Abstract
Spores and pollen of plants were used as flow cytometric materials to efficiently infer genome sizes. Given this advantage, they hold great potential for various flow cytometric applications, particularly as plant genome size standards. To develop such novel standards, we investigated conditions of pretreatment (bead vortex), buffer, and reliable genome sizes of three fern spore collections—Cibotium taiwanense “Kuo4395”, Sphaeropteris lepifera “Tang0001”, and Alsophila metteniana “Lee s.n.”. Additionally, up to 30 year-old spore collections were obtained from herbarium specimens and from samples stored at 4 °C; their spore nuclei were extracted, and the quality and quantity of these nucleus extractions through storage ages were examined. Nuclear extractions with a longer bead vortex duration or lower spore/bead ratio generally resulted in a higher recovered quantity but a lower quality or purity. For each spore standard, the protocol optimization was determined by their performance in bead vortex conditions, and a 1C genome size was further inferred by linear regression (C. taiwanense “Kuo4395” = 5.058 pg; S. lepifera “Tang0001” = 7.117 pg; and A. metteniana “Lee s.n.” = 19.379 pg). Spore nucleus quality and quantity are significantly negatively correlated with storage ages. Nuclear extractions of 10-year-old refrigerated spores remained qualified as a genome size standard; however, none of the herbarium spore collections fit such criteria. Our study is the first to develop and apply dried and refrigerated spores for genome size standards. These standards are ready to use, easy to manipulate, and feature long-term storage in comparison with traditionally used standards of fresh leaves.
1. Introduction
With the advances of genome sequencing technologies, exploration of genomic diversity outside of model organisms is no longer restricted. Meanwhile, prior knowledge of genome size across the tree of life became increasingly important. Genome size information is critical for biologists to select candidate species or strains that are cost-effective for whole-genome sequencing. Additionally, this information provides a basis for further works to study genomic organization, regulation, and evolution [1,2]. This is particularly true for plants because these organisms evolved varied genome size, over 2000-fold, and were marked as the largest among eukaryotes [3,4,5]. The significant variation in genome size among land plants is predominantly driven by whole-genome doubling, thus revealing rounds of polyploidization in their evolutionary histories [6]. Compared to diploids, polyploids, due to their enlarged genome size, might cost double (or more) to complete whole genome sequencing, and are thereby undesirable for such projects. By far, flow cytometry (FCM) proves the most efficient and cost-effective approach to infer accurate genome size in plants [7,8]. Such flow cytometric investigation requires extractions of nuclei from standard(s) with a known genome size (or C-value). Plant standards of this purpose started being developed in the 1980s [9], and several well-established C-value standards belong to certain named strains of crops (e.g., [10,11,12]). In addition to the features of being easy and fast growing, genomic contents are believed to be almost identical from generation to generation in these crop strains. However, the maintenance of “living” standards is necessary in order to retrieve fresh leaves from the same plants for nuclear extraction.
Within the past few decades, FCM methods for plant C-value estimates were not only applied to fresh leaf tissue, but also succeeded in other types of organs, such as seeds [13,14,15], pollen [16,17,18,19], and spores [20,21]. The FCM experiments for pollen and spores were demonstrated to be more efficient than using fresh leaves [18,20]. For example, the bead vortex approach by Kuo and Huang [22] can complete nuclear extractions of a dozen or more spore samples within 2 min, whereas it usually takes more time to chop tissue from a leaf sample. Such methodology of bead vortex continuously succeeded, especially in fern spores [21,23,24,25]. Another great advantage of such “dried” fern spore material is that neither fresh tissue nor a living plant is required for C-value estimation [22]. Moreover, a very small amount of spores is usually required for a FCM experiment, and such an amount can be easily harvested from most fern species [26]. For example, hundreds of millions of spores (>10 g) can be collected from a single leaf of a fern tree [27]. For a single FCM reaction, only 7 mg of spores is required [22], thus a spore collection from a single fern leaf can support thousands of FCM reactions. Last, selfing lines can be generated in most ferns [28,29,30], and are therefore prone to be genetically conserved across generations. This advantage raises the repeatability of genome size estimates. In sum, because of being methodologically efficient, abundantly harvested, dry-stored, and genetically stable, spores of these plants hold a great potential to be developed as a new kind of “ready-to-use” C-value standard.
In addition to the above-mentioned considerations, requirements such as range and precision of genome size estimates should be taken into account when developing an ideal C-value standard [7]. First, an applied standard must have a genome size close enough to that of the target species in order to reduce the risk of estimate error [12,31,32]. Second, chemical interaction coming from buffers and tissue extraction is prone to affect the degree of DNA staining within nuclei, and to have subsequent influence on further estimation of genome size [33]. In addition, some other factors should be considered, particularly for a fern/lycophyte “spore” standard. Kuo et al. [34], for instance, adjusted vortex duration, spore amount, and bead amount for bead vortex pretreatment in order to optimize conditions for nuclear extraction. Last, storage age is one of the most important factors affecting spore viability, and, likewise, quality of spore nuclei. Spore viability in some ferns is known to be strikingly decreasing through time [35,36], while it is still unknown whether spore nuclear quality declines in a similar manner.
To develop spores as new C-value standards for plants, we chose ferns for our study because a high quantity of spores per individual is easily accessible in these plants [26]. We first examined spore traits, including their diameter, mass, nucleus size, and genome size in different fern species, and selected three collections—Cibotium taiwanense C.M.Kuo “Kuo4395”, Sphaeropteris lepifera R.M.Tryon “Tang0001”, and Alsophila metteniana Hance “Lee s.n.” (Table 1) as candidates for the downstream assessment (using the species epithets as their names in the following text). Further, in order to optimize extraction protocol of their spore nuclei, we tested different bead vortex conditions, and determined the best acquisition with either a considerable quality or quantity for spore nuclei among them. Then, to obtain a reliable C-value for future application, we designed several sets of buffers and internal standards. Last, to check integrity of spore nuclei through storage age and conditions, we used additional spore collections of S. lepifera, which were either stored at 4 °C for up to 20 years, or in a herbarium at room temperature for up to 30 years. With the above experiments, three promising and easy-to-use spore standards of C-value estimation are presented in this study.

