Abstract
The aquaporin (AQP) family, also called water channels or major intrinsic proteins, facilitate water transport. AQPs also transport low-molecular-weight solutes, including boric acid, glycerol, urea, and ammonia. Since plants are sessile, water homeostasis is crucial. Therefore, plants have developed diverse AQP variants at higher expression levels than animals. For example, 35 and 33 AQPs have been identified in Arabidopsis and rice, respectively. In the present study, we identified AQPs in morning glory (Ipomoea nil), which has been widely used as a model plant in research on flowering and floral morphology. The importance of AQPs in the opening of morning glory flowers has been reported. In the morning glory genome, 44 AQPs were identified, and their characteristics were analyzed. A phylogenetic analysis revealed five AQP subfamilies in morning glory: plasma membrane-intrinsic proteins (PIPs), tonoplast-intrinsic proteins (TIPs), nodulin 26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs), and X-intrinsic proteins (XIPs). Further, transport substrates of morning glory AQPs were estimated based on their homology to the known AQPs in other plant species and their corresponding amino acid motifs that possess permeability pores. It was expected that PIPs are likely to transport water, carbon dioxide, and hydrogen peroxide; TIPs are likely transport water, hydrogen peroxide, ammonia, urea, and boric acid; NIPs are likely transport water, boric acid, ammonia, glycerol, and formamide; and XIPs are likely to transport water, hydrogen peroxide, and glycerol. Overall, these results suggest that AQPs are involved in water and nutrient transport in Japanese morning glory. An in silico gene expression analysis suggested the importance of AQPs in flower opening, water or nutrient uptakes from the soil to roots, and photosynthesis in morning glory. Our findings provide fundamental information that enables further study into the importance of AQPs in morning glory, including their roles in flower opening and other physiological events.
1. Introduction
Water is essential for plant growth and various other physiological events, including photosynthesis. Plants maintain cellular functions and growth by absorbing water into cells by taking up water from the roots and transporting it to various organs. Moreover, water transport is important for the distribution of nutrients and photoassimilates in plants. Unlike animals, plants are unable to search for water. Therefore, they have developed mechanisms to maintain water homeostasis and adapted to different water conditions. In these processes, aquaporins (AQPs) play indispensable roles; consequently, plants have developed a large gene family of AQPs to adapt to various water conditions.
AQPs are membrane proteins that transport many water molecules with high selectivity. AQPs exhibit the fastest transport speed among various transporters, including ion channels, and they can transport over one billion water molecules per second under the appropriate conditions [1]. In addition to water, AQPs transport small neutral molecules, such as glycerol, urea, ammonia, carbon dioxide, hydrogen peroxide, and boric acid [2,3,4,5].
AQPs are composed of several conserved and characteristic structures, including six transmembrane domains (TMDs) and five connecting loops (loops A–E). Loop B and loop E form a half-helix, and inside each of the half-helices, two asparagine–proline–alanine (NPA) motifs are positioned facing each other (Figure 1). The NPA motifs form a pore with a diameter of 3 Å, through which water molecules permeate. The ar/R filter, comprising four amino acid residues, prevents the passage of molecules other than the substrate; therefore, this structure is important for substrate selectivity. Meanwhile, the Froger’s position, comprising five amino acid residues, has been postulated to selectively transport substrates other than water [6].
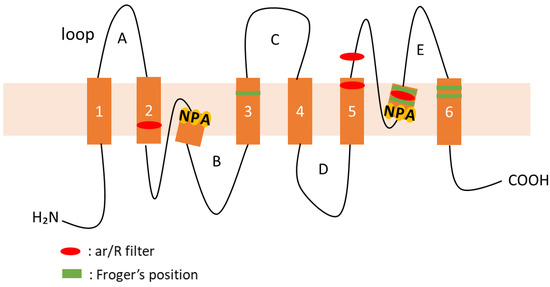
Figure 1.
Schematic representation of the classical structure of AQPs. An AQP monomer showing the six transmembrane helices (1–6) connected by two intracellular (loop B and D) and three extracellular (loop A, C, and E) loops. The conserved residues from the first selective filter (NPA motifs) and the second filter (ar/R filter) are shown in yellow and red, respectively. Froger’s positions are shown in green, respectively.
AQPs are universal in all organisms, including bacteria, plants, and animals [7,8,9]. Thirteen AQPs are present in humans [10], 35 in Arabidopsis [11], 33 in rice [12], and 47 in tomato [13]. AQP expression is higher in plants than in animals, indicating their importance and diverse roles in the former. As previously discussed, plants cannot move in the search for water. Therefore, they possess a highly developed water transport system with diverse AQP families. AQPs in higher plants are classified into five subfamilies based on homology: plasma membrane-intrinsic proteins (PIPs), tonoplast-intrinsic proteins (TIPs), nodulin 26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs), and X-intrinsic proteins (XIPs) [14,15].
Morning glory (Ipomoea nil) is cultivated as an ornamental plant in Japan. In addition, it is used as a model plant in flower research because of the high sensitivity of its flowering to day length, alongside the presence of various mutants of flower colors and the peculiar morphologies of flowers and other organs. These mutants are collected and distributed by the National BioResource Project (NBRP) morning glory (https://shigen.nig.ac.jp/asagao/index.jsp (accessed on 20 August 2021)). The whole genome of Japanese morning glory was sequenced by Hoshino et al. (2016) [16].
AQPs play pivotal roles in the opening and closing of the flowers of plants [17,18], including morning glory [1,19]. Therefore, in the present study, we performed a genome-wide analysis of AQPs in morning glory to predict their corresponding roles. First, the genes encoding AQPs were identified in the morning glory genome. Then, their gene and protein sequences predicted subcellular localization, predicted substrates, and then the gene expression profiles were annotated. Furthermore, based on this information, the putative roles of AQPs in morning glory were discussed.
2. Results
2.1. Genome-Wide Analysis of AQPs in Japanese Morning Glory
To identify AQPs in morning glory, BLASTP was used to search against the Japanese morning glory genome database. Specifically, a BLASTP search was conducted using the amino acid sequences of Arabidopsis and tomato AQPs [11,13] as queries. In total, 45 AQP candidates were identified (Table S1). The annotation of the best hit homolog in NCBI was AQP, and the numbers of amino acid residues in the candidates were similar to those in AQPs from other plant species (100–350 residues). An analysis of genes encoding the 45 AQP candidates revealed that INIL04g32909 and INIL04g32910 were identical to XM_019307143.1 in the Gnomon annotation (Figures S1 and S2). In addition, mapping of the RNA-Seq reads demonstrated that INIL04g32909 lacked RNA-Seq reads in most parts of the second exon; nonetheless, the reads of the coding region of INIL04g32909 still matched the coding region of XM_019307143.1 (Figure S1). Therefore, we concluded that INIL04g32909 and INIL04g32910 represented the same gene and used the Gnomon annotation XM_019307143.1 (InSIP1;1), which was implemented in further analyses. Finally, we identified 44 AQP genes in the Japanese morning glory genome (Table 1).
In a phylogenetic analysis based on the amino acid sequences of the candidate morning glory, Arabidopsis and tomato AQPs, the forty-four morning glory AQPs were classified into five families: thirteen PIPs, eleven TIPs, fifteen NIPs, two SIPs, and three XIPs (Figure 2 and Figure S3). These families were further classified into subfamilies, and the morning glory AQPs were named according to the order of the gene ID (chromosome number and position on the chromosome; Table 1, Figure 2). The InPIPs were classified into four InPIP1s and nine InPIP2s. The InTIPs were classified into six InTIP1s, two InTIP2s, one InTIP3, one InTIP4, and 1 one (Figure 2). The sequence of INIL03g01865 (InTIP3;1) contained an unknown sequence X, which was absent in the corresponding gene in the Gnomon annotation (XM_019346257.1). Furthermore, its RNA-Seq read matched the entire XM_019346257.1 sequence (Figure S4). Therefore, the sequence of XM_019346257 was used as the InTIP3;1 sequence in further analyses. InNIPs were divided into seven subfamilies: three InNIP1s, three InNIP2s, one InNIP3, three InNIP4s, three InNIP5s, one InNIP6, and one InNIP7 (Figure 2). A phylogenetic analysis indicated that INIL06g37831 (InNIP4;2) was highly similar to tomato SlNIP2;2 (Figure S3), although its ar/R filter, which is important for substrate selectivity, was similar to that of NIP4s (Table 2). Therefore, INIL06g37831 was classified as an InNIP4. INIL08g31098 (InNIP4;3) and INIL06g37646 (InNIP5;2) carried deletions in their amino acid sequences, and the second half of the AQP motif sequence was absent, which had no deletions; the AQP motif sequence was present in the corresponding Gnomon annotations (XM_019304577.1 and XM_019314787.1, respectively). Therefore, the sequences of XM_019304577.1 and XM_019314787.1 were used as InNIP4;3 and InNIP5;2, respectively, in further analyses.
