Abstract
Bacterial Leaf Spot (BLS) is a serious bacterial disease of chilli (Capsicum spp.) caused by at least four different Xanthomonas biotypes: X. euvesicatoria pv. euvesicatoria, X. euvesicatoria pv. perforans, X. hortorum pv. gardneri, and X. vesicatoria. Symptoms include black lesions and yellow halos on the leaves and fruits, resulting in reports of up to 66% losses due to unsalable and damaged fruits. BLS pathogens are widely distributed in tropical and subtropical regions. Xanthomonas is able to survive in seeds and crop residues for short periods, leading to the infections in subsequent crops. The pathogen can be detected using several techniques, but largely via a combination of traditional and molecular approaches. Conventional detection is based on microscopic and culture observations, while a suite of Polymerase Chain Reaction (PCR) and Loop-Mediated Isothermal Amplification (LAMP) assays are available. Management of BLS is challenging due to the broad genetic diversity of the pathogens, a lack of resilient host resistance, and poor efficacy of chemical control. Some biological control agents have been reported, including bacteriophage deployment. Incorporating stable host resistance is a critical component in ongoing integrated management for BLS. This paper reviews the current status of BLS of chilli, including its distribution, pathogen profiles, diagnostic options, disease management, and the pursuit of plant resistance.
1. Introduction
Chilli (Capsicum spp.) is an important spice or condiment which is grown in most countries in the world [1]. It is also iconic and has made significant impacts in popular and traditional culture. Also called capsicum, bell pepper and chilli pepper, it originated in tropical regions of Latin America [2]. Chilli is believed to be the first domesticated spice crop [3]. Although it is a quintessential element of the cuisine in many European and Asian nations, chilli is only a relatively recent acquisition.
The year 1492 saw the voyage of Christopher Columbus which opened an exchange of population, concepts, and food crops between what was termed the New World and the Old World. A significant part of the Columbian Exchange, chilli was one of the first crops introduced into Europe from the Americas [4]. At the time there were significant economic interests in black pepper, which saw its price exceed that of gold [5]. Thus, with the poor supply of black pepper, the flavor impact of chilli saw it become one of the world’s most popular condiments [6].
The most recognizable attribute of chilli is its spicy sensation. This sensation is considered one of six main tastes, along with sweet, bitter, sour, salty, and umami [4]. The spiciness of chilli is conferred by its natural alkaloids, known as capsaicinoids, which give the sensation of heat. There are two major capsaicinoids: capsaicin and dihydrocapsaicin, representing approximately 80–90% of the total capsaicinoids in most chilli [7]. It is thought that the capsaicinoids evolved as a way to prevent mammals from eating the fruit, while allowing birds, with their greater potential for seed transport, to be the primary agents for dispersal [8]. It is ironic that this chemical means to dissuade mammals has delivered a sought-after relish which has seen chillis transported and cultivated worldwide by humans.
While chilli fruits are primarily considered a spice, they have many other uses. They can be used for fresh and processed fruits, medicinal ingredients [9], food dyes [10], and even as insecticides [11]. Apart from its flavor, chilli has rich nutritional values, specifically high levels of vitamins A, B, C, and E [12,13]. The popularity of chilli has seen global production steadily increase. The Food and Agriculture Organization [14] reported that chilli production has increased from 29.6 million tons/year in 2010 to 40.3 million tons/year in 2020. Chilli was estimated to have approximately US $30 billion and US $3.8 billion in global value for fresh and dried chilli crops, respectively [15].
2. Bacterial Leaf Spot
A wide range of biotic and abiotic factors affect chilli production. Abiotic factors, such as temperature, drought, flooding, and salinity [16], largely define where chillis are optimally propagated. The presence of biotic factors, such as diseases and pests, can have major effects on chilli yields across cultivars. One of the most economically threatening diseases of chilli is Bacterial Leaf Spot (BLS). BLS is caused by at least four different Xanthomonas biotypes: X. euvesicatoria pv. euvesicatoria, X. euvesicatoria pv. perforans, X. hortorum pv. gardneri and X. vesicatoria. However, it is possible that undiscovered species may also be present. These Xanthomonas biotypes infect plants from the Solanaceae family, including capsicum (Capsicum annuum) and tomato (Lycopersicon esculentum) [17]. The Gram-negative, motile, aerobic, short rod-shaped bacteria can infect leaves, fruits, and stems, causing necrotic lesions and defoliation [18], as shown in Figure 1.