Table 1.
Information and spore traits of the three fern spore collections. All voucher specimens are kept in Herbarium of Taiwan Forestry Research Institute (TAIF). * Living collection of Dr. Cecilia Koo Botanic Conservation Center, no. K017423. # Living collection of National Tsing Hua University, no. TSK20180610.
2. Results
2.1. Spore Traits and FCM Performances
The spore traits are summarized in Table 1. The spores of all the three species are similar in shape and their equatorial diameters. The nucleus sizes positively correlate with their genome sizes, and both change in the same order of A. metteniana > S. lepifera > C. taiwanense (Table 1); however, the changes of spore size (i.e., equatorial diameter) and mass are shown with a different trend that does not correspond to their nucleus or genome size. Specifically, their intracellular space occupied by a nucleus tends to be different in proportion. For example, spores of C. taiwanense have the smallest nuclei among the three while both its spore sizes are the largest. As a result, a spore nucleus occupies a relatively small space within a C. taiwanense spore compared to that in the other two species.
Generally, most trends for the resulting quality and quantity match with our expectations (Figure 1 and Figure 2; Figures S1–S3), and vortex duration is an important contribution to bead vortex results (Table 2). The results are similar between S. lepifera and A. metteniana, in which longer vortex duration was usually accompanied by higher coefficient variation (CV), non-nuclear particles (np), and relative recovery rate (rrr) values (Figure 2 and Figure S3; see “4.2. Protocol optimization” for the calculation of these values), several of which are also significantly supported by the ANOVA results (Table 2). However, the rrr value decreased during the higher spore/bead ratio, and showed a clear opposite trend against our expectation (Figure 2). This pattern instead implies that an increase in spore amount in a tube could not proportionally increase the quantity of nucleus particles. The peak of the rrr value might appear in a low spore–bead ratio, which is likely beyond the ratio values surveyed in the current study. Last, patterns in C. taiwanese suggest that none of the treatments can prevent a high np result under bead vortexing (Figure 3). The pattern coordinates with its low ratio of spore nuclear size and spore size, and implies that properties of infra-sporic structures in certain species might affect the quality of nuclear extraction.
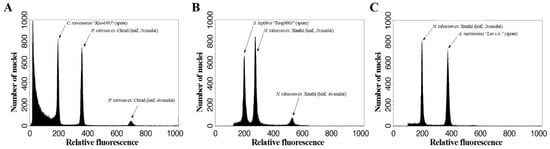
Figure 1.
Estimation of (A) Cibotium taiwanense “Kuo4395”, (B) Sphaeropteris lepifera “Tang0001”, and (C) Alsophila metteniana “Lee s.n.” spore genome sizes by flow cytometry using the internal standards of leaf nuclei of Pisum sativum cv. Ctirad and Nicotiana tabacum cv. Xanthi.
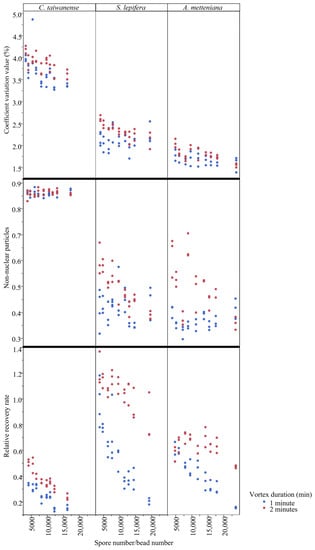
Figure 2.
Coefficient variation (CV) value, non-nuclear particles (np), and relative recovery rate (rrr) of the three spore collections among vortex duration and spore–bead ratios.

Table 2.
ANOVA statistics of cytometric performance using different bead vortex conditions of the three spore collections. * p-value < 0.05.
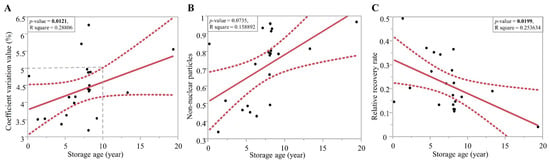
Figure 3.
Linear regression of time and (A) coefficient variation value, (B) non-nuclear particles value, and (C) relative recovery rate of 4 °C-stored Sphaeropteris lepifera spore collections.
Based on these performances, shorter vortex duration and higher spore/bead ratio is apparently best for yielding high quality, while longer vortex duration and lower spore/bead ratio is best for yielding high quantity (i.e., high extraction efficiency). In C. taiwanese, given that all treatments consistently resulted in high np values and similar CV values, access to a high relative recovery rate becomes the most important consideration. Regarding this, a 2 min-vortex with a 3.5 mg-spore and 20 beads (the set of 2, 3.5, and 20) is recommended because it is shown to receive the highest relative recovery rate (Figure S1). In S. lepifera, both great quality and quantity can be maintained under the set of (1, 3.5, and 20), which is shown to receive a high relative recovery rate and a moderate proportion of non-nuclear particles (Figure S1). In A. metteniana, the condition of (1, 3.5, and 16) performed moderately in both quality and quantity of nuclear extraction, where the rrr was high while the np was low (Figure S1).