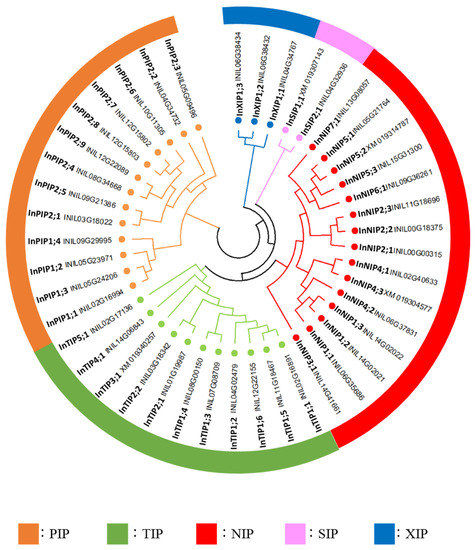
Figure 2.
Phylogenetic tree of the AQPs of Japanese morning glory. Phylogenetic tree was generated by the neighbor-joining method derived from a CLUSTAL alignment of the AQP amino acid sequences of Japanese morning glory.
2.2. Exon–Intron Structure
The gene structures of morning glory AQPs were schematized based on their genome and coding sequences (CDSs) using the Gene Structure Display Server 2.0. Most morning glory AQP genes carry 2–6 exons, and the corresponding gene structures vary across AQP families (Figure 3). Most InPIPs possess three or four exons; most InTIPs possess three exons; most InNIPs possess four or five exons; InSIPs possess three exons; and InXIPs possess two or three exons.
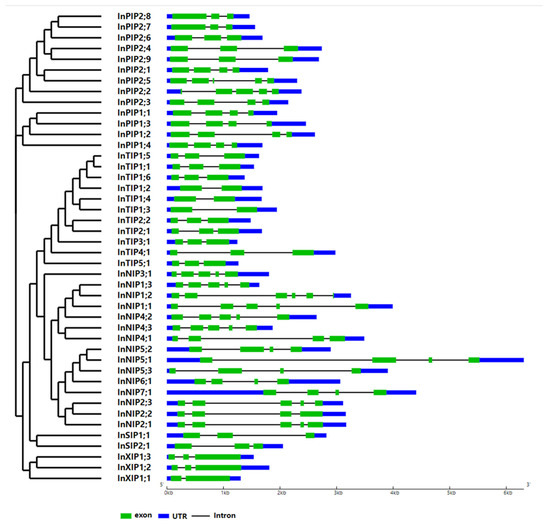
Figure 3.
Exon–intron structure of the AQP genes of Japanese morning glory. A graphic representation of the gene models of all 44 AQPs identified in this study. Gene models are based on genome and CDS sequences.
2.3. TMDs
Typical plant AQPs comprise 200–300 amino acid residues and six TMDs [20] (Figure 1). Here, the TMDs of morning glory AQPs were predicted using two different prediction programs, TMHMM and SOSUI (Table 1). Predictions using both programs were generally consistent, although there were differences in some predicted TMDs. Among the 44 morning glory AQPs, 40 AQPs, except InNIP2;3, InNIP5;3, InSIP1;1, and InSIP2;1, were predicted to have six TMDs by TMHMM or SOSUI (Table 1). The alignment of InNIP2;3 with other NIPs suggested that InNIP2;3 lacks the amino acid sequence from the second NPA motif in the sixth TMD (Figure S5). Although no amino acid deletions were detected in InNIP5;3, InSIP1;1, and InSIP2;1, the corresponding numbers of TMDs were four, five, and seven, respectively (Table 1). Large amino acid sequence deletions in AQPs, such as InNIP2;3, may be caused by the misprediction of their gene structure.

Table 1.
AQP genes identified in the Japanese morning glory genome.
Table 1.
AQP genes identified in the Japanese morning glory genome.
| Name | Gene ID | Location | Amino Acid No. | Molecular Weight (kDa) | TMDs ◇ | pI § | Subcellular Localization | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUGUSTUS | Gnomon (NCBI) | Chromosome No. | Start | End | TMHMM | SOSUI | Plant-mPLoc | Wolf PSORT | ||||
| InPIP1;1 | INIL02g16994 | XM_019337031.1 | chr2 | 41500196 | 41501995 | 284 | 30.4 | 6 | 5 | 8.8 | cell membrane | plas: 9, cyto: 3, pero: 1 |
| InPIP1;2 | INIL05g23971 | XM_019295874.1 | chr5 | 35829017 | 35831432 | 287 | 31.0 | 6 | 6 | 9.0 | cell membrane | plas: 8, cyto: 3, pero: 2 |
| InPIP1;3 | INIL05g24206 | XM_019295755.1 | chr5 | 39507889 | 39510158 | 285 | 30.4 | 6 | 5 | 7.7 | cell membrane | plas: 10, cysk: 2, cyto: 1 |
| InPIP1;4 | INIL09g29995 | XM_019303783.1 | chr9 | 197300 | 198859 | 285 | 30.4 | 6 | 6 | 8.8 | cell membrane | plas: 7, cyto: 3, cysk: 2, vacu: 1 |
| InPIP2;1 | INIL03g18022 | XM_019337682.1 XM_019337681.1 | chr3 | 36083595 | 36085245 | 284 | 30.3 | 6 | 5 | 9.0 | cell membrane | plas: 11, golg: 2 |
| InPIP2;2 | INIL04g34732 | XM_019309792.1 △ | chr4 | 273291 | 275487 | 289 | 30.8 | 6 | 6 | 9.1 | cell membrane | plas: 8.5, cyto_plas: 5, E.R.: 2, chlo: 1, mito: 1 |
| InPIP2;3 | INIL05g09496 | XM_019327277.1 | chr5 | 1980051 | 1982031 | 270 | 28.5 | 6 | 4 | 9.1 | cell membrane | plas: 11, vacu: 2 |
| InPIP2;4 | INIL08g34668 | XM_019311043.1 | chr8 | 29166734 | 29169262 | 282 | 30.2 | 6 | 5 | 8.8 | cell membrane | plas: 10, cysk: 3 |
| InPIP2;5 | INIL09g21386 | XM_019343259.1 XM_019343260.1 | chr9 | 4620360 | 4622484 | 282 | 30.0 | 6 | 5 | 7.7 | cell membrane | plas: 9, cysk: 3.5, cysk_nucl: 2.5 |
| InPIP2;6 | INIL10g11305 | XM_019329230.1 | chr10 | 35274307 | 35275867 | 284 | 30.3 | 6 | 5 | 7.6 | cell membrane | plas: 12, cyto: 1 |
| InPIP2;7 | INIL12g15802 | XM_019335920.1 | chr12 | 5285444 | 5286882 | 285 | 30.3 | 6 | 5 | 8.5 | cell membrane | plas: 12, golg: 2 |
| InPIP2;8 | INIL12g15803 | XM_019335914.1 | chr12 | 5277886 | 5279235 | 285 | 30.3 | 6 | 5 | 8.5 | cell membrane | plas: 13 |
| InPIP2;9 | INIL12g22089 | XM_019343855.1 | chr12 | 60062545 | 60065025 | 287 | 30.8 | 6 | 5 | 7.7 | cell membrane | plas: 11, cysk: 3 |
| InTIP1;1 | INIL02g16891 | XM_019336315.1 | chr2 | 40786840 | 40788260 | 252 | 25.8 | 6 | 6 | 5.8 | vacuole | vacu: 6, plas: 3, chlo: 2, cyto: 2 |
| InTIP1;2 | INIL04g02479 | XM_019296873.1 | chr4 | 12385183 | 12386743 | 246 | 24.9 | 6 | 6 | 6.3 | vacuole | cyto: 9, vacu: 4 |
| InTIP1;3 | INIL07g08709 | XM_019326170.1 | chr7 | 14480111 | 14481905 | 245 | 25.1 | 6 | 6 | 6.1 | vacuole | vacu: 6, cyto: 4, plas: 4 |
| InTIP1;4 | INIL08g00150 | XM_019331552.1 | chr8 | 26850052 | 26851598 | 251 | 25.9 | 4 | 5 | 6.0 | vacuole | plas: 4, vacu: 4, cysk: 3, chlo: 1, cyto: 1 |
