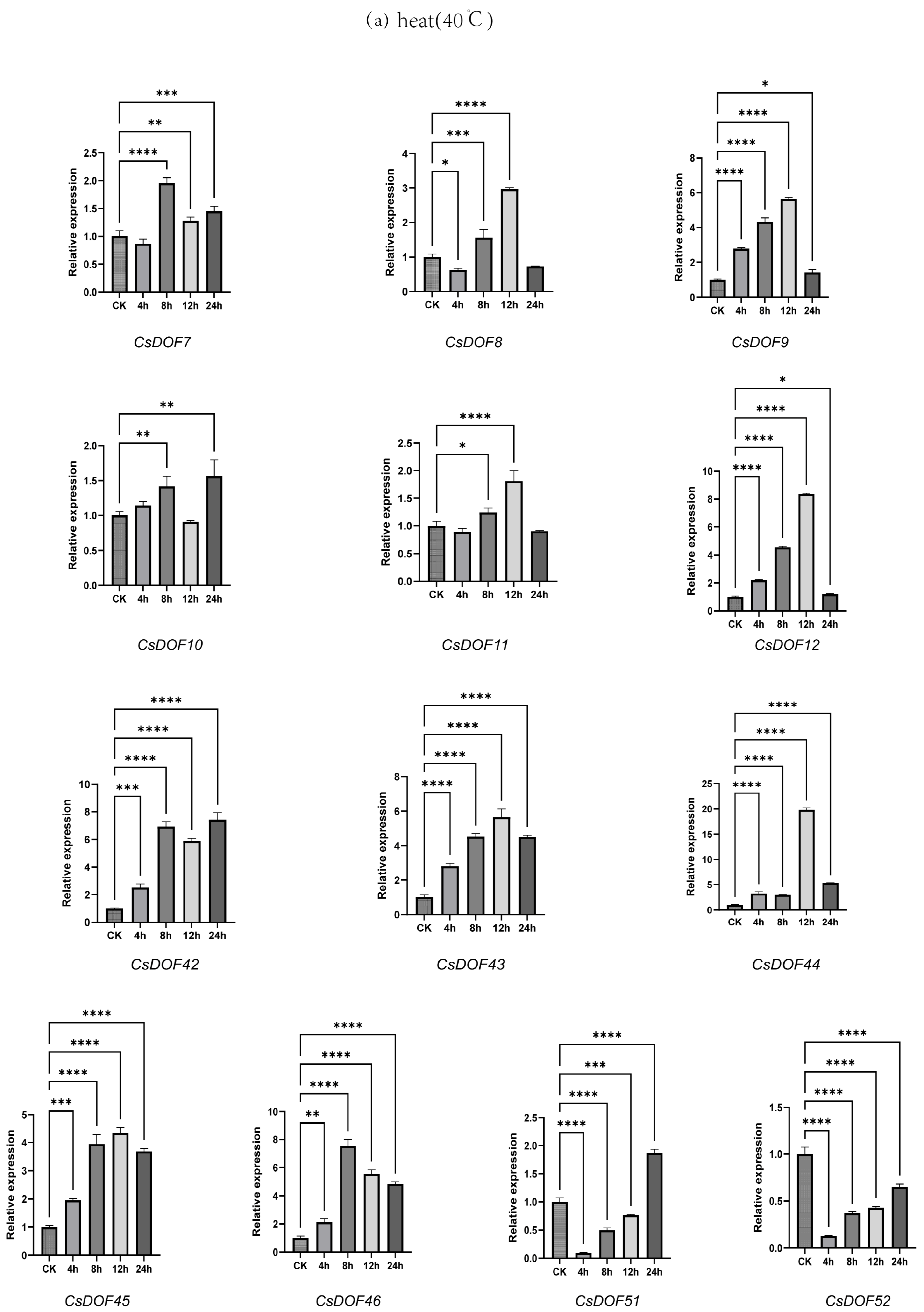
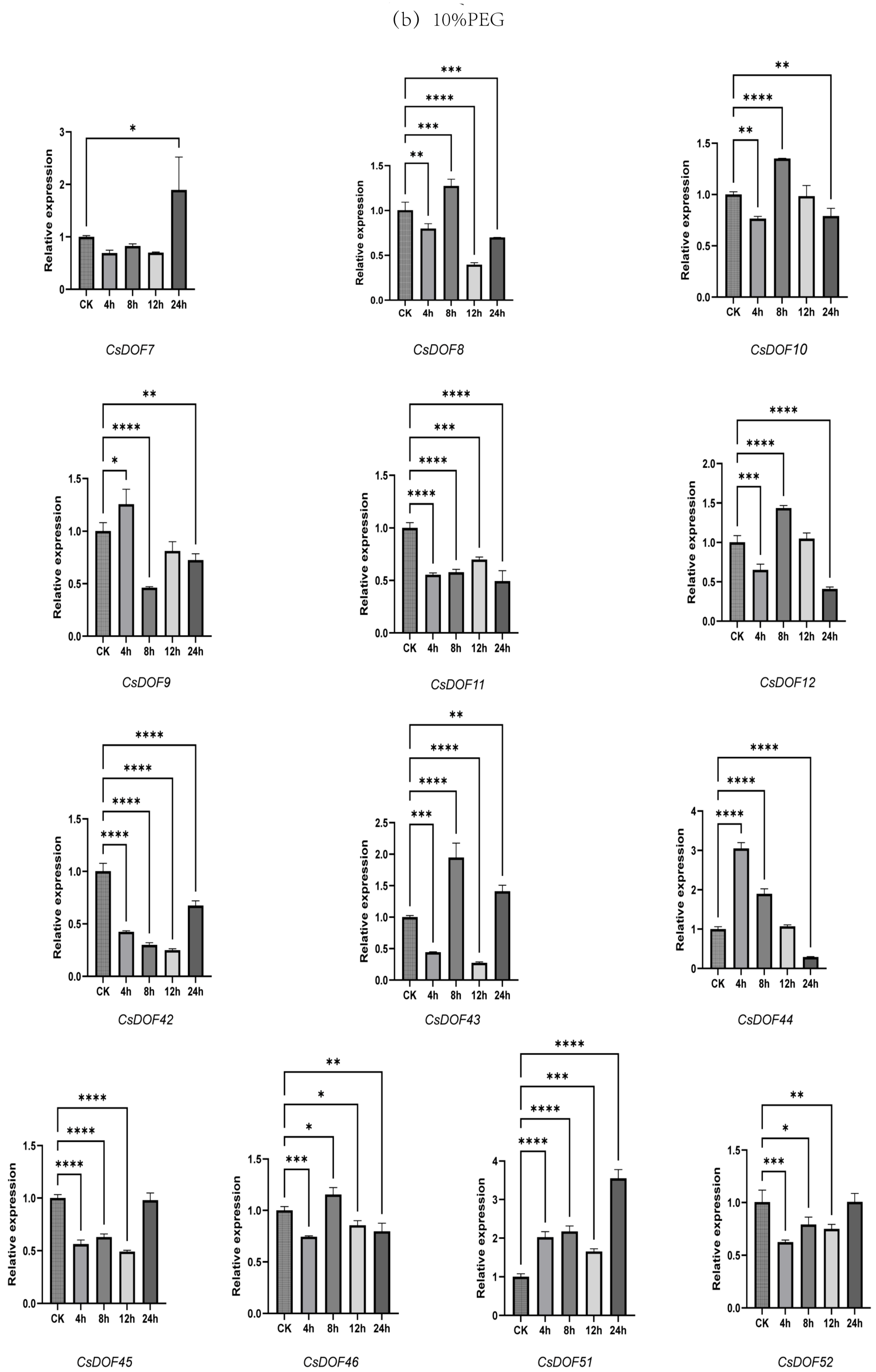
2.9. Expression Analysis of DOF Family Genes in Tieguanyin Under Drought and Heat Treatments
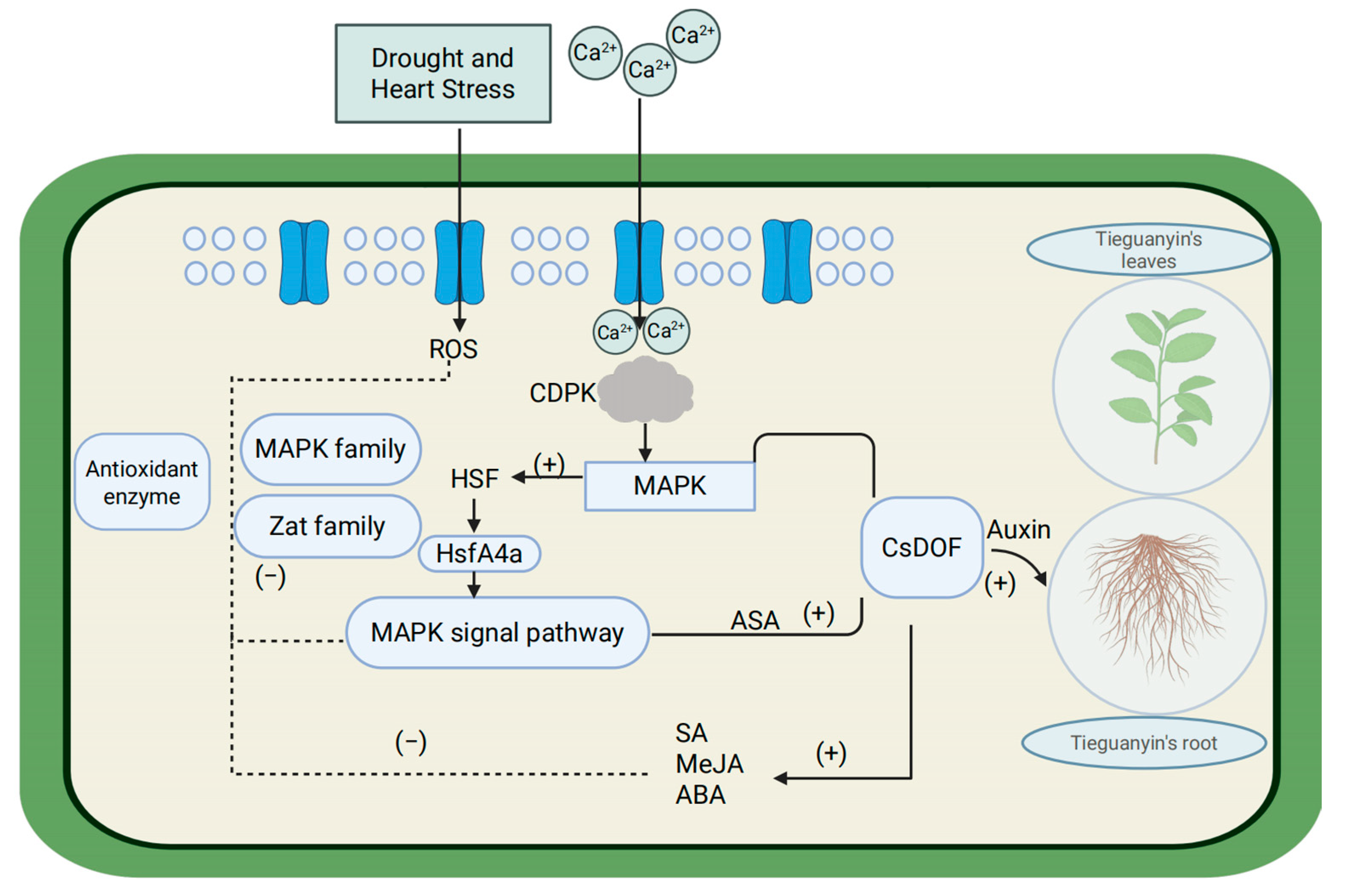
3. Discussion
4. Materials and Methods
4.1. Data Sources
4.2. Identification and Physicochemical Properties of DOF Gene Family
4.3. Analysis of Gene Structure and Conserved Motifs
4.4. Chromosome Mapping and Collinearity Analysis
4.5. Phylogenetic Tree Construction
4.6. Promoter Analysis
4.7. Expression Pattern Analysis (Heat Map Construction)
4.8. Protein Structure Prediction
4.9. Sources of Plant Materials and Abiotic Stress Treatments
4.10. RNA Extraction and qRT-PCR Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cai, C.; Wang, W.; Ye, S.; Zhang, Z.; Ding, W.; Xiang, M.; Wu, C.; Zhu, Q. Overexpression of a novel arabidopsis gene SUPA leads to various morphological and abiotic stress tolerance alternations in Arabidopsis and Poplar. Front. Plant Sci. 2020, 11, 560985. [Google Scholar] [CrossRef]
- Gupta, B.; Shrestha, J. Abiotic stress adaptation and tolerance mechanisms in crop plants. Front. Plant Sci. 2023, 14, 1278895. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.L. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Orphanides, G.; Reinberg, D. A unified theory of gene expression. Cell 2002, 108, 439–451. [Google Scholar] [CrossRef]
- Song, L.; Li, W.; Chen, X. Transcription factor is not just a transcription factor. Trends Plant Sci. 2022, 27, 1087–1089. [Google Scholar] [CrossRef]
- Wang, Q.; Zhu, Z. Transcription factors in the regulation of plant heat responses. Crit. Rev. Plant Sci. 2023, 42, 385–398. [Google Scholar] [CrossRef]
- Wang, X.; Song, Q.; Liu, Y.; Brestic, M.; Yang, X. The network centered on ICEs play roles in plant cold tolerance, growth and development. Planta 2022, 255, 81. [Google Scholar] [CrossRef]
- Wei, H.; Wang, X.; Wang, K.; Tang, X.; Zhang, N.; Si, H. Transcription factors as molecular switches regulating plant responses to drought stress. Physiol. Plant. 2024, 176, e14366. [Google Scholar] [CrossRef]
- Price, L.; Han, Y.; Angessa, T.; Li, C. Molecular pathways of WRKY genes in regulating plant salinity tolerance. Int. J. Mol. Sci. 2022, 23, 10947. [Google Scholar] [CrossRef]
- Liu, C.; Wen, L.; Cui, Y.; Ahammed, G.; Cheng, Y. Metal transport proteins and transcription factor networks in plant responses to cadmium stress. Plant Cell Rep. 2024, 43, 218. [Google Scholar] [CrossRef]
- Qian, Y.; Zhang, T.; Yu, Y.; Gou, L.; Yang, J.; Xu, J.; Pi, E. Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses. Front. Plant Sci. 2021, 12, 677611. [Google Scholar] [CrossRef]
- Jia, H.; Suzuki, M.; McCarty, D.R. Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Wiley Interdiscip. Rev.-Dev. Biol. 2014, 3, 135–145. [Google Scholar] [CrossRef]
- Ballester, P.; Martinez-Godoy, M.A.; Ezquerro, M.; Navarrete-Gomez, M.; Trigueros, M.; Rodriguez-Concepcion, M.; Ferrandiz, C. A transcriptional complex of NGATHA and bHLH transcription factors directs stigma development in Arabidopsis. Plant Cell 2021, 33, 3645–3657. [Google Scholar] [CrossRef]
- Gambhir, P.; Raghuvanshi, U.; Kumar, R.; Sharma, A.K. Transcriptional regulation of tomato fruit ripening. Physiol. Mol. Biol. Plants 2024, 30, 289–303. [Google Scholar] [CrossRef]
- Gupta, S.; Malviya, N.; Kushwaha, H.; Nasim, J.; Bisht, N.C.; Singh, V.K.; Yadav, D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 2015, 241, 549–562. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. Cell Mol. Biol. 1999, 17, 209–214. [Google Scholar] [CrossRef]