2.2. Genome Size Estimate
Intriguingly, C-value estimates of A. metteniana and C. taiwanese deviate between spores and leaves (Table 3 and Table 4). These deviations were likely caused by tissue-specific effects (see the detailed explanation in Discussion). Nonetheless, with the use of spore materials, C-value estimates using different buffers and internal standards are mostly consistent, except when using Isoetes taiwanensis De Vol for S. lepifera (Table 3). Aside from this extreme value, the estimates inferred by linear regression (Table 5) are median among all estimates, and show less than 5% deviation from the estimates with marginal ones (Table 3). Regarding the CV values, <5% were found among three spore collections, which implies the universality of buffer application for spore FCM application. However, buffers seemed to perform slight differences among spore collections (Table 3). For instance, the LB01 buffer resulted in the lowest CV values in C. taiwanense and S. lepifera, but highest in A. metteniana, which implies that buffer performance of genome size estimates is also affected by an interaction between buffer and species. In the future, we recommend the use of 1C-values by linear regression (Table 5) as the main genome size references for these spore standards, and referencing the buffer results (Table 3) if any slight difference of buffer performance is relevant.

Table 3.
C-value estimates of the three spore collections using different buffers and internal standards and inferred by interpolation.

Table 4.
C-value estimation of the three spore collections using different buffers and internal standards by interpolation.

Table 5.
Recommended C-values of the three spore collections inferred by linear regression.
2.3. Effects of Storage Age
All spore collections from herbarium specimens failed to detect an unambiguous fluorescence signal of nuclei, so only the 4 °C-stored samples were included in the following correlation analyses (Figure 3). We found that older spores resulted in significantly higher CV values and lower relative recovery rate. Non-nuclear particles are positively, but not significantly, correlated with storage ages. It is suggested that 4 °C storage age should not exceed ten years in order to get an acceptable CV value (<5%) and relative recovery rate. The oldest sample satisfied with these criteria has a storage age of 13 years. Although it resulted in a CV value of 4.28%, nuclear extraction of a single extraction was sufficient for the requirement of the FCM standard.
3. Discussion
3.1. How Does Fern Spore Standard Perform Better?
Our study demonstrates how fern spore standards perform well and even outperform plant leaf standards in several aspects. First, the fast and easy-to-use bead vortex method for fern spore materials is more efficient than chopping methods for leaf tissue. This feature additionally allows operators to minimize bias during destruction of the materials. Second, in terms of accessibility and availability, sufficient intact nuclei can be easily extracted from long-term preserved materials (Figure 3). In our tests, even the fern spores which were dry-stored under 4 °C for more than ten years were still workable, and the resulting quality and quantity of these nucleus extractions satisfied criteria for C-value estimation. This allows fern spores to serve as ready-to-use and easy-to-maintain standards. In comparison, breeding and maintenance of living plants required for leaf standards is labor intensive, and also requires certain environmental conditions and a specific space for growing plants. Importantly, most of these previously established standards are annual crops favoring the temperate climate. Sustainable accesses to fresh leaves from these plants, therefore, need a well-controlled environment to cultivate them for generations. Once lacking a temperature-controlled growing chamber or a green house, it is difficult for a non-temperate place to maintain such breeding conditions, particularly for their flowering and fruiting. Third, these fern spores feature stable DNA stain results by crossing mixtures with various leaf tissue extractions (i.e., the internal standards here) and various buffers (Table 3). In addition, the genome size estimate seems more accurate when using fern spores instead of conspecific leaf tissue. In the case of C. taiwanese, the estimates from genome sequencing results (L.-Y. Kuo pers. comm.) are much closer to the sporic 1C-value than the leaves. As for leaf materials, previous studies suggest that 1C-value estimates using leaf materials are usually lower than those using spores, and seemingly underestimated [18,20,21,23,24]. Deviations of genome size estimates were revealed from leaves with different tissue conditions [37,38,39], and are presumably caused by different cell stages and different levels of chromatin condensation within nuclei [39]. In particular, chromatin is found increasingly decondensed in plant nuclei during the transition from a vegetative fate into a reproductive one; for example, sporogenesis of spore mother cells [40], and thus become more accessible to DNA staining chemicals.
3.2. Extending Candidates of Spore Standards, and Protocol Optimization
Ideally, the genome size range of emerging spore standards is able to cover the range of land plants. The three new spore standards developed here have 1C-values between 5 and 20 pg. In comparison, common leaf standards’ are between 0.15 and 15 pg [31,41]. Spore-bearing vascular plants with a smaller genome size (1C DNA content < 2 pg), such as Lycopodiaceae, Selaginellaceae, and Gleicheniaceae [25,42,43], and ones with a larger size, such as Plagiogyriaceae, Pteridaceae, or Ophioglossaceae, whose 1C DNA content are about 20 to 40 pg [21,25,44], are promising standards in the future in order to fill in the current gaps in genome size range. When it comes to accessibility and available quantities, species that can be easily cultivated or have a high spore yield are prioritized over heterosporous species or those with low spore yields, such as Selaginellaceae [26]. When considering the possibility that genome sizes can vary moderately among different individuals or even generations within a species [38,45,46,47,48,49] (Supplementary Table S1), initially identifying a single selfed or clone-able strain/individual is critical. Such selection can greatly benefit the reliability of C-value estimation. For instance, the Dryopteris varia Kuntze collection mentioned in Materials and Methods is apomictic and fits this perspective well.
Spore traits could also be important factors, and are particularly sensitive to bead vortex protocols. For instance, green spores such as Osmunda japonica Thunb., whose nuclei are less protected from physical damage by thin spore perine, are fragile under bead vortexing [20]. Among the cases demonstrated here, a C. taiwanense spore nucleus occupies a relatively small space within a spore, which is very different from spores of the other two species (Table 1). This sporic property is likely associated with a high proportion of non-nuclear particles (Figure 2). As a result, when seeking and optimizing protocols for future candidates, we suggest also considering their infra-sporic structure as well as other uninvestigated traits (e.g., spore shape difference, trilete vs. monolete).