| InTIP1;5 | INIL11g18467 | XM_019337154.1 | chr11 | 24689421 | 24690925 | 249 | 25.7 | 6 | 6 | 5.9 | vacuole | plas: 6, vacu: 5, cyto: 2 |
| InTIP1;6 | INIL12g22155 | XM_019343462.1 | chr12 | 60755671 | 60756940 | 252 | 25.9 | 6 | 7 | 5.6 | vacuole | plas: 5, vacu: 5, cyto: 2, chlo: 1 |
| InTIP2;1 | INIL01g19987 | XM_019341113.1 | chr1 | 35538537 | 35540087 | 248 | 25.1 | 7 | 6 | 5.6 | vacuole | vacu: 6, cyto: 4, plas: 4 |
| InTIP2;2 | INIL03g18342 | XM_019338205.1 XM_019338204.1 | chr3 | 3827838 | 38279749 | 248 | 25.1 | 7 | 6 | 5.6 | vacuole | plas: 6, vacu: 6, chlo: 1 |
| InTIP3;1 * | INIL03g01865 | XM_019340257.1 | chr3 | 444425 | 445682 | 262 | 27.7 | 6 | 6 | 6.8 | vacuole | mito: 6, cyto: 2, plas: 2, vacu: 2, nucl: 1 |
| InTIP4;1 | INIL14g06843 | XM_019323579.1 | chr14 | 39466956 | 39469705 | 247 | 25.6 | 7 | 6 | 5.8 | vacuole | plas: 7, vacu: 4, cyto: 2 |
| InTIP5;1 | INIL02g17136 | XM_019336883.1 XM_019336884.1 | chr2 | 42529270 | 42530437 | 253 | 26.2 | 6 | 6 | 6.0 | cell membrane and vacuole | Cyto: 7, chlo: 2.5, vacu: 2, chlo_mito: 2, nucl: 1 |
| InNIP1;1 | INIL06g35686 | XM_019311095.1 | chr6 | 14388531 | 14392215 | 276 | 29.3 | 6 | 6 | 8.9 | cell membrane | plas: 8, vacu: 3, cysk: 2 |
| InNIP1;2 | INIL14g02021 | XM_019343044.1 | chr14 | 8114134 | 8117138 | 249 | 26.2 | 5 | 6 | 5.9 | cell membrane | plas: 12, E.R.: 2 |
| InNIP1;3 | INIL14g02022 | XM_019343057.1 | chr14 | 8098734 | 8100242 | 256 | 27.1 | 6 | 6 | 6.5 | cell membrane | plas: 12, vacu: 1 |
| InNIP2;1 | INIL00g00315 # | XM_019318254.1 | scaffold0125 | 6546 | 9471 | 292 | 31.1 | 6 | 6 | 9.3 | cell membrane | plas: 10, golg: 3 |
| InNIP2;2 | INIL00g18375 # | XM_019339178.1 | scaffold1424 | 4008 | 6927 | 314 | 34.2 | 5 | 5 | 9.6 | cell membrane | plas: 9, E.R.: 2, golg: 2 |
| InNIP2;3 | INIL11g18696 | XM_019339456.1 | chr11 | 8328345 | 8331219 | 247 | 26.8 | 4 | 4 | 9.8 | cell membrane | plas: 7.5, cyto_plas: 6, cyto: 3.5, E.R.: 2 |
| InNIP3;1 | INIL14g41661 | XM_019319645.1 | chr14 | 56617068 | 56618733 | 266 | 28.9 | 5 | 6 | 6.4 | cell membrane | plas: 10, E.R.: 2, vacu: 1 |
| InNIP4;1 | INIL02g40633 | XM_019317591.1 | chr2 | 8659937 | 8663157 | 261 | 27.8 | 6 | 6 | 7.7 | cell membrane | plas: 9, vacu: 3, nucl: 1 |
| InNIP4;2 | INIL06g37831 | XM_019313478.1 | chr6 | 4421259 | 4423701 | 295 | 31.9 | 6 | 6 | 6.1 | cell membrane | plas: 7.5, cyto_plas: 4.5, golg: 3, extr: 1, vacu: 1 |
| InNIP4;3 * | INIL08g31098 | XM_019304577.1 | chr8 | 6161219 | 6162577 | 270 | 29.0 | 6 | 7 | 8.3 | cell membrane | plas: 7, nucl: 3, cyto: 2, chlo: 1 |
| InNIP5;1 | INIL05g21764 | XM_019343441.1 | chr5 | 27557396 | 27563220 | 296 | 30.8 | 5 | 6 | 8.3 | cell membrane | plas: 7, vacu: 5, extr: 1 |
| InNIP5;2 * | INIL06g37646 | XM_019314787.1 | chr6 | 6126438 | 6128483 | 300 | 31.3 | 5 | 6 | 6.6 | cell membrane | plas: 11, vacu: 2 |
| InNIP5;3 | INIL15g31300 | XM_019304934.1 | chr15 | 7534705 | 7538310 | 250 | 25.5 | 5 | soluble | 9.0 | cell membrane | plas: 6, cyto: 4, vacu: 3 |
| InNIP6;1 | INIL09g36261 | XM_019311905.1 | chr9 | 8779488 | 8782317 | 220 | 22.8 | 6 | 5 | 9.0 | cell membrane | plas: 6, vacu: 6, nucl: 1 |
| InNIP7;1 | INIL13g08057 | XM_019325163.1 XM_019325162.1 | chr13 | 2555745 | 2559812 | 235 | 25.0 | 6 | 6 | 6.4 | cell membrane and vacuole | Vacu: 10, plas: 3 |
| InSIP1;1 ○ | INIL04g32909 | XM_019307143.1 | chr4 | 7444038 | 7444872 | 239 | 25.3 | 5 | 7 | 9.6 | cell membrane and vacuole | Vacu: 5, chlo: 2, cyto: 2, extr: 2, nucl: 1, mito: 1 |
| + | ||||||||||||
| INIL04g32910 | chr4 | 7444938 | 7446809 | |||||||||
| InSIP2;1 | INIL04g32936 | XM_019307287.1 | chr4 | 7664318 | 7666212 | 241 | 26.5 | 4 | 4 | 9.1 | cell membrane | vacu: 7, plas: 3, E.R.: 3 |
| InXIP1;1 | INIL04g34767 | XM_019309584.1 | chr4 | 432408 | 433612 | 321 | 34.8 | 6 | 6 | 7.0 | cell membrane | plas: 10, golg: 3 |
| InXIP1;2 | INIL06g38432 | XM_019313788.1 | chr6 | 308805 | 310475 | 339 | 36.3 | 6 | 7 | 7.7 | cell membrane | plas: 9.5, cyto_plas: 5.5, E.R.: 2, vacu: 1 |
| InXIP1;3 | INIL06g38434 | XM_019313789.1 XM_019313790.1 | chr6 | 301085 | 302502 | 335 | 35.8 | 6 | 6 | 7.2 | cell membrane | plas: 8.5, cyto_plas: 5, E.R.: 2, golg: 2 |
* Since the amino acid sequence of INIL03g01865 contained X and INIL08g31098 and INIL06g37646 had deletions in the amino acid sequence, the molecular weight, transmembrane domains, isoelectric point, and subcellular localization were predicted using the amino acid sequence of Gnomon annotations; # the chromosome number is unknown; therefore, scaffold is shown; ◇ transmembrane domains; § isoelectric point; ○ InSIP1;1 is expected to consist of two genes (INIL04g32909, INIL04g32910); △ and this gene is expected to encode something other than an AQP; therefore, this is considered to be an annotation error.