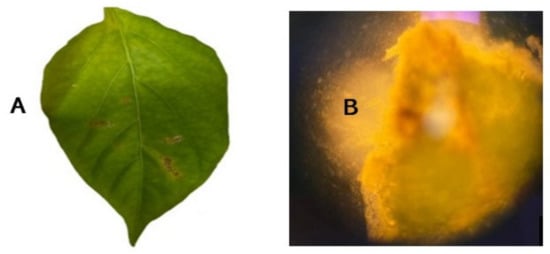
Figure 1.
Diseased chilli leaf infected with X. euvesicatoria pv. euvesicatoria (A), and bacterial oozing from lesion viewed under phase contrast (100× magnification) (B).
BLS-associated leaf lesions are often surrounded by a yellow halo of variable size. Approximately 24 h post-inoculation, hypersensitive cultivars exhibit a tissue collapse reaction from dark green to grey, while susceptible reactions lead to the appearance of water-soaked lesions, 3 days after inoculation [17]. On some cultivars, leaves infected with Xanthomonas may display several small lesions (1 mm) which can cover >80% of the leaf area, whereas in other cultivars, fewer large lesions (>5 mm) may be found. BLS symptoms on chilli leaves and fruits are presented in Figure 2. BLS causes a reduction in photosynthetic area and can potentially lead to infection of the fruit, causing crop failures and economic losses [18]. BLS may not reduce the fruit quantity, but the economic loss is mainly due to lower market value of the unwanted damaged fruit [19].
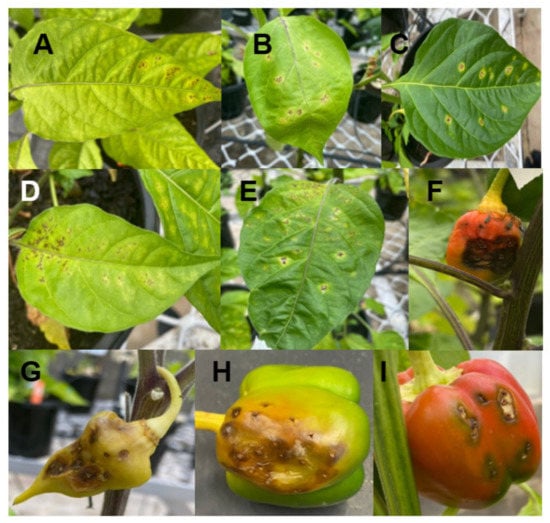
Figure 2.
BLS symptoms on leaves (A–E) and fruits of different chilli cultivars, Capsicum chinense Jacq (F,G) and Capsicum annuum (H,I).
Xanthomonas biotypes causing BLS are widely distributed (Figure 3). Of these, X. euvesicatoria pv. euvesicatoria, X. euvesicatoria pv. perforans, X. hortorum pv. gardneri and X. vesicatoria biotypes causing BLS have been found in five continents (Africa, Asia, Europe, North America, and South America), while in Australia, X. euvesicatoria pv. euvesicatoria, X. euvesicatoria pv. perforans and X. vesicatoria have been isolated [20]. In the USA, it was found that predominant strains can change over time [21]. This has also been seen with other bacterial pathosystems, where pathogen populations appear to change in response to the release of host cultivars that were bred to be resistant to an earlier predominant strain [22].
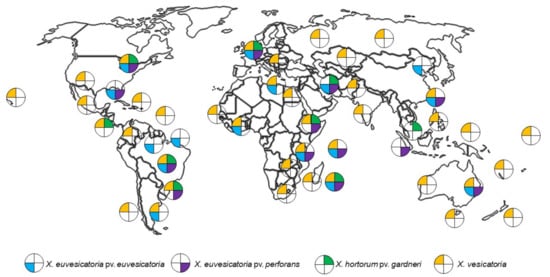
Figure 3.
Current distribution of Xanthomonas spp. causing Bacterial Leaf Spot of Chilli. The database represents records between 2010 to 2020 of Xanthomonas was obtained from European and Mediterranean Plant Protection Organization (EPPO) via https://gd.eppo.int/taxon/XANTEU; https://gd.eppo.int/taxon/XANTGA; https://gd.eppo.int/taxon/XANTPF; https://gd.eppo.int/taxon/XANTVE accessed on 1 April 2022. The world map was purchased from https://www.etsy.com/au accessed on 1 April 2022.