3.3. Storage Method of Spore for FCM
In this study, spore collections of S. lepifera stored for over ten years are available for estimating its genome size. In contrast, regardless of age (8–30 years old), all the herbarium spores of S. lepifera used in this study failed to be extracted with sufficient nuclei, and there was no detection of an obvious signal peak corresponding to spore nuclei. These results imply that the storage method of the spore is critical for the success of FCM. Nonetheless, Roberts [26] successfully received clear, blunt fluorescent signals of Rosa canina L. pollen and leaf tissue collected from herbarium specimens. Though these materials were relatively fresh (24 months old), they indicated that nuclei of some plants or organs can bear harsh treatments, and will not be completely destroyed shortly after specimen vouchering. In addition to age and (pre)treatments for herbarium specimens, taxon-specific properties seem to be the major factors for FCM results (see below). For instance, eusporangiate structures in some ferns appear to better protect their spores as well as the spore nuclei inside. Even being stored in a herbarium over 20 years, intact spore nuclei can be easily accessed from specimens of such fern taxa [21].
For fern spores, though it is difficult to examine the tissue condition directly, spore viability can be a key indicator—a higher germination rate means a higher proportion of intact nuclei. Previous studies suggest that spore storage conditions can be taxon-dependent in herbaria. For instance, some taxa that contain a sporocarp or live in fire-prone environments have a spore viability of a month to nearly 100 years [50,51,52]. Under dry storage, spore germination rate declines through age, e.g., [53,54], and viability can be better maintained at the lower temperatures [35,55,56,57]. In consequence, we suggest that fern spores used as FCM materials should be refrigerated for long-term preservation. Although a few plant species or their specific organs might be vulnerable to drying procedures and maintenance methods among different herbaria [53], keeping refrigerated duplicates for FCM experiments is highly recommended prior to preparation of herbarium specimens.
4. Materials and Methods
4.1. Spore Sample Source
In order to develop broadly applicable standards, we reviewed previously reported fern genome sizes [8,25,42], and selected candidates after preliminary tests by quickly checking their flow cytometric performance. Four species were chosen: Cibotium taiwanense (1n nuclei = 4.51 pg) [25], Sphaeropteris lepifera (1n nuclei = 6.45 pg) [42], Alsophila metteniana [1n nuclei = 17.00 pg (this study)], and Dryopteris varia (1n nuclei = 24~26 pg) [22,58]. However, because the triploid apomictic individual of D. varia yielded insufficient spores, we were unable to complete a full assessment for this species. Three spore collections for the three remaining candidate species are detailed in Table 1. These three collections were revealed with the genome sizes (Figure 1) matching that from previous conspecific or congeneric reports with ploidy inferences [42,43]. As a result, C. taiwanense “Kuo4395”, S. lepifera “Tang0001”, and A. metteniana “Lee s.n.” are assumed to be, respectively, a diploid, diploid, and tetraploid. These ploidies are also identical with previous cytological reports of the same species known from the same source region (i.e., Taiwan) [59]. These spore collections were stored at 4 °C less than 6 months before any further experiments. Currently, living plants and spore sources of the three collections are maintained at the Dr. Cecilia Koo Botanic Conservation Center (KBCC), Taiwan Forestry Research Institute (TFRI), and National Tsing Hua University (NTHU). These spore sources can be shared under legal process for academic use, and requested from the corresponding authors (L.-Y.K. and Y-.M.H.) and the Dr. Cecilia Koo Botanic Conservation Center (the fern collection manager, Chun-Ming Chen; email: forestaray@gmail.com). To examine spore nuclear quality through storage age (see below in “Effects of storage age”), additional spore collections of S. lepifera were used. These samples included those also stored in a 4 °C fridge within a dark room, but with different ages up to 20 years (Table S1), and that from 8-to-30-year-old specimens stored at the TAIF herbarium under room temperature (air conditioned ~20 °C) (Table S2). Methods of spore collection and isolation were based on Huang et al. [60].
Spore size, mass, and nucleus size were examined. More than 60 spores for each collection were photographed under a light microscope (WILD M8; Leica, Wetzlar, Germany), and their equatorial diameter was measured using Image Pro plus 5.0.0 (Media Cybernetics, Silver Spring, MD, USA). To measure spore mean mass (MX), we first weighted spores at quantum satis (MT) using a sensitive microbalance (MX5; Mettler Toledo, Columbus, OH, USA), and diluted them with a fixed magnification (CT) of distilled water; then counted the concentration of spores (NX) under a microscope with 10 replicates for inferring Mx. To mix each spore solution (in a tube) well, 5-second vortexing was applied before counting.
For spore nucleus size, we used 2.3 mm stainless steel beads (BioSpec, Bartlesville, OK, USA) to break the spores for their nuclear extraction. The spore nuclei were dyed with acetocarmine for 10 to 30 min, and then photographed under a light microscope. For each species, 30 spore nuclei were measured by Image Pro plus 5.0.0 (Media Cybernetics, Silver Spring, MD, USA).
4.2. Protocol Optimization
Our protocol optimization for spore nuclear extraction mainly followed the designs of Kuo et al. [20], with some further modifications. In total, 18 bead vortex treatments were applied with three spore amounts (3.5, 7, and 10.5 mg per tube), three bead amounts (12, 16, 20 beads ea.; 2.3 mm stainless steel beads; BioSpec, Bartlesville, MD, USA), and two vortex durations (1 or 2 min). Three replicates were performed for each bead vortex treatment. Through these treatments, we sought the condition(s) most effective and productive for spore nuclear extraction. The spores and beads were added with 0.25 mL GPB buffer [34] [0.5 mM spermine, 30 mM sodium citrate, 20 mM MOPS, 80 mM KCl, 20 mM NaCl, 0.5% (v/v) Triton X-100, pH = 7.0; with 0.5% (v/v) 2-mercaptoethanol, 40 mg mL−1 PVP-40, and 0.1 mg mL−1 RNaseA added before using] in a 1.5 mL microcentrifuge tube (Gunster Biotech, Taipei, Taiwan). The GPB buffer was chosen for this initial test because it is was known to be widely applicable across fern taxa [25,42,43]. The GPB buffer mixtures were then vortexed at 1900 rpm for either 1 or 2 min. After vortexing, extractions from three different spore collections were filtered through different sieve sizes depending on their spore traits (i.e., spore size and spore nuclear size; Table 1) in order to efficiently remove non-nuclear particles, including intact spores, while allowing spore nuclei to pass through, 20 µm circular nylon filters (Sysmex Partec, Kobe, Hyogo, Japan) were chosen for the extractions of A. metteniana and S. lepifera, while 10 µm circular nylon filters (Sysmex Partec, Hyogo, Japan) were used for those of C. taiwanense. Then, each filtered extraction was divided into two aliquots: one was mixed with the nuclear extraction of internal standard [(Nicotiana tabacum ‘Xanthi’; 2C = 10.04 pg] [12] under a fixed volume ratio, while another was not. All of the solutions were finally dyed with a 1:50 volume of propidium iodide (PI) solution (2.04 mg mL−1). After incubating at 4 °C in the dark for 1 h, all samples were run on a BD FACSCan system (BD Biosciences, Franklin Lakes, NJ, USA). For each treatment, over 1300 particles were collected for both spore and 2n nuclei of the internal standard, and all measured peaks of signal had coefficient variation (CV) values less than 5%.