2.4. Conserved Motifs and Subcellular Localization
Plant AQPs possess several conserved and characteristic protein motifs, including two highly conserved NPA motifs, the ar/R filter (H2, H5, LE1, and LE2), and the Froger’s position (P1–P5) (Figure 1). These motifs are important for the selectivity of the substrate transport [2,6]. These motifs in morning glory AQPs were confirmed in this study and are summarized in Table 2.

Table 2.
Amino acid composition of the NPA motifs, ar/R filters (H2, H5, LE1, and LE2), and Froger’s positions (P1–P5) in Japanese morning glory AQPs.
Table 2.
Amino acid composition of the NPA motifs, ar/R filters (H2, H5, LE1, and LE2), and Froger’s positions (P1–P5) in Japanese morning glory AQPs.
| Name | NPA Motif 1 | ar/R Filter 2 | Froger’s Positions 3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| First | Second | H2 | H5 | LE1 | LE2 | P1 | P2 | P3 | P4 | P5 | |
| InPIP1;1 | NPA | NPA | F | H | T | R | M | S | A | F | W |
| InPIP1;2 | NPA | NPA | F | H | T | R | M | S | A | F | W |
| InPIP1;3 | NPA | NPA | F | H | T | R | M | S | A | F | W |
| InPIP1;4 | NPA | NPA | F | H | T | R | M | S | A | F | W |
| InPIP2;1 | NPA | NPA | F | H | T | R | A | S | A | F | W |
| InPIP2;2 | NPA | NPA | F | H | T | R | A | S | A | F | W |
| InPIP2;3 | NPA | NPA | F | H | T | R | A | S | A | F | W |
| InPIP2;4 | NPA | NPA | F | H | T | R | A | S | A | F | W |
| InPIP2;5 | NPA | NPA | F | H | T | R | A | S | A | F | W |
| InPIP2;6 | NPA | NPA | F | H | T | R | A | S | A | F | W |
| InPIP2;7 | NPA | NPA | F | H | T | R | A | S | A | F | W |
| InPIP2;8 | NPA | NPA | F | H | T | R | A | S | A | F | W |
| InPIP2;9 | NPA | NPA | F | H | T | R | A | S | A | F | W |
| InTIP1;1 | NPA | NPA | H | I | A | V | T | S | A | Y | W |
| InTIP1;2 | NPA | NPA | H | I | A | V | T | A | A | Y | W |
| InTIP1;3 | NPA | NPA | H | I | A | V | T | A | S | Y | W |
| InTIP1;4 | NPA | NPA | H | I | A | V | T | S | A | Y | W |
| InTIP1;5 | NPA | NPA | H | I | A | V | T | A | A | Y | W |
| InTIP1;6 | NPA | NPA | H | I | A | V | T | S | A | Y | W |
| InTIP2;1 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| InTIP2;2 | NPA | NPA | H | I | G | R | T | S | A | Y | W |
| InTIP3;1 | NPA | NPA | H | V | A | R | L | A | A | Y | W |
| InTIP4;1 | NPA | NPA | H | I | A | R | T | S | A | Y | W |
| InTIP5;1 | NPA | NPA | N | V | G | Y | T | A | A | Y | W |
| InNIP1;1 | NPA | NPA | W | V | A | R | F | S | A | Y | L |
| InNIP1;2 | NPA | NPA | W | V | A | R | F | S | A | Y | M |
| InNIP1;3 | NPA | NPA | W | V | A | R | F | S | A | Y | M |
| InNIP2;1 | NPA | NPA | G | I | - | R | L | T | A | Y | I |
| InNIP2;2 | NPA | NPA | G | S | G | R | L | T | A | Y | I |
| InNIP2;3 | NPA | - | G | S | - | - | L | - | - | - | - |
| InNIP3;1 | NPA | NPA | W | V | A | R | F | S | A | F | V |
| InNIP4;1 | NPA | NPA | W | V | A | R | F | S | A | Y | I |
| InNIP4;2 | NPA | NPA | W | S | A | R | F | T | A | Y | M |
| InNIP4;3 | NPA | NPA | W | V | A | R | L | T | A | Y | L |
| InNIP5;1 | NPS | NPV | A | I | G | R | F | T | A | Y | L |
| InNIP5;2 | NPS | NPV | A | I | G | R | F | T | A | Y | L |
| InNIP5;3 | NPS | NPV | A | I | G | R | F | S | A | Y | L |
| InNIP6;1 | NPA | NPV | T | I | A | R | F | T | A | Y | L |
| InNIP7;1 | NPS | NPA | A | V | A | R | Y | S | A | Y | F |
| InSIP1;1 | NPT | NPA | I | T | P | N | F | A | A | Y | W |
| InSIP2;1 | NPL | NPA | F | K | G | S | I | V | A | Y | W |
| InXIP1;1 | NPI | NPA | I | T | A | R | V | C | A | F | W |
| InXIP1;2 | NPT | NPA | I | T | A | R | V | C | A | F | W |
| InXIP1;3 | NPI | NPA | I | T | A | R | V | C | A | F | W |
1 Asparagine–proline–alanine motifs in loops B (LB) and E (LE); 2 aromatic/arginine filters by four residues, with one in helix 2 (H2) and helix 5 (H5), and two in loop E (LE1,LE2); and 3 five key positions in the protein sequence (P1–P5) associated with function and specific physicochemical properties for each subgroup.
The motifs in InPIPs are highly conserved: all InPIPs carry two NPA motifs; the ar/R filter is F-H-T-R in all InPIPs; and the Froger’s position is M-S-A-F-W in InPIP1s and A-S-A-F-W in InPIP2s (Table 2, Figure S6). Likewise, InTIPs have two highly conserved NPA motifs (Table 2, Figure S7); the ar/R filter is H-I/V-A/G-V/R/Y in all InTIPs (Table 2). Additionally, the Froger’s position is T/L-S/A-A/A/S-Y-W in InTIPs (Table 2).
In contrast, InNIPs carry diverse NPA motifs, ar/R filters, and Froger’s position (Table 2). The first NPA motif in InNIP5;1, InNIP5;2, InNIP5;3, and InNIP7;1 is NPS; additionally, the second NPA motif in InNIP5;1, InNIP5;2, InNIP5;3, and InNIP6;1 is NPV (Table 2, Figure S5). InNIP2;3 lacks the C-terminal region, which includes the second NPA motif, ar/R filter (except H2 and H5), and Froger’s position (except P1) (Table 2, Figure S5). NIPs can be classified into three subgroups according to their ar/R filter sequence: NIPI, NIPII, and NIPIII [2]. The ar/R filter is W-V-A-R for NIP1s, NIP3s, and NIP4s and W-V/S-A-R for InNIP1s, InNIP3;1, and InNIP4s (Table 2, Figure S5); therefore, these were classified as NIPI. The ar/R filter is A/T-I/V-G/A-R for InNIP5s, InNIP6s, and InNIP7s (Table 2, Figure S5); thus, these were classified as NIPII. Except for InNIP2;3, which lacked the C-terminal amino acid sequence (Table 2, Figure S5), the ar/R filter for InNIP2s was determined to be G-I/S-G-R; hence, these were classified as NIPIII. Among the InNIPs, the Froger’s position is generally conserved to F/L-S/T-A-Y/F-L/M/I/V in NIPI, F/Y-S/T-A-Y-L/F in NIPII, and L-T-A-Y-I in NIPIII (Table 2, Figure S5).
The first NPA motif of InSIPs is N-P-/T/L, with some variations (Table 2, Figure S8). The first NPA motif of InXIPs is NP-I/T (Table 2, Figure S9). The second NPA motif extends to the NPARC motif in XIPs of other plant species [21], and all NPA motifs in InXIPs are also NPARC motifs (Figure S9). The ar/R filter and Froger’s position in InXIPs are highly conserved to I-T-A-R and V-C-A-F-W, respectively (Table 2, Figure S9).