Bacterial taxonomy has always been complicated, and Xanthomonas stands out as a seminal example of the challenges of bacterial nomenclature. A bacterium causing BLS was formally identified in South Africa as Bacterium vesicatoria [23]. In the same year, in Indiana, USA, a pathogen causing BLS was isolated from a tomato plant and named as B. exitiosum [24]. This was later reclassified as X. vesicatoria [25]. The pathovar convention was instituted in response to the poor biochemical resolution of multiple species of Xanthomonas, with the fact that their key distinguishing features were their host range. Thus, biochemically indistinguishable strains were placed into individual species, which were further demarcated by a pathovar epithet depending on the host [26]. Following this consolidation, the main known BLS pathogen was reclassified as X. campestris pv. vesicatoria.
Later DNA homology studies found that X. campestris pv. vesicatoria could be resolved into two distinct species: X. axonopodis pv. vesicatoria as type A and X. vesicatoria as type B [27]. Earlier still, another BLS pathogen on tomato, originally described as Pseudomonas gardneri [28] was reclassified as X. gardneri [29]. A fourth pathogen, provisionally placed in Group C, was formally described as X. perforans [29]. Thereafter, reclassification of the Xanthomonas species which causes BLS resulted in four species: X. euvesicatoria as group A; X. vesicatoria as group B; X. perforans as group C, and X. gardneri for group D [29]. The strain of X. gardneri was proposed for reclassification and named as X. cynarae pv. gardneri after similarity results of whole genome sequences between X. gardneri and X. cynarae were found to be above the 95–96% threshold [30]. The whole-genome based phylogeny, overall genome-related indices calculations and biochemical phenotypic profiling were obtained to reclassify the former pathovar of X. cynarae as X. hortorum, which resulted in a new name as X. hortorum pv. gardneri [31]. Thus, currently, four distinct Xanthomonas biotypes cause similar BLS symptoms on a range of solanaceous plants, and it is important to identify the specific strain to understand the epidemiology of the problem.
There are further pathotype divisions among the BLS strains. This is based on hypersensitive or susceptible reactions associated with four host resistance genes Bs1-4 [32] and corresponding bacterial effectors [33], which are presented in Table 1. While eleven pathotypes have been identified for strains within X. euvesicatoria pv. euvesicatoria, this assessment does not appear to have been completed for X. euvesicatoria pv. perforans, X. hortorum pv. garnderi, and X. vesicatoria. It is important to know what races are present in order to effectively manage them using genetic means.

Table 1.
Races and species of Xanthomonas causing Bacterial Leaf Spot.
3. Infection Process and Life Cycle
There are two critical infection processes for Xanthomonas. The first is the epiphytic phase where bacterial cells are introduced to aerial surfaces such as leaves and fruits [41]. After initial contact, the bacteria penetrate through natural openings such as stomata, wounds or hydathodes [42]. They then continue by moving to the center veins to multiply [43]. The second phase is the endophytic phase where the bacteria multiply within the host tissue. Subsequently, a high bacterial cell population develops, and the bacteria re-emerge onto the surface of the leaf and through rain and wind dispersal are transmitted into a new host and a new infection cycle is initiated [44]. When the bacteria reach a sufficient population, they invade the mesophyll tissues causing the disease symptoms on the leaves [43].
Xanthomonas employ important strategies to attach to the host plant and infect a leaf or fruit surface. These include the use of specific adhesins such as the XadM protein and lipopolysaccharides (LPS) [45]. These adhesins facilitate the attachment to the surface of the plant and have an important role in virulence expression of Xanthomonas [44]. The LPS provides protection from antimicrobial compounds and unfavorable environmental conditions, such as drying [46]. After attaching to the leaf, Xanthomonas forms a biofilm that is important for its survival [47]. Biofilms are three-dimensional structures composed of a bacterial community and its extracellular matrix, which attaches to substrates. The extracellular matrix consists of exopolysaccharides (EPS), extracellular DNA, protein, and lipids [48]. In Xathomonas species, the biofilm matrix also contains xanthan gum, production of which is directed by an operon of genes, including gumB to gumM [49,50]. The extracellular matrix is critical to the infection process.