In addition to the CV values of peaks of spore nuclei, we also applied two additional indicators, “non-nuclear particles (np)” and “relative recovery rate (rrr)”, respectively, to infer purity and quantity of spore nuclear extractions. A lower CV value represents higher quality of nuclei, while greater np implies a higher proportion of non-nuclear particles, which likely resulted from over-disruption during bead vortexing and caused noisy signals for flow cytometric inference. For np values, the aliquots without internal standard were used for inference. Relative recovery rate (rrr) stands for the number of spore nuclei in proportion to its initial input of spore (mg). In other words, a higher rrr value means a higher efficiency of extracting spore nuclei per unit of spores. For rrr values, the aliquots mixed with internal standard were used. Because the nuclear extraction of the internal standard was prepared from the same stock as for each spore collection, the 2n nuclear amount of the internal standard should be constant across the same experimental batch.
Together with the CV values, significance of these indicators among different bead vortex treatments was examined using three-way ANOVA and Tukey’s honest significant difference (HSD) test in JMP13 software (SAS Institute Inc., Cary, NC, USA). We expect that longer vortex duration will contribute to higher CV, np and rrr values. We also expect that the higher spore/bead ratio will lead to higher rrr values and lower CV and np values ―that is, with consistent bead amounts, the more spores we use, the more nuclei we can acquire. The ideal treatments, with either high quantity (i.e., extraction efficiency), quality, or purity for nucleus acquisition, were decided for each spore collection based on the above statistics.
4.3. Genome Size Estimates
To reveal a reliable range of C-value in each of the three spore collections, different buffers and internal standards were used for genome size estimation. The buffers included GPB [34], Beckman [61] [50 mM Na2SO3, 50 mM Tris-HCl, 1.0% (v/v) Triton X-100, pH = 7.5], and LB01 buffer [32] [0.5 mM spermine, 80 mM KCl, 20 mM NaCl, 2 mM Na2EDTA, 15 mM Tris-HCl, 0.1% (v/v) Triton X-100, pH = 7.5]. Additionally, 0.5% (v/v) 2-mercaptoethanol, 40 mg mL−1 PVP-40, and 0.1 mg mL−1 RNaseA were added to these buffers before use. The conditions of bead vortexing and applied filters for spore nuclei of each species were based on suggestions in “Protocol optimization” (see in the previous section). In addition, genome sizes were estimated using the leaf nuclei from the same parental plants, but only performed under the GPB buffer. For the chopping method of leaf materials, we followed Kuo and Huang [22]. Five internal standards were chosen for genome size estimation of spore or leaf nuclei: Vicia faba L. cv. Inovec (2C = 26.90 pg), Secale cereale L. cv. Dankovske (2C = 16.19 pg), Nicotiana tabacum L. cv. Xanthi (2C = 10.04 pg), Pisum sativum L. cv. Ctirad (2C = 9.09 pg), and Isoetes taiwanensis (2C = 3.368 pg) [12,32,62]. Six replicates were performed for each buffer/standard combination in each collection, and the particle number per peak was set to over 10,000. Other flow cytometric conditions were the same as described (see in the previous section “Protocol optimization”). The lowest CV values were reported for buffer selection in each species.
We used two approaches to infer C-value: (1) simply using ratio of fluorescence intensity (i.e., the particles’ mean FL-area) to multiply the known genome size of the internal standard [31,32], and (2) a linear regression with results of fluorescence intensities coming from three different internal standards [31]. The second approach relied on the known genome sizes of the internal standards and their ratios of fluorescence intensities to that of a certain sample. We applied these plots to infer a linear regression line and its formula for each candidate. The resulting formula returned the sample’s genome size value with the ratio of fluorescence intensities set to 1. For this approach, we used only the data coming from the same buffer, and only the GPB buffer was applied.
Notably, because the nuclei of internal standards from leaf tissue and that of our spore samples isolated by different destructive methods (i.e., chopping for leaves and bead-vortexing for spores), these nucleus extractions had to be prepared separately. Standardization from such sample preparation is defined as a pseudo-internal one [31], despite different nucleus extractions that are first mixed to create a homogeneous condition before staining. However, no evidence so far was revealed that the staining property of the nuclei per se can be affected by the different pretreatments [21]. Our preliminary test showed that the estimates, whether the spore nuclei of two samples were extracted together in the same tube or not, are not significantly different. As a result, different processes of nucleus extractions and sample pretreatments were unlikely concerns for the standardization, as well as for the precision of genome size estimation.