Furthermore, the subcellular localization of morning glory AQPs was predicted using two programs, Plant-mPLoc and Wolf PSORT II. Both programs predicted that InPIPs and InXIPs would localize to the plasma membrane, which is consistent with previous experimental results [1,15]. TIPs have been reported to be localized to the vacuolar membrane [1], and our predictions using Plant-mPLoc were consistent with this report. However, the predicted localization of most TIPs using Wolf PSORT was to the plasma membrane (Table 1), which was considered to be a misprediction. Although NIPs have been reported to localize to the endoplasmic reticulum membrane and rhizobial peribacteroid membrane, most were predicted to be localized to the plasma or vacuolar membrane (Table 1). Nonetheless, the most likely localization of NIPs occurs in the plasma membrane and endoplasmic reticulum [5]. InSIPs were predicted to be localized to the plasma or vacuolar membrane. However, SIPs have been reported to be localized to intracellular membrane systems, with the endoplasmic reticulum membrane as the most likely localization candidate [22].
2.5. Gene Expression in Various Organs of Japanese Morning Glory
To estimate the physiological functions of InAQPs in morning glory, gene expression data, indicated by corresponding reads per kilobase million (RPKM) values, for each organ (embryo, flower, leaf, root, seed coat, and stem) were obtained from the morning glory RNA-Seq database (Table 3). As discussed previously, the sequences of XM_019346257, XM_019304577.1, and XM_019314787.1 were used as InTIP3;1, InNIP4;3, and InNIP5;2, respectively, in further analyses. However, for the gene expression data analysis, the INIL03g01865, INIL08g31098, and INIL06g37646 sequences were used because the RPKM values of the predicted INIL03g01865, INIL08g31098, and INIL06g37646 transcripts, but not the XM_019346257, XM_019304577.1, and XM_019314787.1 transcripts, were registered in this database. Most InPIPs and InTIPs showed high expression; additionally, several of these AQPs, including InPIP1;1, InPIP1;2, InPIP1;4, InPIP2;1, InPIP2;2, InPIP2;9, InTIP1;1, InTIP1;3, InTIP1;4, InTIP1;6, InTIP2;1 and InTIP2;2, exhibited a constitutive expression in all the analyzed organs. Among these, InPIP1;2 showed a particularly high expression in all the organs. In contrast, most InNIPs, InSIPs, and InXIPs exhibited low expression in all the analyzed organs (Table 3). The detailed gene expression patterns of morning glory AQPs, along with their predicted transport substrates, are discussed below.

Table 3.
Gene expression of AQPs in Japanese morning glory.
3. Discussion
3.1. AQPs Have Been Highly Conserved throughout Evolution
In the present study, 44 AQPs were identified in the morning glory genome. This number observed is similar to that in other plant genomes, such as 35 found in Arabidopsis [11], 33 in rice [12], 41 in potato [23], 47 in tomato [13], and 51 in flax [24]. The exon–intron structures in each AQP subfamily in morning glory (Figure 3) are similar to those in other plants, including tomatoes [13] and citrus plants [25]. Therefore, the gene structure of each AQP subfamily is conserved in plants. XIPs represent a novel clade of AQPs, first described in the moss Physcomitrella patens [21]. Three XIPs were identified in morning glory: InXIP1;1, InXIP1;2, and InXIP1;3. Interestingly, XIPs are present in a diverse range of plants, including tomato, potato, citrus, and flax, but not Arabidopsis or rice [11,12,13,23,24,25]. XIPs form a clade independent of other subfamilies and have been proposed to play specific roles in substrate transport. In morning glory, InXIPs formed an independent clade from other subfamilies (Figure 2). A phylogenetic analysis of morning glory, tomato, potato, tobacco, cotton, and citrus XIPs revealed that XIPs in morning glory are similar to those of plants belonging to Solanaceae (tomato, potato, and tobacco), which is closely related to the Convolvulaceae, to which morning glory belongs (Figure 4).
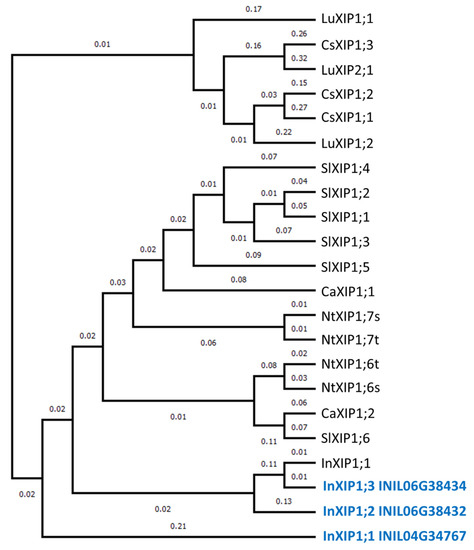
Figure 4.
Phylogenetic tree of XIP family members. A phylogenetic tree was generated by the neighbor-joining method derived from a CLUSTALW alignment of the AQP amino acid sequences of Japanese morning glory InXIPs from this study (indicated in blue) and InXIP1;1 (HM475296) [15], alongside tobacco NtXIPs [26], tomato SlXIPs [13], pepper CaXIPs [27], citrus CsXIPs [25], and flax LuXIPs [24]. Numbers on the phylogenetic tree indicate branch length.
3.2. Prediction of AQP Transport Substrates in Morning Glory
AQPs possess several characteristic conserved motifs, including two highly conserved NPA motifs, the ar/R filter (H2, H5, LE1, and LE2), and the Froger’s position (P1–P5) (Figure 1). These motifs contribute to the selective transport of water and other substrates via AQPs. Based on the similarities of motif sequences between morning glory (Table 2) and other plants AQPs, substrates for which were identified (Figure S3), and substrates for morning glory AQPs were predicted.
PIPs are classified into PIP1 and PIP2 subgroups. PIP2s are characterized by a shorter N-terminus and a longer C-terminus than PIP1s [1]; these PIP characteristics were also observed in InPIPs. The phosphorylation of N-terminal serine residues in PIP2 positively regulates its water transport activity [1,5]. Recently, calcium-dependent protein kinase (CDPK) was shown to mediate this serine phosphorylation in gentian [18]. These serine residues are also conserved in InPIP2s (Figure S6), implying the occurrence of phosphorylation regulation of the water transport activity in these AQPs. In radish AQPs, amino acid residues I (PIP1s) and V (PIP2s), located after the second NPA motif in PIP1s and PIP2s, respectively, are important for the water transport activity; corresponding differences in these residues can explain the higher water transport activity of PIP2s compared to that of PIP1s [28]. In morning glory PIPs, the amino residue V was detected in all InPIP2s, while residue I was detected in all InPIP1s (Figure S6). Therefore, InPIP2s have a higher water transport activity than InPIP1s. The ar/R filter and Froger’s position of InPIP1s were the same as those of NtAQP1, which transports carbon dioxide and urea [2,29]. Therefore, InPIP1s are likely to transport carbon dioxide, urea, and water. Arabidopsis AtPIP2;1 and AtPIP2;4 transport hydrogen peroxide [2]. Therefore, considering their high similarly to AtPIP2;1 and AtPIP2;4 (Figure S3), InPIP2s are likely to transport hydrogen peroxide.
TIPs transport water, ammonia, hydrogen peroxide, and urea [1,2]. InTIP1s and InTIP2s showed high similarity to Arabidopsis AtTIP1;1, AtTIP1;2, and AtTIP2;3, which transport hydrogen peroxide (Figure S3). Therefore, InTIP1s and InTIP2s may transport hydrogen peroxide. The ar/R filters and Froger’s positions of the ammonia-transporting TIPs are H-I-G-R and T-S-A-Y-W, respectively [2]. The ar/R filters and Froger’s positions of InTIP2;1 and InTIP2;2 were the same (Table 2), suggesting that InTIP2;1 and InTIP2;2 transport ammonia. For TIPs that transport urea, the ar/R filters are H-I-A-V or H-I-A-R and the Froger’s positions are T-S/A-A-Y-W and T-S-A-Y-W [2]. The ar/R filters and Froger’s positions of InTIP1;1, InTIP1;2, InTIP1;4, InTIP1;5, InTIP1;6, and InTIP4;1 were consistent with these urea-associated sequences (Table 2, Figure S7). Therefore, these AQPs are predicted to transport urea. In the ar/R filters of InTIP1;1, InTIP1;2, InTIP1;4, InTIP1;5, and InTIP1;6, an amino acid residue at LE2 is V, which has a smaller molecular size than R. Thus, their pore size may be wider, and these InTIPs may be able to transport urea, which has a molecular size larger than that of water [2].