- Zou, X.; Sun, H. DOF transcription factors: Specific regulators of plant biological processes. Front. Plant Sci. 2023, 45, 386–391. [Google Scholar] [CrossRef]
- Zou, Z.; Zhu, J.; Zhang, X. Genome-wide identification and characterization of the Dof gene family in cassava (Manihot esculenta). Gene 2019, 687, 298–307. [Google Scholar] [CrossRef]
- Yanagisawa, S. The transcriptional activation domain of the plant-specific Dof1 factor functions in plant, animal, and yeast cells. Plant Cell Physiol. 2001, 42, 813–822. [Google Scholar] [CrossRef]
- Ruta, V.; Longo, C.; Lepri, A.; De Angelis, V.; Occhigrossi, S.; Costantino, P.; Vittorioso, P. The Dof transcription factors in seed and seedling development. Plants 2020, 9, 218. [Google Scholar] [CrossRef]
- Le Hir, R.; Bellini, C. The plant-specific Dof transcription factors family: New players involved in vascular system development and functioning in Arabidopsis. Front. Plant Sci. 2013, 4, 164. [Google Scholar] [CrossRef]
- da Silva, D.C.; Falavigna, V.S.; Fasoli, M.; Buffon, V.; Porto, D.D.; Pappas, G.J.; Pezzotti, M.; Pasquali, G.; Revers, L.F. Transcriptome analyses of the Dof-like gene family in grapevine reveal its involvement in berry, flower and seed development. Hortic. Res. 2016, 3, 16042. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, P.; Wang, W.; Kong, L.; Tian, S.; Qin, G. Genome-wide binding analysis of the tomato transcription factor SlDof1 reveals its regulatory impacts on fruit ripening. Mol. Hortic. 2021, 1, 9. [Google Scholar] [CrossRef]
- Yu, H.; Ma, Y.; Lu, Y.; Yue, J.; Ming, R. Expression profiling of the Dof gene family under abiotic stresses in spinach. Sci. Rep. 2021, 11, 14429. [Google Scholar] [CrossRef]
- Lucas-Reina, E.; Romero-Campero, F.J.; Romero, J.M.; Valverde, F. An evolutionarily conserved Dof-constans module controls plant photoperiodic signaling. Plant Physiol. 2015, 168, 561. [Google Scholar] [CrossRef]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Wang, S.; Bai, Y.; Li, P.; Yang, L.; Wang, X. Genome-wide identification and expression analysis of the Dof (DNA binding with one finger) protein family in monocot and dicot species. Physiol. Mol. Plant Pathol. 2019, 108, 101431. [Google Scholar] [CrossRef]
- Vicente-Carbajosa, J.; Moose, S.P.; Parsons, R.L.; Schmidt, R.J. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. USA 1997, 94, 7685–7690. [Google Scholar] [CrossRef]
- Yue, Y.; Du, J.; Li, Y.; Thomas, H.R.; Frank, M.H.; Wang, L.; Hu, H. Insight into the petunia Dof transcription factor family reveals a new regulator of male-sterility. Ind. Crops Prod. 2021, 161, 113196. [Google Scholar] [CrossRef]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality30. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef]
- He, C.; Zhou, J.; Li, Y.; Zhang, D.; Ntezimana, B.; Zhu, J.; Wang, X.; Xu, W.; Wen, X.; Chen, Y.; et al. The aroma characteristics of oolong tea are jointly determined by processing mode and tea cultivars. Food Chem.-X 2023, 18, 100730. [Google Scholar] [CrossRef]
- Lou, W.; Sun, K.; Zhao, Y.; Deng, S.; Zhou, Z. Impact of climate change on inter-annual variation in tea plant output in Zhejiang, China. Int. J. Climatol. 2021, 41, E479–E490. [Google Scholar] [CrossRef]
- Kang, H.; Zhou, H.; Ye, Y.; Yang, J.; Liu, Z.; He, P.; Li, B.; Wu, Y.; Wang, Y.; Tu, Y. Tieguanyin oolong tea extracts alleviate behavioral abnormalities by modulating neuroinflammation in App/Ps1 mouse model of alzheimer’s disease. Foods 2022, 11, 81. [Google Scholar] [CrossRef]