4.4. Effects of Storage Age
In this test, we used only S. lepifera spore collections. The condition of bead vortexing and applied filter were based on suggestions in “Protocol optimization” (see in the previous section). LB01 buffer [with 0.5% (v/v) 2-mercaptoethanol, 40 mg mL−1 PVP-40, and 0.1 mg mL−1 RNaseA added] was used because in comparison with the other two, this buffer resulted in the lowest CV value for S. lepifera spore nuclei, as revealed by the results from “Genome size estimation”. Nicotiana tabacum cv. Xanthi was used as the internal standard, and added into each sample (same as in “Protocol optimization”). Other flow cytometric conditions or experimental details were the same as in “Protocol optimization”. CV values, non-nuclear particles (np), and relative recovery rate (rrr) (see the equations in “Protocol optimization”) were applied as indicators of spore nuclear quality. Linear regression against storage age was performed individually for these three parameters using JMP13 software (SAS Institute Inc., Cary, NC, USA).
5. Conclusions
This study is the first to develop fern spores at genome size standards for a flow cytometric purpose. The convenience of these new spore standards against traditional leaf ones are emphasized here. To encourage use of these standards, we also provided the protocols and guidelines for them (detailed in Optimization and suggestion of bead vortex condition), including their genome size estimate (detailed in Genome size estimate). We are looking forward to more fern or even lycophyte spore standards developed in the near future in order to broaden the applicability of this emerging method. Importantly, we hope this convenient tool can also facilitate investigation of C-value diversity in plants that is insightful for plant genomic research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12010140/s1, Table S1: Flow cytometric results and collection information of additional S. lepifera spore collections. Table S2: Collection information of additional S. lepifera spore collections from Herbarium of Taiwan Forestry Research Institute (TAIF). Table S3: FCM results with different bead, spore amounts, and bead-vortxing conditions. Figure S1: Coefficient variation values of the three spore collections. Figure S2: Non-nuclear particle values of the three spore collections. Figure S3: Relative recovery rate values of the three spore collections.
Author Contributions
L.-Y.K. and S.-K.T. designed the study, carried out the experiments, and prepared the manuscript. S.-K.T. performed the statistical analyses. L.-Y.K., S.-K.T., P.-H.L., W.-T.L., C.-H.L. and Y.-M.H. collected the material used in the study. L.-Y.K., S.-K.T. and Y.-M.H. discussed the experimental design, the results, and the writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Ministry of Education and Ministry (Higher Education Sprout Project) of Science and Technology of Taiwan and Ministry of Science and Technology of Taiwan (MOST-109-2621-B-007-001-MY3).
Data Availability Statement
All the data are available at the Supplementary Materials of this article.
Acknowledgments
We thank KBCC (Cecilia Koo Botanic Conservation Center) and TAIF (Herbarium of Taiwan Forestry Research Institute) for providing and preserving fern spore collections; Alexandria Quinlan for help with English editing; Weng-Liang Chiou for providing comments on the manuscript; three anonymous reviewers for their comments on the manuscript; Ing. Jaroslav Doležel for providing seeds of genome size standards; Chuan-Ya Lin and Wen-Yuan Kao for providing the sensitive microbalance; Yun Chen for assistance with the measurement of spore traits.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dufresne, F.; Jeffery, N. A Guided tour of large genome size in animals: What we know and where we are heading. Chromosom. Res. 2011, 19, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Kersey, P.J. Plant genome sequences: Past, present, future. Curr. Opin. Plant Biol. 2019, 48, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, O.; Pellicer, J.; Christenhusz, M.J.M.; Gateway, P.; Street, T.; Sg, H. Genomic gigantism in the whisk-fern family (Psilotaceae): Tmesipteris obliqua challenges record holder Paris japonica. Bot. J. Linn. Soc. 2017, 183, 509–514. [Google Scholar] [CrossRef]
- Hidalgo, O.; Pellicer, J.; Christenhusz, M.; Schneider, H.; Leitch, A.R.; Leitch, I.J. Is there an upper limit to genome size? Trends Plant Sci. 2017, 22, 567–573. [Google Scholar] [CrossRef]
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I.J. Genome size diversity and its impact on the evolution of land plants. Genes 2018, 9, 88. [Google Scholar] [CrossRef]
- One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679. [Google Scholar] [CrossRef]
- Doležel, J.; Bartoš, J.A.N. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005, 95, 99–110. [Google Scholar] [CrossRef]
- Pellicer, J.; Leitch, I.J. The plant DNA C-values database (release 7.1): An updated online repository of plant genome size data for comparative studies. New Phytol. 2020, 226, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Greilhuber, J.; Volleth, M.; Loidl, J. Genome size of man and animals relative to the plant Allium cepa. Can. J. Genet. Cytol. 1983, 25, 554–560. [Google Scholar] [CrossRef]
- Doležel, J.; Greilhuber, J.; Lucretti, S.; Meister, A.; Lysák, M.A.; Nardi, L.; Obermayer, R. Plant genome size estimation by flow cytometry: Inter-laboratory comparison. Ann. Bot. 1998, 82, 17–26. [Google Scholar] [CrossRef]