The ar/R filter of InTIP5;1 differed from that of the other InTIPs (Table 2, Figure S7). This is also the case for tomato [13] and potato [23] TIPs. Meanwhile, in Arabidopsis, AtTIP5;1 is highly expressed in pollen and has been reported to transport urea and water [30]. Furthermore, AtTIP5;1 is expected to transport boric acid, because the expression of AtTIP5;1 was induced under high boron conditions, and AtTIP5;1 overexpression enhanced high boron tolerance [31]. InTIP5;1 is highly similar to AtTIP5;1 (Figure S3) and may, therefore, transport urea and boric acid, in addition to water.
NIPs transport boric acid, urea, glycerol, formamide, and silicic acid [1,2], suggesting that InNIPs transport ammonia, glycerol, and formamide. NIPII transports glycerol, urea, and boric acid [2]. Arabidopsis AtNIP5;1, AtNIP6;1, and AtNIP7;1 contribute to boron transport [32,33,34]. Expanded NPA motifs of InNIP5;1, InNIP5;2, InNIP5;3, InNIP6;1, and InNIP7;1 were NPV or NPS, similar to those of AtNIP5;1, AtNIP6;1, and AtNIP7;1. Furthermore, the ar/R filters and Froger’s positions of InNIP5;1, InNIP5;2, InNIP5;3, InNIP6;1, and InNIP7;1 were similar to those of AtNIP5;1, AtNIP6;1, and AtNIP7;1. Therefore, InNIP5;1, InNIP5;2, InNIP5;3, InNIP6;1, and InNIP7;1, which are highly similar to AtNIPs (Figure S3), may also transport boric acid.
Urea, which has a molecular size similar to that of boric acid, is also transported by NIPII [2]; therefore, InNIP5;1, InNIP5;2, InNIP5;3, InNIP6;1, and InNIP7;1 may transport urea. NIPIII transports urea, boric acid, and silicates [2]. The ar/R filters and Froger’s positions of silicic acid transporting NIPs are G-S-G-R and L-T-A-Y-F, respectively [2], whereas the Froger’s position of InNIP2;2 differs slightly (Table 2, Figure S5). Therefore, it is unclear whether InNIP2;2 transports silicic acid.
AtSIPs are localized to the endoplasmic reticulum membrane; moreover, while AtSIP1;1 and AtSIP1;2 transport water, AtSIP2;1 did not confer a water transporting function in a yeast heterologous expression system [22]. Therefore, whether InSIPs possess water transport activity remains unclear.
The first three amino acid residues (H2, H5, and LE1) in the ar/R filter of InXIPs are hydrophobic amino acids (Table 2, Figure S9); thus, InXIPs may contribute to the transport of molecules other than water [2]. Solanales XIPs transport hydrogen peroxide, glycerol, urea, and boric acid, as evidenced by the measurements of transport activity using Xenopus laevis oocytes or a yeast heterologous expression system [15]. InXIPs are highly similar to Solanales XIPs (Figure 4); thus, InXIPs may transport hydrogen peroxide, glycerol, urea, and boric acid.
3.3. Physiological Roles of AQPs in Morning Glory
In plants, AQPs maintain water homeostasis and play diverse roles in stress response, cell elongation and expansion, stomatal opening and closing, flower opening and closing, fertilization, germination, and fruit enlargement [1,19,35]. The physiological functions of AQPs in morning glory are discussed below based on organ-specific gene expression patterns and predicted substrates.
PIPs and TIPs have been reported to possess a potent water transport capacity [1]. InPIP1;2 was expressed in all analyzed organs, and its expression level (RPKM value) was higher than that of the other AQPs (Table 3). Therefore, InPIP1;2 may be the major AQP in morning glory. Further, the expression of InPIP1;2 was particularly high in roots. Additionally, InPIP1;3, InPIP2;6, InPIP2;7, InPIP2;9, InTIP1;1, InTIP1;4, InTIP1;6, InTIP2;1, and InTIP4;1 were also highly expressed in roots (Table 3). Therefore, these AQPs may play important roles in water uptake from the soil to the roots. Wang et al. (2020) [35] reported that PIPs and TIPs were abundant in roots and were important for root growth, suggesting that InAQPs also play an important role in root growth.
InPIP1;2, InPIP2;3, InTIP1;3, and InTIP2;1 were highly expressed in leaves (Table 3); thus, these AQPs are expected to contribute to water transport in leaves. Additionally, some PIPs transport carbon dioxide [29]. In tobacco, the overexpression of NtAQP1, which transports carbon dioxide, increased photosynthetic activity and promoted leaf growth [35]. Therefore, InPIP1;2 and InPIP2;3, which are highly similar to NtAQP, may play vital roles in photosynthesis through carbon dioxide transport.
In the current study, InPIP1;1, InPIP1;2, InPIP1;4, InPIP2;1, InPIP2;9, InTIP1;3, InTIP1;4, and InTIP2;1 were highly expressed in stems (Table 3). Arabidopsis AtTIP1;1 is highly expressed in the hypocotyl, and its expression pattern is related to cell elongation [36]. In rice, PIP and TIP transcripts are concentrated at the internodes of growing plants [37]. Therefore, InAQPs contribute to stem elongation.
InPIP1;1, InPIP1;2, InPIP2;1, InPIP2;9, InTIP1;3, InTIP1;4, and InTIP2;1 were highly expressed in flowers (Table 3). AQPs have been reported to contribute to the opening and closing of flowers in several plants, including Japanese morning glory [1], tulips [17], roses [38,39], and barley [19]. In tulips, TgPIP1;1, TgPIP1;2, TgPIP2;1, and TgPIP2;2 were expressed in petals; in particular, TgPIP2;2 was highly expressed, indicating its significance to flower opening within this plant [17]. In roses, RhPIP2;1 and RhTIP1;1 were involved in flower opening [38,39]. Additionally, in barley, it was suggested that HvTIP1;1, HvTIP1;2, HvTIP2;3 and HvPIP2;1 are important in the flowering process [19]. In Japanese morning glory, flower opening occurs within a few hours and it requires abundant and rapid water uptake into petal cells and their vacuoles. Therefore, InPIP1;1, InPIP1;2, InPIP2;1, InPIP2;9, InTIP1;3, InTIP1;4, and InTIP2;1 may play critical roles in the uptake of water into petal cells and their vacuoles. In fact, mercury, an AQP inhibitor, has been reported to suppress flower opening in morning glory [1]
In tulips, PIP2 phosphorylation regulates light- and temperature-dependent flower opening and closing [40,41]. Recently, CDPK, which is involved in PIP2 phosphorylation, was identified in gentians [18]. CDPK phosphorylates serine residues at the C-terminus of GsPIP2s; this phosphorylation is essential for the light-dependent opening of gentian flowers. The phosphorylated serine residue at the C-terminus is conserved in InPIP2s, including InPIP2;1 and InPIP2;9 (Figure S6). Thus, the phosphorylation of InPIP2s, including InPIP2;1 and InPIP2;9, may be important for the opening of Japanese morning glory flowers. Further, using an anti-phosphorylated serine antibody, which was used in the previous study on gentian [18], we confirmed the phosphorylation of the serine residue at the C-terminus of InPIP2s during the opening of the Japanese morning glory flowers (unpublished). Therefore, InPIP2 phosphorylation is also important for flower opening in Japanese morning glory.
InPIP1;2, InPIP1;4, InPIP2;1 and InPIP2;9 were highly expressed in the seed coat, whereas InPIP1;2 and InTIP3;1 were highly expressed in the embryos (Table 3). These AQPs may be involved in the water uptake or dehydration in seed coats or embryos during the seed coat’s maturation or germination. InTIP3;1 was specifically expressed in embryos (Table 3). TIP3 was originally identified as αTIP—a dominant TIP in the storage vacuoles of seed coats [42]. In Vicia faba, TIP3 promotes water uptake at the early stages of germination [35]. Therefore, InTIP3;1 may be involved in the water uptake during the seed germination of morning glory.
Overall, the expression levels of InNIPs in Japanese morning glory were low, which is consistent with findings in other plant species [24,43]. Nonetheless, InNIP1;1 was expressed in all organs, InNIP2;1 was expressed in leaves, and InNIP5;1 was expressed in roots (Table 3). InNIP5;1 may contribute to boric acid and urea uptake in the roots. In fact, the homolog of InNIP5;1 in Arabidopsis, AtNIP5;1, has been reported to be involved in the uptake of boric acid in roots [44].