- Wang, Y.; Kong, D.; Gao, Y.; Ying, L.; Huang, Q.; Xu, P. Chemical characterization and bioactivity of phenolics from Tieguanyin oolong tea. J. Food Biochem. 2019, 43, e12894. [Google Scholar] [CrossRef]
- Zheng, C.; Wang, Y.; Ding, Z.; Zhao, L. Global transcriptional analysis reveals the complex relationship between tea quality, leaf senescence and the responses to cold-drought combined stress in Camellia sinensis. Front. Plant Sci. 2016, 7, 1858. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, C.; Deng, W. CsICE1 and CsCBF1: Two transcription factors involved in cold responses in Camellia sinensis. Plant Cell Rep. 2012, 31, 27–34. [Google Scholar] [CrossRef]
- Das, A.; Das, S.; Mondal, T.K. Identification of differentially expressed gene profiles in young roots of tea [Camellia sinensis (L.) O. Kuntze] subjected to drought stress using suppression subtractive hybridization. Plant Mol. Biol. Report. 2012, 30, 1088–1101. [Google Scholar] [CrossRef]
- Chen, M. The tea plant leaf cuticle: From plant protection to tea quality. Front. Plant Sci. 2021, 12, 751547. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, H.; Mischke, S.; Meinhardt, L.W.; Zhang, D.; Zhu, X.; Li, X.; Fang, W. Exogenous abscisic acid significantly affects proteome in tea plant (Camellia sinensis) exposed to drought stress. Hortic. Res. 2014, 1, 14029. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y.; Sun, K.; Chen, Y.; Chen, X.; Li, X. Exogenous melatonin enhances cold, salt and drought stress tolerance by improving antioxidant defense in tea plant (Camellia sinensis (L.) O. Kuntze). Molecules 2019, 24, 1826. [Google Scholar] [CrossRef]
- Upadhyaya, H.; Panda, S.K. Abiotic stress responses in tea [Camellia sinensis L (O) Kuntze]: An overview. Rev. Agric. Sci. 2013, 1, 1–10. [Google Scholar] [CrossRef]
- Nalina, M.; Saroja, S.; Rajkumar, R.; Radhakrishnan, B.; Chandrashekara, K.N. Variations in quality constituents of green tea leaves in response to drought stress under south Indian condition. Sci. Hortic. 2018, 233, 359–369. [Google Scholar]
- Rahimi, M.; Kordrostami, M.; Mortezavi, M. Evaluation of tea (Camellia sinensis L.) biochemical traits in normal and drought stress conditions to identify drought tolerant clones. Physiol. Mol. Biol. Plants 2019, 25, 56–69. [Google Scholar] [CrossRef]
- Cao, L.; Ye, F.; Fahim, A.M.; Ma, C.; Pang, Y.; Zhang, X.; Zhang, Q.; Lu, X. Transcription factor ZmDof22 enhances drought tolerance by regulating stomatal movement and antioxidant enzymes activities in maize (Zea mays L.). Theor. Appl. Genet. 2024, 137, 132. [Google Scholar] [CrossRef]
- Zhao, C.; Bai, H.; Li, C.; Pang, Z.; Xuan, L.; Lv, D.; Niu, S. Genome-wide identification of the Dof gene family in kiwifruit (Actinidia chinensis) and functional validation of Acdof22 in response to drought stress. Int. J. Mol. Sci. 2024, 25, 9103. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Tong, Q.; Xu, G.; Xu, M.; Li, H.; Fan, P.; Li, S.; Liang, Z. Transcriptomic analysis of grapevine Dof transcription factor gene family in response to cold stress and functional analyses of the VaDof17d gene. Planta 2021, 253, 55. [Google Scholar] [CrossRef]
- He, X.; Zhang, M.; Huang, Y.; Yu, J.; Zhao, X.; Zheng, Q.; Liu, Z.; Lan, S. Genome-Based identification of the Dof gene family in three cymbidium species and their responses to heat stress in Cymbidium goeringii. Int. J. Mol. Sci. 2024, 25, 7662. [Google Scholar] [CrossRef]
- Chen, G.; Xu, Y.; Gui, J.; Huang, Y.; Ma, F.; Wu, W.; Han, T.; Qiu, W.; Yang, L.; Song, S. Characterization of Dof transcription factors and the heat-tolerant function of PeDof-11 in passion fruit (Passiflora edulis). Int. J. Mol. Sci. 2023, 24, 12091. [Google Scholar] [CrossRef]