Formation of the Xanthomonas biofilm is governed by intricate genetic processes. Responses of the bacteria to their population density, known as quorum sensing, are known to give better protection against biotic and abiotic factors [51]. Although quorum sensing aspects for BLS-inducing Xanthomonas species are not well characterized, those of the closely related X. axonopodis pv. citri, causal agent of citrus canker [52], provide insights. Quorum sensing in this pathogen induces flagellum production, triggering both swimming and swarming motility, followed by attachment of biofilm from planktonic cells resulting in a mature biofilm [52]. This strengthens the colonization of host tissue in canker induction. Given the broad utility of quorum sensing in closely related bacterial pathosystems, it is likely these also operate in BLS interactions.
Xanthomonas strains are reportedly able to survive from season to season in chilli and tomato seed, dead tomato debris, soil, and in the rhizospheres of soybean, tomato, and wheat [53]. BLS epidemiology is influenced by environmental factors which promote the broader dispersal of pathogens. This leads to the question of why susceptible plants are infected in some areas, while plants grown from the same seed in another area may not develop symptoms [41]. It was reported that X. euvesicatoria pv. euvesicatoria can survive in plant debris, inoculated sandy loam soil, and the rhizosphere of chilli and non-host plants. In addition, the pathogen was found to survive at least 18 months either in soil or in seed in a field previously planted with chilli diseased with BLS [53]. The pathogen can also be spread from infected plants to healthy plants by rain and wind. The life cycle of Xanthomonas causing BLS of chilli is presented in Figure 4.
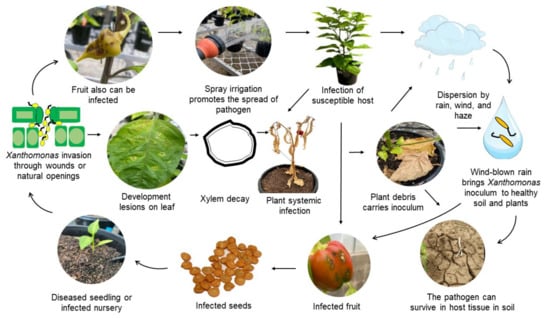
Figure 4.
Disease cycle of Bacterial Leaf Spot of Chilli. Adapted from An et al. [41] and Osdaghi et al. [54] with some modification and addition.
4. Disease Detection
Several conventional and advanced methods have been used to diagnose BLS. Conventional detection is mainly based on visual identification of BLS symptoms, presence of bacterial oozing, and culture-based confirmation (Figure 1). Xanthomonas species can grow on a range of media, including Wilbrink’s media, Yeast-Glucose-Calcium Carbonate Agar (YGCA), Nutrient Broth-Yeast Extract Agar (NBYA), Adenine-supplemented Yeast Peptone Glucose Agar (YPGA), King’s B media, Nutrient Agar (NA) and Nutrient Dextrose (ND) [55]. While most Xanthomonas species are typically readily cultured, differentiating them can present challenges.
Xanthomonas species can be difficult to identify based on direct observations of cultures. With few exceptions, such as the pigmentless X. axonopodis pv. mangiferaeindicae of mango, and X. fragariae, which is more cream yellow than bright yellow [56], a given Xanthomonas species looks like any other Xanthomonas species. While host could be useful for pathovar determination in other plants, this cannot be done for BLS, as four distinct biotypes may occur on a single host. Protein profiles visualized using Sodium Dodecylsulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) were historically used to differentiate strains. X. euvesicatoria pv. euvesicatoria has a specific protein around 32–35 kDa, while X. euvesicatoria pv. perforans, X. hortorum pv. gardneri, and X. vesicatoria have one at 25–27 kDa [57]. Serological assays using a monoclonal antibody were also commonly used to detect X. euvesicatoria pv. euvesicatoria [58]. Fatty Acid Methyl Ester (FAME) analysis can also distinguish the Xanthomonas but requires specialized equipment and expertise [59]. Pectolytic and amylolytic activities are relatively simple methods that can distinguish BLS strains. X. euvesicatoria pv. euvesicatoria and X. hortorum pv. gardneri both have negative results for amylolytic and pectolytic activities, while X. euvesicatoria pv. perforans and X. vesicatoria strains strongly digest starch and pectic substrates [29]. The features of Xanthomonas species causing BLS of chilli are provided in Table 2.

Table 2.
Differentiation of Xanthomonas spp. causing BLS of chilli.