InSIP1;1 was expressed in all organs, albeit at low levels. However, InSIP2;1 was not expressed in any organ (Table 3). No or low expression of InXIPs was detected in the analyzed organs. Low expression levels of InXIP1;2 in leaves and very low expression levels of InXIP1;3 in stems were detected (Table 3). These results are consistent with the low expression of XIPs observed in other plants [43,45]. The expression levels of some genes with low initial expression are altered depending on the growth stage and external environment. Therefore, gene expression should be further analyzed at different growth stages and under various environmental conditions.
4. Materials and Methods
4.1. Identification of AQPs in Japanese Morning Glory
Initially, a BLASTP search of the Japanese morning glory genome (http://viewer.shigen.info/asagao/download.php (accessed on 30 January 2021)) was performed using the AQP amino acid sequences from Arabidopsis [11] and tomato [13], as queries under the default setting (e-value: 1 × e−10). Among the candidate Japanese morning glory AQPs, those with fewer than 100 or more than 350 amino acid residues were excluded. Moreover, candidates without an expanded NPA motif (NPA/I/L/S/T/V) were excluded. Gene structure, specifically the structure of the untranslated regions, exons, and introns, was visualized using the Gene Structure Display Server 2.0 (http://gsds.gao-lab.org/ (accessed on 22 August 2022)).
4.2. Multiple Sequence Alignment and Phylogenetic Analysis
Multiple sequence alignments of amino acid sequences were performed using CLUSTALW (https://www.genome.jp/tools-bin/clustalw (accessed on 19 October 2021)) with the default settings. Phylogenetic trees were constructed using the neighbor-joining algorithm in CLUSTALW and visualized using MEGAX [46].
4.3. Prediction of TMD, Molecular Weight, Isoelectric Point (pI), and Subcellular Localization
TMDs were predicted using the TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/ (accessed on 29 April 2021)) and SOSUI (https://harrier.nagahama-i-bio.ac.jp/sosui/sosui_submit.html (accessed on 29 April 2021)). The molecular weight (kDa) and pI of proteins were predicted using ProtParam (http://web.expasy.org/protparam (accessed on 29 April 2021)). Subcellular localization was predicted using Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/ (accessed on 28 May 2021)) and WoLF PSORT II (https://www.genscript.com/wolf-psort.html (accessed on 28 May 2021)).
4.4. Prediction of Gene Expression Profile
Gene expression data, expressed as RPKM values for different organs and tissues of Japanese morning glory, were obtained from the Japanese morning glory RNA-Seq database (http://viewer.shigen.info/asagao/jbrowse.php?data=data/Asagao_1.2 (accessed on 30 October 2021)). Details of the samples used for expression analysis have been reported previously (Table S21 in Hoshino et al., 2016 [16]).
5. Conclusions
The present study offers a comprehensive overview of AQPs in morning glory. A total of 44 AQPs were identified in the morning glory genome, and their details were summarized. Morning glory has been widely used as a model plant for flowering and floral organ development. In the present study, our findings suggest that morning glory AQPs are involved in flower opening. Overall, the information presented here can help clarify the physiological functions of AQPs in this plant.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants12071511/s1: Figure S1. Schematic diagram of the gene structure and gene expression of InSIP1;1 on JBrowse.; Figure S2. Alignment of genome sequences of INIL04g32909, INIL04g32910, and XM_019307143; Figure S3. Phylogenetic trees of AQPs of Arabidopsis, tomato, and Japanese morning glory; Figure S4. Schematic diagram of the gene structure and gene expression of InTIP3;1 on JBrowse; Figure S5. Alignment of amino acid sequences of InNIP subfamily members; Figure S6. Alignment of amino acid sequences of InPIP subfamily members; Figure S7. Alignment of amino acid sequences of InTIP subfamily members; Figure S8. Alignment of amino acid sequences of InSIP subfamily members; and Figure S9. Alignment of amino acid sequences of InXIP subfamily members; Table S1. Homology of AQP in morning glory with AQP in Arabidopsis and tomato.
Author Contributions
Conceptualization, K.S.; formal analysis, T.I.; funding acquisition, K.S.; methodology, A.H.; project administration, K.S.; supervision, K.S.; visualization, T.I.; writing—original draft, T.I.; writing—review and editing, A.H., S.O., S.M. and K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported partially by the Grant-in-Aids for Scientific Research (KAKENHI: 15H04449, 18H03950, 20K20372, 21K19111, 21H02184) from the Japan Society for the Promotion of Science (JSPS).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data can be requested from the corresponding authors.
Acknowledgments
We thank the National Bioresource Project (NBRP) moring glory for the discussion about the genome data and the reconfirmation of the annotations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Katsuhara, M.; Hanba, Y.T.; Shiratake, K.; Maeshima, M. Review: Expanding roles of plant aquaporins in plasma membranes and cell organelles. Funct. Plant Biol. 2008, 35, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hove, R.M.; Bhave, M. Plant aquaporins with non-aqua functions: Deciphering the signature sequences. Plant Mol. Biol. 2011, 75, 413–430. [Google Scholar] [CrossRef]
- Maurel, C.; Boursiac, Y.; Luu, D.T.; Santoni, V.; Shahzad, Z.; Verdoucq, L. Aquaporins in plants. Physiol. Rev. 2015, 95, 1321–1358. [Google Scholar] [CrossRef]
- Pommerrenig, B.; Diehn, T.A.; Bienert, G.P. Metalloido-porins: Essentiality of Nodulin 26-like intrinsic proteins in metalloid transport. Plant Sci. 2015, 238, 212–227. [Google Scholar] [CrossRef]
- Singh, R.K.; Deshmukh, R.; Muthamilarasan, M.; Rani, R.; Prasad, M. Versatile roles of aquaporin in physiological processes and stress tolerance in plants. Plant Physiol. Biochem. 2020, 149, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Froger, A.; Tallur, B.; Thomas, D.; Delamarche, C. Prediction of functional residues in water channels and related proteins. Protein Sci. 1998, 7, 1458–1468. [Google Scholar] [CrossRef]
- Borgnia, M.J.; Kozono, D.; Calamita, G.; Maloney, P.C.; Agre, P. Functional reconstitution and characterization of AqpZ, the E. coli water channel protein. J. Mol. Biol. 1999, 291, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Johansson, I.; Karlsson, M.; Johanson, U.; Larsson, C.; Kjellbom, P. The role of aquaporins in cellular and whole plant water balance. Biochim. Biophys. Acta Biomembr. 2000, 1465, 324–342. [Google Scholar] [CrossRef] [PubMed]
- Tyerman, S.D.; Niemietz, C.M.; Bramley, H. Plant aquaporins: Multifunctional water and solute channels with expanding roles. Plant Cell Environ. 2002, 25, 173–194. [Google Scholar] [CrossRef]
- Gonen, T.; Walz, T. The structure of aquaporins. Q. Rev. Biophys. 2006, 39, 361–396. [Google Scholar] [CrossRef] [PubMed]
- Johanson, U.; Karlsson, M.; Johansson, I.; Gustavsson, S.; Sjovall, S.; Fraysse, L.; Weig, A.R.; Kjellbom, P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 2001, 126, 1358–1369. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, J.; Ishikawa, F.; Yamaguchi, T.; Uemura, M.; Maeshima, M. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 2005, 46, 1568–1577. [Google Scholar] [CrossRef]