- Gou, C.; Li, J.; Chen, B.; Cheng, G.; Zheng, Z.; Peng, H.; El—Sappah, A.H. Genome wide identification of Dof transcription factors in Carmine radish reveals RsDof33 role in cadmium stress and anthocyanin biosynthesis. Sci. Rep. 2025, 15, 4766. [Google Scholar] [CrossRef]
- Cao, X.; Wan, W.; Mao, H.; Yin, D.; Deng, X.; Yan, H.; Ren, L. Genome-Wide identification and expression analysis of Dof transcription factors in Lotus (Nelumbo nucifera Gaertn.). Plants 2022, 11, 2057. [Google Scholar] [CrossRef]
- Xiang, L.; Liu, C.; Luo, J.; He, L.; Deng, Y.; Yuan, J.; Wu, C.; Cai, Y. A tuber mustard AP2/ERF transcription factor gene, BjABR1, functioning in abscisic acid and abiotic stress responses, and evolutionary trajectory of the ABR1 homologous genes in Brassica species. PeerJ 2018, 6, e6071. [Google Scholar] [CrossRef]
- Mariyam Shafiq, M.; Haseeb, M.; Atif, R.M.; Naqvi, S.A.A.A.A.; Ali, N.; Javed, M.A.; Gillani, F.; Haider, M.S. Genome-wide identification and characterization of a plant-specific Dof transcription factor gene family in olive (Olea europaea) and its comparison with Arabidopsis. Hortic. Environ. Biotechnol. 2021, 62, 949–968. [Google Scholar] [CrossRef]
- Negi, J.; Moriwaki, K.; Konishi, M.; Yokoyama, R.; Nakano, T.; Kusumi, K.; Hashimoto—Sugimoto, M.; Schroeder, J.I.; Nishitani KYanagisawa, S.; Iba, K. Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 2013, 23, 479–484. [Google Scholar] [CrossRef]
- Mu, Z.; Xu, M.; Manda, T.; Chen, J.; Yang, L.; Hwarari, D. Characterization, evolution, and abiotic stress responses of leucine-rich repeat receptor-like protein kinases (LRR-RLK) in Liriodendron chinense. BMC Genom. 2024, 25, 748. [Google Scholar] [CrossRef]
- Vila-Aiub, M.M.; Goh, S.S.; Gaines, T.A.; Han, H.P.; Busi, R.; Yu, Q.; Powles, S.B. No fitness cost of glyphosate resistance endowed by massive EPSPS gene amplification in Amaranthus palmeri. Planta 2014, 239, 793–801. [Google Scholar] [CrossRef]
- Cregut, D.; Civera, C.; Macias, M.J.; Serrano, L. A tale of two secondary structure elements: When a beta-hairpin becomes an alpha-helix. J. Mol. Biol. 1999, 292, 389–401. [Google Scholar] [CrossRef]
- Hewage, K.A.H.; Yang, J.; Wang, D.; Hao, G.; Yang, G.; Zhu, J. Chemical manipulation of abscisic acid signaling: A new approach to abiotic and biotic stress management in agriculture. Adv. Sci. 2020, 7, 2001265. [Google Scholar] [CrossRef]
- Bao, A.; Zhao, Z.; Ding, G.; Shi, L.; Xu, F.; Cai, H. The stable level of glutamine synthetase 2 plays an important role in rice growth and in carbon-nitrogen metabolic balance. Int. J. Mol. Sci. 2015, 16, 12713–12736. [Google Scholar] [CrossRef]
- Ji, Y.; Li, Q.; Liu, G.; Selvaraj, G.; Zheng, Z.; Zou, J.; Wei, Y. Roles of cytosolic glutamine synthetases in Arabidopsis development and stress responses. Plant Cell Physiol. 2019, 60, 657–671. [Google Scholar] [CrossRef]
- Wang, J.; Chen, W.; Wang, H.; Li, Y.; Wang, B.; Zhang, L.; Wan, X.; Li, M. Transcription factor CsDOF regulates glutamine metabolism in tea plants (Camellia sinensis). Plant Sci. 2021, 302, 110720. [Google Scholar] [CrossRef]
- Liu, M.; Jiao, Z.; Lou, H.; Tang, D.; Tian, X.; Zhou, B.; Ruan, J.; Fernie, A.R.; Zhang, Q. A glutamine synthetase-Dof transcription factor module regulates nitrogen remobilization from source to sink tissues in tea plants. Plant Physiol. 2024, 197, kiae644. [Google Scholar] [CrossRef]