Molecular methods have increasingly been adopted for BLS diagnosis. A range of PCR primers targeting different genes have been designed to detect different BLS Xanthomonas species. Since the 1980s, scientists have been using PCR for the detection of plant diseases. PCR has considerable advantages over other detection methods because it does not require isolation and cultivation of pathogens and is highly sensitive and faster than cultured-based methods. As a result, many PCR-based diagnostic methods for the identification of BLS pathogens have been reported. In 1994, the hrp gene clusters which determine the hypersensitivity and pathogenicity responses were selected to detect and identify X. euvesicatoria pv. euvesicatoria [62]. Oligonucleotide primers specific for hrp genes were used to detect the bacterium by PCR, and it was determined that as few as 10 CFU/mL could be detected. Furthermore, the pairs of Xeu2.4 and Xeu2.5 primers have been successfully developed to detect X. euvesicatoria pv. euvesicatoria and validated using 64 strains of the bacterium from different hosts and regions of origin. The assay sensitivity was up to 1 ng/mL of DNA, which made this protocol suitable for detecting the target in symptomless plants [63]. Primers XCVF and XCVR designed from the rhs family of proteins from X. euvesicatoria pv. euvesicatoria were developed with consistent results of amplification of a 517 bp fragment from various PCR machines and Taq polymerase enzymes provided by different manufacturers [64]. Amplified Fragment Length Polymorphism (AFLP) is another PCR-based technique which involves restriction of genomic DNA to detect polymorphisms in DNA. In 2009, four specific different primers pairs, Bs-XeF/Bs-XeR; Bs-XvF/Bs-XvR; Bs-XgF/Bs-XgR; and Bs-XpF/Bs-XpR, were developed to detect and differentiate all four Xanthomonas biotypes causing BLS using an AFLP based approach [65]. All strains from different geographic origins were identified and discriminated by the four different primer sets.
Even though PCR-based methods have become the gold standard for disease detection, including for Xanthomonas species, these all require a laboratory facility. Rapid field diagnosis can be achieved using Recombinase Polymerase Amplification (RPA) or Loop-Mediated Isothermal Amplification (LAMP). RPA has been developed and successfully used for field detection of BLS pathogens, including X. euvesicatoria pv. euvesicatoria, X. euvesicatoria pv. perforans, X. hortorum pv. gardneri, and X. vesicatoria [66]. Likewise, LAMP protocols have been adopted because they are quick, simple to perform and highly sensitive [67,68]. The sensitivity of LAMP was reported to be up to 1 pg/µL of genomic DNA, however, by calculation, this represents approximately 0.17 genomes per µL, so may be overstated. However, with a 30-min processing time and utility for field diagnostics, LAMP could become an important management tool for BLS [68]. A summary of primers which have been used to detect Xanthomonas causing Bacterial Leaf Spot of chilli is presented in Table 3.

Table 3.
Summary of Molecular Detection on Xanthomonas causing Bacterial Leaf Spot Disease of Chilli. PCR = Polymerase Chain Reaction; LAMP = Loop-Mediated Isothermal Amplification; RPA = Recombinase Polymerase Amplification; AFLP = Amplified Fragment Length Polymorphism; MLSA = MultiLocus Sequence Analysis.
5. Management and Control
Management for BLS is complicated because of an absence of resilient host resistance. Control mainly revolves around cultural practices, chemical control, host genetic resistance and, to a lesser extent, biological control. Xanthomonas species that cause BLS are known to remain in the seed, therefore the use of disease-free-seeds and transplants is one of the key management tools. Diagnostic testing of seeds and seed treatments are typically used to detect the level of seed infection [75]. After seed testing, a combination of planting resistant varieties, strict phytosanitation and bactericidal applications are control options for BLS disease [76].
5.1. Chemical Control
Copper products have historically been used as the main chemical prevention measure for BLS. The use of copper was started by Professor Millardet in 1882, who developed Bordeaux mixture (copper sulphate and lime), which was effective at controlling grape downy mildew [77]. The mixtures of copper and mancozeb were significantly effective for control of BLS compared to only basic copper sulphate [78]. In addition, copper-tolerant strains were killed by the combination of copper and mancozeb, while the sprays of fixed copper controlled only sensitive strains [79]. The addition of mancozeb was known to increase solubility of copper and led to improvements in the effectiveness of copper against tolerant Xanthomonas.