- Reuscher, S.; Akiyama, M.; Mori, C.; Aoki, K.; Shibata, D.; Shiratake, K. Genome-wide identification and expression analysis of aquaporins in tomato. PLoS ONE 2013, 8, e79052. [Google Scholar] [CrossRef]
- Johanson, U.; Gustavsson, S. A new subfamily of major intrinsic proteins in plants. Mol. Biol. Evol. 2002, 19, 456–461. [Google Scholar] [CrossRef]
- Bienert, G.P.; Bienert, M.D.; Jahn, T.P.; Boutry, M.; Chaumont, F. Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 2011, 66, 306–317. [Google Scholar] [CrossRef]
- Hoshino, A.; Jayakumar, V.; Nitasaka, E.; Toyoda, A.; Noguchi, H.; Itoh, T.; Shin-I, T.; Minakuchi, Y.; Koda, Y.; Nagano, A.J.; et al. Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat. Commun. 2016, 7, 13295. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Katsuhara, M.; Sawa, Y.; Ishikawa, T.; Shibata, H. Characterization of four plasma membrane aquaporins in tulip petals: A putative homolog is regulated by phosphorylation. Plant Cell Physiol. 2008, 49, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, K.; Niinae, T.; Goto, F.; Sugiyama, N.; Watanabe, A.; Shimizu, M.; Shiratake, K.; Nishihara, M. Calcium-dependent protein kinase 16 phosphorylates and activates the aquaporin PIP2;2 to regulate reversible flower opening in Gentiana scabra. Plant Cell. 2022, 34, 2652–2670. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Tong, T.; Jiang, W.; Cheng, J.H.; Deng, F.L.; Wu, X.J.; Chen, Z.H.; Ouyang, Y.A.; Zeng, F.R. Highly conserved evolution of aquaporin PIPs and TIPs confers their crucial contribution to flowering process in plants. Front. Plant Sci. 2022, 12, 761713. [Google Scholar] [CrossRef]
- Tornroth-Horsefield, S.; Wang, Y.; Hedfalk, K.; Johanson, U.; Karlsson, M.; Tajkhorshid, E.; Neutze, R.; Kjellbom, P. Structural mechanism of plant aquaporin gating. Nature 2006, 439, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Danielson, J.A.H.; Johanson, U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 2008, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, F.; Suga, S.; Uemura, T.; Sato, M.H.; Maeshima, M. Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett. 2005, 579, 5814–5820. [Google Scholar] [CrossRef]
- Venkatesh, J.; Yu, J.W.; Park, S.W. Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiol. Biochem. 2013, 73, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Shivaraj, S.M.; Deshmukh, R.K.; Rai, R.; Belanger, R.; Agrawal, P.K.; Dash, P.K. Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Sci. Rep. 2017, 7, 46137. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.J.; Ma, Q.L.; Ma, Z.Z.; Zhou, G.F.; Feng, F.F.; Le, S.; Lei, C.Y.; Gu, Q.Q. Genome-wide identification and characterization of sweet orange (Citrus sinensis) aquaporin genes and their expression in two citrus cultivars differing in drought tolerance. Tree Genet. Genomes 2019, 15, 17. [Google Scholar] [CrossRef]
- De Rosa, A.; Watson-Lazowski, A.; Evans, J.R.; Groszmann, M. Genome-wide identification and characterisation of aquaporins in Nicotiana tabacum and their relationships with other Solanaceae species. BMC Plant Biol. 2020, 20, 266. [Google Scholar] [CrossRef] [PubMed]
- Uppuluri, L.S.; Motukuri, S.R.K.; Kumar, D. Genome-wide identification, characterization of aquaporin gene family and understanding aquaporin transport system in hot pepper (Capsicum annuum L.). Sci. Hortic. 2021, 286, 110260. [Google Scholar] [CrossRef]
- Suga, S.; Maeshima, M. Water channel activity of radish plasma membrane aquaporins heterologously expressed in yeast and their modification by site-directed mutagenesis. Plant Cell Physiol. 2004, 45, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Flexas, J.; Ribas-Carbo, M.; Hanson, D.T.; Bota, J.; Otto, B.; Cifre, J.; McDowell, N.; Medrano, H.; Kaldenhoff, R. Tobacco aquaporin NtAQP1 is involved in mesophyll conductance to CO2 in vivo. Plant J. 2006, 48, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Soto, G.; Fox, R.; Ayub, N.; Alleva, K.; Guaimas, F.; Erijman, E.J.; Mazzella, A.; Amodeo, G.; Muschietti, J. TIP5;1 is an aquaporin specifically targeted to pollen mitochondria and is probably involved in nitrogen remobilization in Arabidopsis thaliana. Plant J. 2010, 64, 1038–1047. [Google Scholar] [CrossRef]
- Pang, Y.Q.; Li, L.J.; Ren, F.; Lu, P.L.; Wei, P.C.; Cai, J.H.; Xin, L.G.; Zhang, J.A.; Chen, J.; Wang, X.C. Overexpression of the tonoplast aquaporin AtTIP5;1 conferred tolerance to boron toxicity in Arabidopsis. J. Genet. Genom. 2010, 37, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Wallace, I.S.; Takano, J.; Roberts, D.M.; Fujiwara, T. NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 2008, 20, 2860–2875. [Google Scholar] [CrossRef]
- Miwa, K.; Tanaka, M.; Kamiya, T.; Fujiwara, T. Molecular mechanisms of boron transport in plants: Involvement of Arabidopsis NIP5;1 and NIP6;1. Adv. Exp. Med. Biol. 2010, 679, 83–96. [Google Scholar] [PubMed]
- Li, T.; Choi, W.G.; Wallace, I.S.; Baudry, J.; Roberts, D.M. Arabidopsis thaliana NIP7;1: An anther-specific boric acid transporter of the aquaporin superfamily regulated by an unusual tyrosine in helix 2 of the transport pore. Biochemistry 2011, 50, 6633–6641. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Z.J.; Liu, F.; Sun, L.R.; Hao, F.S. Versatile roles of aquaporins in plant growth and development. Int. J. Mol. Sci. 2020, 21, 9485. [Google Scholar] [CrossRef]
- Ludevid, D.; Hofte, H.; Himelblau, E.; Chrispeels, M.J. The expression pattern of the tonoplast intrinsic protein gamma-tip in Arabidopsis-thaliana is correlated with cell enlargement. Plant Physiol. 1992, 100, 1633–1639. [Google Scholar] [CrossRef]
- Muto, Y.; Segami, S.; Hayashi, H.; Sakurai, J.; Murai-Hatano, M.; Hattori, Y.; Ashikari, M.; Maeshima, M. Vacuolar proton pumps and aquaporins involved in rapid internode elongation of deepwater rice. Biosci. Biotechnol. Biochem. 2011, 75, 114–122. [Google Scholar] [CrossRef]
- Ma, N.; Xue, J.Q.; Li, Y.H.; Liu, X.J.; Dai, F.W.; Jia, W.S.; Luo, Y.B.; Gao, J.P. Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiol. 2008, 148, 894–907. [Google Scholar] [CrossRef]
- Xue, J.Q.; Yang, F.; Gao, J.P. Isolation of Rh-TIP1;1, an aquaporin gene and its expression in rose flowers in response to ethylene and water deficit. Postharvest Biol. Technol. 2009, 51, 407–413. [Google Scholar] [CrossRef]
- Azad, A.K.; Sawa, Y.; Ishikawa, T.; Shibata, H. Characterization of protein phosphatase 2a acting on phosphorylated plasma membrane aquaporin of tulip petals. Biosci. Biotechnol. Biochem. 2004, 68, 1170–1174. [Google Scholar] [CrossRef]
- Azad, A.K.; Sawa, Y.; Ishikawa, T.; Shibata, H. Phosphorylation of plasma membrane aquaporin regulates temperature-dependent opening of tulip petals. Plant Cell Physiol. 2004, 45, 608–617. [Google Scholar] [CrossRef]
- Johnson, K.D.; Herman, E.M.; Chrispeels, M.J. An abundant, highly conserved tonoplast protein in seeds. Plant Physiol. 1989, 91, 1006–1013. [Google Scholar] [CrossRef]
- Yaguinuma, D.H.; dos Santos, T.B.; de Souza, S.G.H.; Vieira, L.G.E.; Ribas, A.F. Genome-wide identification, evolution, and expression profile of aquaporin genes in Coffea canephorain response to water deficit. Plant Mol. Biol. Reporter. 2021, 22, 7269. [Google Scholar] [CrossRef]
- Takano, J.; Wada, M.; Ludewig, U.; Schaaf, G.; von Wiren, N.; Fujiwara, T. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 2006, 18, 1498–1509. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Zaplana, A.; Nicolas-Espinosa, J.; Carvajal, M.; Barzana, G. Genome-wide analysis of the aquaporin genes in melon (Cucumis melo L.). Sci. Rep. 2020, 10, 22240. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).