- Xia, E.; Tong, W.; Wu, Q.; Wei, S.; Zhao, J.; Zhang, Z.; Wei, C.; Wan, X. Tea plant genomics: Achievements, challenges and perspectives. Hortic. Res. 2020, 7, 7. [Google Scholar] [CrossRef]
- Waldvogel AMFeldmeyer, B.; Rolshausen, G.; Exposito-Alonso, M.; Rellstab, C.; Kofler, R.; Mock, T.; Schmid, K.; Schmitt, I.; Bataillon, T.; Savolainen, O.; et al. Evolutionary genomics can improve prediction of species’ responses to climate change. Evol. Lett. 2020, 4, 4–18. [Google Scholar] [CrossRef]
- Xia, E.H.; Tong, W.; Hou, Y.; An, Y.; Chen, L.; Wu, Q.; Liu, Y.; Yu, J.; Li, F.; Li, R.; et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol. Plant 2020, 13, 1013–1026. [Google Scholar] [CrossRef]
- Storz, J.F.; Opazo, J.C.; Hoffmann, F.G. Gene duplication, genome duplication, and the functional diversification of vertebrate globins. Mol. Phylogenetics Evol. 2013, 66, 469–478. [Google Scholar] [CrossRef]
- Yu, Q.; Li, C.; Zhang, J.; Tian, Y.; Wang, H.; Zhang, Y.; Zhang, Z.; Xiang, Q.; Han, X.; Zhang, L. Genome-wide identification and expression analysis of the Dof gene family under drought stress in tea (Camellia sinensis). PeerJ 2020, 8, e9269. [Google Scholar] [CrossRef]
- Li, H.; Huang, W.; Liu, Z.; Wang, Y.; Zhuang, J. Transcriptome-based analysis of Dof family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis). Int. J. Genom. 2016, 2016, 5614142. [Google Scholar] [CrossRef]
- Lijavetzky, D.; Carbonero, P.; Vicente-Carbajosa, J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003, 3, 17. [Google Scholar] [CrossRef]
- Cai, K.; Xie, X.; Han, L.; Chen, J.; Zhang, J.; Yuan, H.; Shen, J.; Ren, Y.; Zhao, X. Identification and functional analysis of the DOF gene family in Populus simonii: Implications for development and stress response. Front. Plant Sci. 2024, 15, 1412175. [Google Scholar] [CrossRef]
- Liu, H.; Lyu, H.M.; Zhu, K.; Van de Peer, Y.; Cheng, Z. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 2021, 105, 1072–1082. [Google Scholar] [CrossRef]
- Movahedi, S.; Van de Peer, Y.; Vandepoele, K. Comparative network analysis reveals that tissue specificity and gene function are important factors influencing the mode of expression evolution in Arabidopsis and rice. Plant J. 2011, 156, 1316–1330. [Google Scholar] [CrossRef]
- Boccaccini, A.; Lorrai, R.; Ruta, V.; Frey, A.; Mercey-Boutet, S.; Marion-Poll, A.; Tarkowská, D.; Strnad, M.; Costantino, P.; Vittorioso, P. The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels. BMC Plant Biol. 2016, 16, 198. [Google Scholar] [CrossRef]
- Ge, A.H.; Wang, E. Exploring the plant microbiome: A pathway to climate—Smart crops. Cell 2025, 188, 1469–1485. [Google Scholar] [CrossRef]
- Jiao, Z.; Tang, D.; Fan, K.; Zhang, Q.; Liu, M.; Ruan, J. Genome—Wide identification of the DNA—Binding one zinc finger (Dof) transcription factor gene family and their potential functioning in nitrogen remobilization in tea plant (Camellia sinensis L.). Sci. Hortic. 2022, 292, 110615. [Google Scholar] [CrossRef]
- Qin, L.; Zhang, Y.; Wu, X.; Sheng, J.; Ma, H.; Geng, Y.; Wang, Y. Onion AcCDF4 ectopia overexpression regulates flowering and abiotic stress response in Arabidopsis. Eur. J. Hortic. Sci. 2023, 88, 1–12. [Google Scholar] [CrossRef]
- Corrales, A.R.; Nebauer, S.G.; Carrillo, L.; Fernandez-Nohales, P.; Marques, J.; Renau-Morata, B.; Granell, A.; Pollmann, S.; Vicente-Carbajosa, J.; Molina, R.; et al. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 2014, 65, 995–1012. [Google Scholar] [CrossRef]
- Chen, M.; Liu, X.; Huan, L.; Sun, M.; Liu, L.; Chen, X.; Gao, D.; Li, L. Genome-wide analysis of Dof family genes and their expression during bud dormancy in peach (Prunus persica). Sci. Hortic. 2017, 214, 18–26. [Google Scholar] [CrossRef]