The use of small molecule compounds is an alternative approach to control BLS. Application of D-leucine and 3-indolylacetonitrile (IAN) effectively inhibits biofilm formation [80]. In addition, the application of IAN reduced BLS on tomato significantly and improved the effectiveness of copper against X. euvesicatoria pv. perforans [81]. Particularly, the regulation of BLS caused by copper-resistant X. euvesicatoria pv. perforans has been substantially improved by IAN. Carvacrol, the major component of essential oils from Zataria multiflora, also reported to be effective against plant pathogens, significantly decreased the severity of BLS on tomato and improves the efficacy of copper against copper-resistant X. euvesicatoria pv. perforans, which provides a more environmentally sustainable approach to control BLS [82]. The application of acibenzolar-S-methyl (ASM) treatment through weekly foliar sprays showed significant reductions of BLS in field-grown pepper [83]. ASM is a plant activator which induces systemic resistance in some plants, including pepper and tomato. The new strategy was developed using a hybrid nanoparticle, copper-zinc, on sensitive strains [84]. Based on this approach, compared to the controls, the hybrid nanoparticle significantly decreased the growth of bacteria in vitro. Several other small molecules also have been reported to inhibit the growth of BLS pathogens [85]. These inhibited the four BLS Xanthomonas biotypes but were also effective against copper- and streptomycin-resistant Xanthomonas strains. Importantly, there appear to have minimal impacts on the beneficial bacteria. These small molecules were composed by several amine-based functional groups, such as indole, imidazole, oxazole, piperidine, pyrazol, pyrimidine, and quinoline, which have been recognized for their antimicrobial activities [85].
5.2. Biological Control
Biological control is a promising alternative in disease management by inhibiting the growth of pathogens, improving plant defenses, or adjusting the environment to foster beneficial microbes. Biological control offers an environmentally sustainable alternative to traditional control methods that does not impact beneficial organisms [86] and is a low-cost strategy that can enjoy sustained utility if applied judiciously.
Biological control agents have been reported in several areas of chilli and tomato farming against Bacterial Leaf Spot, for instance, in vitro methods revealed that Lactobacillus MK3, Trichoderma reseii QM 9414, and Pseudomonas aeruginosa 1128 have showed significant potential as antagonists of X. euvesicatoria pv. euvesicatoria [87]. Bacillus velezensis IP22 was able to reduce the infected pepper leaves by 65% compared to positive control inoculated by X. euvesicatoria pv. euvesicatoria. This impact was related to the production of antimicrobial metabolites, lipopeptides from fengycin and locillomycin families, representing a promising strategy as a biocontrol agent of BLS of chilli [87]. Bacillus pumilus INR7 was confirmed to induce the resistance against X. euvesicatoria pv. euvesicatoria when combined with benzothiadiazole (BTH), a chemical inducer which boosted the expression of defense marker genes of pepper, CaPR1, CaTin1, and CaPR4 [88]. Significant reduction of approximately 50% in the severity of BLS was also obtained using humic acids and suspension (108 CFU/mL) of Herbaspirillum seropedicae in tomato [89]. Paenibacillus elgii was evaluated and effectively suppressed the chilli BLS in pot trials with control values of 67% [90]. The mutant strain of X. euvesicatoria pv. euvesicatoria 75-3S hrpG was also evaluated for BLS under both greenhouse and field conditions and showed significant reductions in disease of 57% compared to control [91].
Bacteriophages are viruses which infect and replicate within bacteria [92]. These been studied as natural antimicrobial agents for BLS of chilli and tomato. A foliar application of bacteriophage decreased the BLS incidence from 40.5% (control) to 0.9% on greenhouse-grown tomato [93]. Ten bacteriophages were successfully isolated and characterized, aiming to contribute to an integrated BLS control strategy of reducing the use of conventional pesticides [94]. The other bacteriophage studies were also reported such as using Bacteriophage KΦ1 [95], XaF13 [96], ΦXaF18 [97] isolated from a pepper field infected by BLS. On a commercial scale, bacteriophage has been used to control BLS of chilli. One of the profitable bacteriophages has been produced by The AgriPhage [98] and showed that this bacteriophage management becomes another choice to deal with the BLS issue. The combination of biological control and other methods, such as systemic acquired resistance (SAR) inducer, was also reported to have promising results on BLS. The integration of bacteriophage, SAR, and copper hydroxide was able to reduce the disease intensity by 96–98% in pepper [99].