- Doležel, J.; Sgorbati, S.; Lucretti, S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 1992, 85, 625–631. [Google Scholar] [CrossRef]
- Johnston, J.S.; Bennett, M.D.; Rayburn, A.L.; Galbraith, D.W.; Price, H.J. Reference standards for determination of DNA content of plant nuclei. Am. J. Bot. 1999, 86, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Bino, R.J.; Lanteri, S.; Verhoeven, H.A.; Kraak, H.L. Flow cytometric determination of nuclear replication stage in seed tissues. Ann. Bot. 1993, 72, 181–187. [Google Scholar] [CrossRef]
- Kolarčik, V.; Kocová, V.; Vašková, D. Flow cytometric seed screen data are consistent with models of chromosome inheritance in asymmetrically compensating allopolyploids. Cytom. Part A 2018, 93, 737–748. [Google Scholar] [CrossRef]
- Sliwinska, E.; Zielinska, E.; Jedrzejczyk, I. Are seeds suitable for flow cytometric estimation of plant genome size? Cytom. Part A 2005, 64, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Van Tuyl, J.M.; de Vries, J.N.; Bino, R.J.; Kwakkenbos, T.A.M. Identification of 2n-pollen producing interspecific hybrids of Lilium using flow cytometry. Cytologia 1989, 54, 737–745. [Google Scholar] [CrossRef]
- Pan, G.; Zhou, Y.; Fowke, L.C.; Wang, H. An efficient method for flow cytometric analysis of pollen and detection of 2n nuclei in Brassica napus pollen. Plant Cell Rep. 2004, 23, 196–202. [Google Scholar] [CrossRef]
- Roberts, A.V. The use of bead beating to prepare suspensions of nuclei for flow cytometry from fresh leaves, herbarium leaves, petals and pollen. Cytom. Part A 2007, 71, 1039–1044. [Google Scholar] [CrossRef]
- Kron, P.; Loureiro, J.; Castro, S.; Čertner, M. Flow cytometric analysis of pollen and spores: An overview of applications and methodology. Cytom. Part A 2021, 99, 348–358. [Google Scholar] [CrossRef]
- Kuo, L.-Y.; Huang, Y.-J.; Chang, J.; Chiou, W.-L.; Huang, Y.-M. Evaluating the spore genome sizes of ferns and lycophytes: A flow cytometry approach. New Phytol. 2017, 213, 1974–1983. [Google Scholar] [CrossRef]
- Kuo, L.-Y.; Tang, S.K.; Kao, T.-T.; Ebihara, A.; Fawcett, S.; Hsiao, M.-C.; Shinohara, W.; Dauphin, B. A dormant resource for genome size estimation in ferns: C-value inference of the Ophioglossaceae using herbarium specimen spores. Appl. Plant Sci. 2021, 9, e11452. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.-Y.; Huang, Y.-M. Determining genome size from spores of seedless vascular plants. Bio-Protocol 2017, 7, e2322. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-W.; Kuo, L.-Y.; Huang, Y.-H.; Hsu, T.-C.; Dang, M.T.; Luu, H.T.; Li, C.-W.; Huang, Y.-M. A new species and a new record of Stegnogramma (Thelypteridaceae; Polypodiales) from southern Vietnam. Syst. Bot. 2019, 44, 768–774. [Google Scholar] [CrossRef]
- Chen, C.-W.; Sundue, M.; Kuo, L.-Y.; Teng, W.-C.; Huang, Y.-M. Phylogenetic analyses place the monotypic Dryopolystichum within Lomariopsidaceae. PhytoKeys 2017, 78, 83–107. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kuo, L.-Y.; Li, F.-W. A roadmap for fern genome sequencing. Am. Fern J. 2019, 109, 212–223. [Google Scholar] [CrossRef]
- Rose, J.P.; Dassler, C.L. Spore production and dispersal in two temperate fern species, with an overview of the evolution of spore production in ferns. Am. Fern J. 2017, 107, 136–155. [Google Scholar] [CrossRef]
- Conant, D.S. A radioisotope technique to measure spore dispersal of the tree fern Cyathea arborea Sm. Pollen and Spores 1978, 20, 580–593. [Google Scholar]
- Soltis, P.; Soltis, D. Evolution of inbreeding and outcrossing in ferns and fern-allies. Plant Species Biol. 1990, 5, 1–11. [Google Scholar] [CrossRef]
- Soltis, D.; Soltis, P. The distribution of selfing rates in homosporous ferns. Am. J. Bot. 1992, 79, 97–100. [Google Scholar] [CrossRef]
- Haufler, C.H.; Pryer, K.M.; Schuettpelz, E.; Sessa, E.B.; Farrar, D.R.; Moran, R.; Schneller, J.J.; Watkins, J.E., Jr.; Windham, M.D. Sex and the single gametophyte: Revising the homosporous vascular plant life cycle in light of contemporary research. Bioscience 2016, 66, 928–937. [Google Scholar] [CrossRef]
- Temsch, E.M.; Koutecký, P.; Urfus, T.; Šmarda, P.; Doležel, J. Reference standards for flow cytometric estimation of absolute nuclear DNA content in plants. Cytom. Part A 2022, 101, 710–724. [Google Scholar] [CrossRef] [PubMed]
- Doležel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 2, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, J.; Rodriguez, E.; Doležel, J.; Santos, C. Comparison of four nuclear isolation buffers for plant DNA flow cytometry. Ann. Bot. 2006, 98, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, J.; Rodriguez, E.; Doležel, J.; Santos, C. Two new nuclear isolation buffers for plant dna flow cytometry: A test with 37 species. Ann. Bot. 2007, 100, 875–888. [Google Scholar] [CrossRef]
- Quintanilla, L.G.; Amigo, J.; Pangua, E.; Pajarón, S. Effect of storage method on spore viability in five globally threatened fern species. Ann. Bot. 2002, 90, 461–467. [Google Scholar] [CrossRef]
- Aragon, C.F.; Pangua, E. Spore Viability under different storage conditions in four rupicolous Asplenium L. taxa. Am. Fern J. 2004, 94, 28–38. [Google Scholar] [CrossRef]
- Suda, J.; Trávníček, P. Reliable DNA ploidy determination in dehydrated tissues of vascular plants by DAPI flow cytometry—New prospects for plant research. Cytom. Part A 2006, 69, 273–280. [Google Scholar] [CrossRef]