- Aloo, B.N.; Dessureault-Rompre, J.; Tripathi, V.; Nyongesa, B.O.; Were, B.A. Signaling and crosstalk of rhizobacterial and plant hormones that mediate abiotic stress tolerance in plants. Front. Microbiol. 2023, 14, 1171104. [Google Scholar] [CrossRef]
- Seminario, A.; Song, L.; Zulet, A.; Nguyen, H.T.; Gonzalez, E.M.; Larrainzar, E. Drought stress causes a reduction in the biosynthesis of ascorbic acid in soybean plants. Front. Plant Sci. 2017, 8, 1042. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald Eduardo Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Lugassi, N.; Yadav, B.S.; Egbaria, A.; Wolf, D.; Kelly, G.; Neuhaus, E.; Raveh, E.; Carmi, N.; Granot, D. Expression of Arabidopsis hexokinase in tobacco guard cells increases water-use efficiency and confers tolerance to drought and salt stress. Plants 2019, 8, 613. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhu, L.; Wang, Q.; Hou, X. Autophagy-Related2 regulates chlorophyll degradation under abiotic stress conditions in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 4515. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef]
- Shi, X.; Bao, J.; Lu, X.; Ma, L.; Zhao, Y.; Lan, S.; Cao, J.; Ma, S.; Li, S. The mechanism of Ca2+ signal transduction in plants responding to abiotic stresses. Environ. Exp. Bot. 2023, 216, 105514. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Jin, L.; Peng, R. Crosstalk between Ca2+ and other regulators assists plants in responding to abiotic stress. Plants 2022, 11, 1351. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, V.; Örvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2002, 31, 629–638. [Google Scholar] [CrossRef]
- Wang, G.; Wei, P.; Tan, F.; Yu, M.; Zhang, X.; Chen, Q.; Wang, X. The transcription factor AtDOF4.7 is involved in ethylene- and IDA-mediated organ abscission in Arabidopsis. Front. Plant Sci. 2016, 7, 863. [Google Scholar] [CrossRef]
- Qu, A.; Ding, Y.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 903–930. [Google Scholar] [CrossRef]
- Kim, Y.; Chung, Y.; Lee, E.; Tripathi, P.; Heo, S.; Kim, K.H. Root response to drought stress in rice (Oryza sativa L.). Int. J. Mol. Sci. 2020, 21, 1513. [Google Scholar] [CrossRef]
- Ma, X.; Zhou, G.; Li, G.; Wang, Q. Quantitative evaluation of the trade-off growth strategies of maize leaves under different drought severities. Water 2021, 13, 1852. [Google Scholar] [CrossRef]
- Yang, X.; Lu, M.; Wang, Y. Response mechanism of plants to drought stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Li, M.; Zhang, C.; Hou, L.; Liu, X.; Zhao, H.; Pang, X.; Bo, W.; Li, Y. Differences in leaf cuticular wax induced by whole-genome duplication in autotetraploid sour jujube. Hortic. Plant J. 2024, 10, 66–76. [Google Scholar] [CrossRef]
- Rhaman, M.S.; Imran, S.; Rauf, F.; Khatun, M.; Baskin, C.C.; Murata, Y.; Hasanuzzaman, M. Seed priming with phytohormones: An effective approach for the mitigation of abiotic stress. Plants 2021, 10, 37. [Google Scholar] [CrossRef]
- Per, T.S.; Khan, M.I.R.; Anjum, N.A.; Masood, A.; Hussain, S.J.; Khan, N.A. Jasmonates in plants under abiotic stresses: Crosstalk with other phytohormones matters. Environ. Exp. Bot. 2018, 145, 104–120. [Google Scholar] [CrossRef]
- Li, N.; Euring, D.; Cha, J.Y.; Lin, Z.; Lu, M.; Huang, L.; Kim, W.Y. Plant hormone-mediated regulation of heat tolerance in response to global climate change. Front. Plant Sci. 2021, 11, 627969. [Google Scholar] [CrossRef]