Some microbes proposed as prospective biocontrol agents behave as opportunistic pathogens which are included in the biosafety level 2 (BLS-2) classification. These pose risk not only to the environment, but to humans. For instance, P. aeruginosa is known to be promising for the control of BLS, but has adverse effects on humans, particularly immunocompromised patients such as those suffering from pneumonia, urinary tract infection and gastrointestinal infections [100,101]. Therefore, appropriate strategies to mitigate risks to humans and the environment need to be undertaken. It is advisable to carry out taxonomic characterization up to species and strain level, including whole genome sequencing. This aims to determine the possibility of the closeness of the target microorganism to BSL-2 or above [101]. Furthermore, the Environmental and Human Safety Index (EHSI), a tool to evaluate the safety of biocontrol candidates before being used in the field, can be adopted for biosafety awareness [102].
6. Host–Pathogen Interactions
There are currently more than 1200 genomes of Xanthomonas available (NCBI website). This will help in understanding taxonomy, pathogenicity, and virulence factors of Xanthomonas [41]. Xanthomonas sp. genomes encode more than 4,000 proteins [44]. The Xanthomonas strains causing BLS rely on some protein effectors to compete with other microorganisms and to colonize the chilli plant. These protein effectors are delivered to their targets by bacterial secretion systems. In X. euvesicatoria pv. euvesicatoria, it is known to have five different secretion systems, including Type I, II, III, V and VI [103]. Type 1 secretion system (T1SS) has a main role in quorum sensing signal [103]; T2SS in cell wall degradation [104]; T3SS maintains the successful of pathogenicity [105]; T5SS in plant attachment [106]; while T6SS is to compete with other microorganisms [107]. In all Xanthomonas species, there are two pathogenesis-correlated gene clusters; the first one is xps, which encodes type II secretion system (T2SS), and rpf, which coordinates the synthesis of pathogenicity factors.
The type II secretion (T2S) system primarily secretes degradative enzymes transported by the Sec or TAT (twin-arginine translocation) systems through the inner bacterial membrane and also contributes to the virulence of plant pathogenic bacteria. Various Xanthomonas spp. contain two T2S systems encoded by homologous gene clusters of xps and xcs. A virulence function has been identified for xps-T2S systems and appears to be dispensable for virulence [108]. The Xps-T2S system from the plant pathogen X. vesicatoria promotes disease and leads to the translocation of the type III secretion (T3S) mechanism of proteins effectors that are released into the plant cell [109]. Thus, understanding the T3 effectors is critical to understanding host and pathogen interaction [110].
The Xps-T2S system secreted degradative enzymes such as cellobiosidases, cellulases, lipases, endoglucanases, polygalacturonases, xylanases, and proteases [111]. The rpf gene is another gene cluster that plays an important role in the pathogenicity of Xanthomonas. Rpf is a regulation of pathogenicity factors present in all Xanthomonads that encodes components that control the synthesis and perception of the signal molecule DSF (diffusible signal factor). The synthesis of the virulence factors and biofilm formation is controlled by the rpf of the DSF systems, and it is necessary for full virulence of X. euvesicatoria, X. oryzae pv. oryzae, X. oryzae pv. oryzicola, X. campestris pv. armoracle [106].
The synthesis of extracellular enzymes, extracellular polysaccharide biofilm dispersal and virulence in X. euvesicatoria pv. euvesicatoria is positively controlled by the rpf gene cluster [112]. Some of the rpf genes have been described in detail. The major X. euvesicatoria pv. euvesicatoria aconitase is encoded by rpfA that involved in iron homeostasis. In the synthesis of a small diffusible signal molecule, rpfB and rpfF are involved [113].
7. Type III Effector Biology and Action
Suppression of plant immunity is an effective virulence strategy to colonize the host by Gram-negative bacteria. The Gram-negative bacteria use a type III secretion mechanism to provide effectors into the host cell to accomplish this. These effectors target the main plant immune signaling pathway modules [114]. This type III secretion system (T3SS) is controlled by the hypersensitive response and pathogenicity (hrp) gene cluster [111]. There are more than 20 genes in this gene cluster, grouped into various transcriptional units [111]. Avirulence proteins are a class of type III effectors in plant pathogens. The term “avirulence” (avr) describes bacterial genes that specify the basic recognition of bacteria by plants with a resistance (R) gene that matches them. The detection of an avr protein mediated by the plant R gene leads to the induction of a plant defense reaction that typically involves hypersensitive response (HR), a rapid localized cell death associated with arresting pathogen ingress. Avirulence protein limits the host range of the pathogen, an effect that is detrimental to the bacteria and is likely not their primary role [115].