- Zoschke, R.; Liere, K.; Börner, T. From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J. 2007, 50, 710–722. [Google Scholar] [CrossRef]
- Kron, P.; Husband, B.C. Using flow cytometry to estimate pollen DNA content: Improved methodology and applications. Ann. Bot. 2012, 110, 1067–1078. [Google Scholar] [CrossRef]
- She, W.; Grimanelli, D.; Rutowicz, K.; Whitehead, M.W.J.; Puzio, M.; Kotliński, M.; Jerzmanowski, A.; Baroux, C. Chromatin reprogramming during the somatic-to-reproductive Cell Fate Transition in Plants. Development 2013, 140, 4008–4019. [Google Scholar] [CrossRef]
- Praça-Fontes, M.M.; Carvalho, C.R.; Clarindo, W.R.; Cruz, C.D. Revisiting the DNA C-values of the genome size-standards used in plant flow cytometry to choose the “best primary standards”. Plant Cell Rep. 2011, 30, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.; Hidalgo, O.; Pellicer, J.; Liu, H.; Marquardt, J.; Robert, Y.; Christenhusz, M.; Zhang, S.; Gibby, M.; Leitch, I.J. Genome evolution of ferns: Evidence for relative stasis of genome size across the fern phylogeny. New Phytol. 2016, 210, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Liu, H.; Meza-Torres, E.I.; Morero, R.E.; Vega, A.J.; Liang, Z.; Ebihara, A.; Leitch, I.J.; Schneider, H. Evolution of genome space occupation in ferns: Linking genome diversity and species richness. Ann. Bot. 2021, mcab094. [Google Scholar] [CrossRef]
- Dauphin, B.; Grant, J.; Mráz, P. Ploidy level and genome size variation in the homosporous ferns Botrychium s.l. (Ophioglossaceae). Plant Syst. Evol. 2016, 302, 575–584. [Google Scholar] [CrossRef]
- Cros, J.; Gavalda, M.-C.; Chabrillange, N.; Récalt, C.; Duperray, C.; Hamon, S. Variations in the total nuclear DNA content in African Coffea species (Rubiaceae). Café Cacao Thé 1994, 38, 3–10. [Google Scholar]
- Doležel, J.; Doleželová, M.; Novák, F.J. Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana). Biol. Plant. 1994, 36, 351–357. [Google Scholar] [CrossRef]
- Greilhuber, J.; Ebert, I. Genome size variation in Pisum sativum. Genome 1994, 37, 646–655. [Google Scholar] [CrossRef]
- Baranyi, M.; Greilhuber, J. Flow cytometric analysis of genome size variation in cultivated and wild Pisum sativum (Fabaceae). Plant Syst. Evol. 1995, 194, 231–239. [Google Scholar] [CrossRef]
- Marhold, K.; Kudoh, H.; Pak, J.-H.; Watanabe, K.; Španiel, S.; Lihová, J. Cytotype diversity and genome size variation in eastern Asian polyploid Cardamine (Brassicaceae) species. Ann. Bot. 2009, 105, 249–264. [Google Scholar] [CrossRef]
- Sussman, A.S. Longevity and resistance of the propagules of bryophytes and pteridophytes. In Differenzierung und Entwicklung/Differentiation and Development; Lang, A., Ed.; Springer: Berlin/Heidelberg, Germany, 1965; pp. 2733–2740. [Google Scholar]
- Johnson, D.M. New records for longevity of Marsilea sporocarps. Am. Fern J. 1985, 75, 30–31. [Google Scholar] [CrossRef]
- Paul, S.K.; Dixon, K.W.; Miller, B.P. The persistence and germination of fern spores in fire-prone, semi-arid environments. Aust. J. Bot. 2015, 62, 518–527. [Google Scholar] [CrossRef]
- Windham, M.D.; Wolf, P.G.; Ranker, T.A. Factors affecting prolonged spore viability in herbarium collections of three species of Pellaea. Am. Fern J. 1986, 76, 141–148. [Google Scholar] [CrossRef]
- Magrini, S.; Olmati, C.; Onofri, S.; Scoppola, A. Recovery of viable germplasm from herbarium specimens of Osmunda regalis L. Am. Fern J. 2010, 100, 159–166. [Google Scholar] [CrossRef]
- Kato, Y. The effect of freezing and organic solvents on viability of chlorophyllous fern spores. Cytologia 1976, 41, 387–393. [Google Scholar] [CrossRef]
- Ballesteros, D.; Hill, L.M.; Lynch, R.T.; Pritchard, H.W.; Walters, C. Longevity of preserved germplasm: The temperature dependency of aging reactions in glassy matrices of dried fern spores. Plant Cell Physiol. 2019, 60, 376–392. [Google Scholar] [CrossRef]
- Ballesteros, D.; Pence, V.C. Fern Conservation: Spore, gametophyte, and sporophyte ex situ storage, in vitro culture, and cryopreservation. In Current Advances in Fern Research; Fernández, H., Ed.; Springer: Cham, Switzerland, 2018; pp. 227–249. [Google Scholar]
- Hori, K.; Tono, A.; Fujimoto, K.; Kato, J.; Ebihara, A.; Watano, Y.; Murakami, N. Reticulate evolution in the apogamous Dryopteris varia complex (Dryopteridaceae, subg. Erythrovariae, sect. Variae) and its related sexual species in Japan. J. Plant Res. 2014, 127, 661–684. [Google Scholar] [CrossRef]
- Chiou, W.-L.; Huang, Y.-M.; Lee, P.-H. Mating systems of Cyatheaceae native to Taiwan. In Pteridology in the New Millennium; Springer: Dordrecht, The Netherlands, 2003; pp. 485–489. [Google Scholar]
- Huang, Y.-M.; Wong, S.-L.; Chiou, W.-L. The collection and storage of pteridophyte spores. Taiwan J. For. Sci. 2003, 18, 75–79. [Google Scholar]
- Ebihara, A.; Ishikawa, H.; Matsumoto, S.; Lin, S.; Iwatsuki, K.; Takamiya, M.; Watano, Y.; Ito, M. Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the Vandenboschia radicans complex (Hymenophyllaceae) in Japan and adjacent areas. Am. J. Bot. 2005, 92, 1535–1547. [Google Scholar] [CrossRef]
- Wickell, D.; Kuo, L.-Y.; Yang, H.-P.; Ashok, A.D.; Irisarri, I.; Dadras, A.; Vries, S.de; Vries, J.de; Huang, Y.-M.; Li, Z.; et al. Underwater CAM photosynthesis elucidated by Isoetes genome. Nat. Commun. 2021, 12, 6348. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).