- Salvi, P.; Manna, M.; Kaur, H.; Thakur, T.; Gandass, N.; Bhatt, D.; Muthamilarasan, M. Phytohormone signaling and crosstalk in regulating drought stress response in plants. Plant Cell Rep. 2021, 40, 1305–1329. [Google Scholar] [CrossRef]
- Altaf, M.A.; Shahid, R.; Kumar, R.; Altaf, M.M.; Kumar, A.; Khan, L.U.; Saqib, M.; Nawaz, M.A.; Saddiq, B.; Bahadur, S.; et al. Phytohormones mediated modulation of abiotic stress tolerance and potential crosstalk in horticultural crops. J. Plant Growth Regul. 2022, 42, 4724–4750. [Google Scholar] [CrossRef]
- Li, H.; Liu, Z.; Wu, Z.; Wang, Y.; Teng, R.; Zhuang, J. Differentially expressed protein and gene analysis revealed the effects of temperature on changes in ascorbic acid metabolism in harvested tea leaves. Hortic. Res. 2018, 5, 65. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, C.; Shu, W.; Ye, Z.; Li, H.; Zhang, Y. The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem. Biophys. Res. Commun. 2016, 474, 736–741. [Google Scholar] [CrossRef]


| Gene Accession | Gene ID | Size/aa | MW /Da | Theoretical pI | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|
| CsTGY15G0001562a | CsDOF1 | 298 | 32,584.53 | 6.58 | 52.30 | 47.55 | −0.804 |
| CsTGY15G0001562b | CsDOF2 | 292 | 32,094.00 | 6.58 | 50.88 | 48.84 | −0.824 |
| CsTGY02G0001308a | CsDOF3 | 322 | 35,039.34 | 7.71 | 48.58 | 52.76 | −0.754 |
| CsTGY15G0000298b | CsDOF4 | 225 | 24,360.63 | 8.61 | 50.88 | 44.71 | −0.829 |
| CsTGY02G0002212b | CsDOF5 | 184 | 20,429.35 | 8.28 | 55.52 | 32.39 | −1.091 |
| CsTGY13G0002427a | CsDOF6 | 333 | 35,723.59 | 9.21 | 62.62 | 66.79 | −0.602 |
| CsTGY04G0002458a | CsDOF7 | 325 | 34,441.49 | 9.20 | 53.28 | 58.18 | −0.517 |
| CsTGY11G0000951b | CsDOF8 | 316 | 33,927.58 | 9.15 | 62.64 | 50.32 | −0.672 |
| CsTGY01G0000579a | CsDOF9 | 349 | 36,960.20 | 9.24 | 55.70 | 57.56 | −0.516 |
| CsTGY04G0003389a | CsDOF10 | 345 | 36,654.78 | 8.93 | 49.35 | 62.43 | −0.488 |
| CsTGY04G0003066a | CsDOF11 | 342 | 36,222.31 | 9.20 | 52.63 | 58.16 | −0.554 |
| CsTGY10G0001612b | CsDOF12 | 341 | 35,505.43 | 9.18 | 59.77 | 50.67 | −0.577 |
| CsTGY10G0001612a | CsDOF13 | 343 | 35,712.61 | 9.18 | 59.70 | 50.93 | −0.573 |
| CsTGY01G0002022b | CsDOF14 | 340 | 36,828.13 | 8.72 | 52.56 | 57.41 | −0.551 |
| CsTGY08G0000732a | CsDOF15 | 328 | 36,331.21 | 8.62 | 51.05 | 56.13 | −0.772 |
| CsTGY08G0000732b | CsDOF16 | 329 | 36,432.32 | 8.62 | 52.10 | 55.96 | −0.772 |
| CsTGY11G0000819b | CsDOF17 | 374 | 40,329.96 | 7.25 | 45.15 | 61.52 | −0.533 |
| CsTGY11G0000819a | CsDOF18 | 306 | 33,187.77 | 6.78 | 46.07 | 50.95 | −0.720 |
| CsTGY04G0002605b | CsDOF19 | 275 | 30,933.40 | 7.29 | 47.37 | 48.55 | −0.878 |
| CsTGY15G0001184a | CsDOF20 | 321 | 34,988.04 | 6.48 | 45.51 | 49.16 | −0.668 |
| CsTGY15G0001184b | CsDOF21 | 321 | 34,961.01 | 6.48 | 47.95 | 49.16 | −0.660 |
| CsTGY06G0002442a | CsDOF22 | 307 | 34,410.47 | 6.10 | 48.78 | 49.87 | −0.738 |
| CsTGY04G0001690a | CsDOF23 | 309 | 34,596.60 | 6.14 | 44.27 | 50.45 | −0.768 |
| CsTGY04G0001690b | CsDOF24 | 309 | 34,582.57 | 6.14 | 43.56 | 50.13 | −0.769 |
| CsTGY14G0001635b | CsDOF25 | 294 | 32,015.60 | 8.11 | 55.97 | 60.31 | −0.594 |
| CsTGY14G0001635a | CsDOF26 | 290 | 31,758.36 | 7.56 | 55.85 | 61.14 | −0.596 |
| CsTGY12G0002088a | CsDOF27 | 268 | 29,609.09 | 8.38 | 62.48 | 55.60 | −0.708 |
| CsTGY12G0002088b | CsDOF28 | 268 | 29,635.17 | 8.38 | 61.04 | 57.05 | −0.691 |
| CsTGY07G0002456a | CsDOF29 | 260 | 28,623.71 | 9.19 | 58.67 | 44.69 | −0.855 |
| CsTGY06G0001769a | CsDOF30 | 281 | 30,751.20 | 8.98 | 44.05 | 55.84 | −0.686 |
| CsTGY14G0002493a | CsDOF31 | 393 | 44,607.07 | 9.15 | 39.90 | 79.87 | −0.338 |
| CsTGY04G0001110b | CsDOF32 | 247 | 27,173.19 | 4.90 | 53.90 | 63.93 | −0.526 |
| CsTGY04G0001110a | CsDOF33 | 247 | 27,120.14 | 4.81 | 52.50 | 63.93 | −0.498 |
| CsTGY09G0001901b | CsDOF34 | 247 | 27,871.06 | 4.84 | 54.69 | 61.94 | −0.644 |
| CsTGY09G0001901a | CsDOF35 | 247 | 27,872.04 | 4.77 | 54.19 | 61.94 | −0.644 |
| CsTGY08G0001892b | CsDOF36 | 309 | 34,364.48 | 9.26 | 73.70 | 37.31 | −1.201 |
| CsTGY08G0001892a | CsDOF37 | 310 | 34,482.58 | 9.15 | 72.87 | 37.19 | −1.204 |
| CsTGY08G0000731a | CsDOF38 | 255 | 28,315.20 | 9.55 | 58.67 | 44.00 | −1.046 |
| CsTGY08G0000731b | CsDOF39 | 254 | 28,121.08 | 9.61 | 59.89 | 47.64 | −1.009 |
| CsTGY08G0000666b | CsDOF40 | 254 | 27,562.42 | 8.13 | 60.72 | 45.35 | −0.78 |
| CsTGY08G0000666a | CsDOF41 | 256 | 27,794.64 | 8.18 | 58.71 | 45.39 | −0.784 |
| CsTGY08G0001840b | CsDOF42 | 290 | 32,016.35 | 6.49 | 53.40 | 54.83 | −0.678 |
| CsTGY08G0001840a | CsDOF43 | 290 | 32,085.46 | 6.75 | 53.38 | 55.17 | −0.683 |
| CsTGY08G0002044b | CsDOF44 | 178 | 19,965.31 | 9.11 | 57.00 | 47.13 | −0.803 |
| CsTGY08G0000894b | CsDOF45 | 161 | 18,141.50 | 8.94 | 43.88 | 49.69 | −0.684 |
| CsTGY08G0000886b | CsDOF46 | 161 | 18,141.50 | 8.94 | 43.88 | 49.69 | −0.684 |
| CsTGY13G0000664a | CsDOF47 | 479 | 52,129.17 | 5.98 | 45.90 | 55.11 | −0.653 |
| CsTGY12G0000884b | CsDOF48 | 463 | 51,378.19 | 5.74 | 61.36 | 56.24 | −0.799 |
| CsTGY12G0000884a | CsDOF49 | 463 | 51,378.19 | 5.74 | 61.36 | 56.24 | −0.799 |
| CsTGY06G0000459a | CsDOF50 | 468 | 51,745.36 | 6.91 | 50.26 | 57.93 | −0.825 |
| CsTGY01G0003400a | CsDOF51 | 463 | 50,075.45 | 5.97 | 57.22 | 51.62 | −0.846 |
| CsTGY15G0001391a | CsDOF52 | 464 | 50,800.58 | 5.52 | 51.39 | 54.91 | −0.844 |
| CsTGY02G0001411b | CsDOF53 | 442 | 48,480.56 | 6.00 | 49.29 | 61.61 | −0.721 |
| CsTGY02G0001411a | CsDOF54 | 442 | 48,310.31 | 6.00 | 48.40 | 61.38 | −0.704 |
| CsTGY13G0002412b | CsDOF55 | 233 | 24,293.06 | 7.67 | 44.78 | 57.00 | −0.358 |
| CsTGY04G0000391a | CsDOF56 | 224 | 23,584.15 | 7.58 | 52.45 | 51.29 | −0.521 |
| CsTGY09G0002539b | CsDOF57 | 239 | 25,202.69 | 6.51 | 62.78 | 47.74 | −0.654 |
| CsTGY09G0002539a | CsDOF58 | 255 | 26,815.42 | 5.75 | 60.72 | 47.41 | −0.652 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, Y.; Tan, C.; Zhang, Y.; Wu, H.; Chen, D.; Yue, H.; Ding, Z.; Cao, S.; Zheng, K. Genome-Wide Characterization and Functional Analysis of CsDOF Transcription Factors in Camellia sinensis cv. Tieguanyin Under Combined Heat–Drought Stress. Plants 2025, 14, 1829. https://doi.org/10.3390/plants14121829
Wen Y, Tan C, Zhang Y, Wu H, Chen D, Yue H, Ding Z, Cao S, Zheng K. Genome-Wide Characterization and Functional Analysis of CsDOF Transcription Factors in Camellia sinensis cv. Tieguanyin Under Combined Heat–Drought Stress. Plants. 2025; 14(12):1829. https://doi.org/10.3390/plants14121829
Chicago/Turabian StyleWen, Yingxin, Cunyi Tan, Yujie Zhang, Hua Wu, Dian Chen, Heng Yue, Zekai Ding, Shijiang Cao, and Kehui Zheng. 2025. "Genome-Wide Characterization and Functional Analysis of CsDOF Transcription Factors in Camellia sinensis cv. Tieguanyin Under Combined Heat–Drought Stress" Plants 14, no. 12: 1829. https://doi.org/10.3390/plants14121829
APA StyleWen, Y., Tan, C., Zhang, Y., Wu, H., Chen, D., Yue, H., Ding, Z., Cao, S., & Zheng, K. (2025). Genome-Wide Characterization and Functional Analysis of CsDOF Transcription Factors in Camellia sinensis cv. Tieguanyin Under Combined Heat–Drought Stress. Plants, 14(12), 1829. https://doi.org/10.3390/plants14121829