The AvrBs3 protein is a broad family of proteins retained in the Xanthomonas genus, which also has a role in both avirulence and virulence. The effector of AvrBs3 from X. vesicatoria on chilli and tomato is a causal agent of BLS. Symptoms of early infection are likely to coincide with the release of water and nutrients that support bacterial multiplication. The ability of X. euvesicatoria pv. euvesicatoria to cause disease in susceptible plants and to induce HR in resistant plants depends on the T3SS encoded by hrp gene cluster [116]. The hrp gene cluster encodes more than 20 proteins, 11 of which are retained in the pathogenic bacteria of plants and animals. These genes have been renamed hrc (hrp conserved) and are assumed to encode the core components of the secretion apparatus. Expression of hrp gene in Xanthomonas is regulated by HrpG and HrpX. As an OmpR family regulator, HrpG triggers the hrcC and hrpX expression in X. vesicatoria or only hrpX in X. euvesicatoria pv. euvesicatoria. The expression of other hrp genes along with some effector genes is regulated by hrpX, an AraC-type transcriptional activator [117]. The mechanistic insight of T3SS in X. euvesicatoria pv. euvesicatoria is explained in Figure 5.
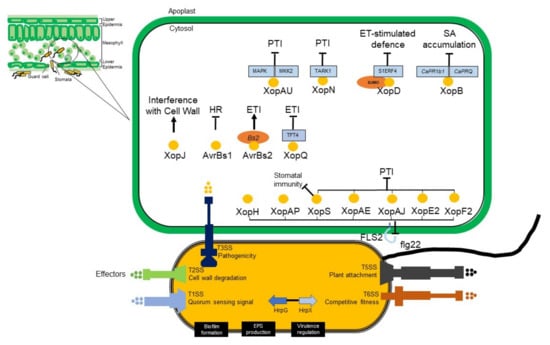
Figure 5.
The mechanistic insight of the Type 3 secretion system in X. euvesicatoria pv. euvesicatoria. T3SS as to suppress the plant immunity, some effectors have been reported to promote the disease colonization. Some effectors have more than one role, such as XopF2, XopE2, XopAJ, XopAE [118], and XopS [119]. These effectors inhibit the flg-22 induced signalling and suppress the pattern-triggered immunity (PTI). Effectors XopAP and XopH are inhibitors of flg22-induced reporter gene activation, but the PTI-related inhibition is not known yet [107]. XopS has been reported also to stabilize pepper to regulate the defense response, stomatal immunity [119], and disease symptoms [120]. While effector AvrBsT suppresses the hypersensitive response (HR) as effect of effector protein AvrBs1 in resistant chilli plant [121]. Chilli plants carrying the Bs2 gene are resistant to X. euvesicatoria pv. euvesicatoria strains, which contain AvrBs2. The interaction between Bs2 and AvrBs2 resulted in Effector-Triggered Immunity (ETI), which led to suppression of effectors into plant cell [122]). During the infection, XopD effector is required for pathogen growth and symptom-development delay. This effector carries small ubiquitin-like modifier (SUMO) and desumoylates S1ERF4 to suppress ethylene levels, which increases susceptibility of host plant to X. euvesicatoria pv. euvesicatoria [123]. To interfere the host immune signalling, XopAU effector manipulates the host MAPK signal and activation of MKK2 [124]), while XopB interferes the PTI and suppresses SA accumulation [125]. XopN interacted with the Tomato Atypical Receptor-Like Kinase1 (TARK1) and suppresses PAMP-triggered immune response [126]. While XopJ is to inhibit the cell wall-based defense response [127].
8. Resistance Genes
Host-plant resistance is a significant component of an integrated management program for BLS. There are five known independent dominant genes (Bs1, Bs2, Bs3, Bs4, and Bs7) and two recessive genes (Bs5 and Bs6) for qualitative resistance controlling BLS resistance [30,106,128]. A combination of Bs1, Bs2, and Bs3 genes gave the lowest disease scores of BLS. A transgenic approach to disease resistance was investigated in tomatoes based on the chilli resistance gene Bs2 [129]. For the Bs2 gene, the avirulence gene avrBs2 was reported to be involved in the fitness of X. euvesicatoria pv. euvesicatoria and is known to be highly conserved among other Xanthomonas species. This finding also suggested that Bs2 gene in other plant species is functional, which might support stable resistance [130].
The Bs3 gene in chilli also conferred resistance to X. vesicatoria. It has been shown in genetic and molecular studies that X. euvesicatoria pv. euvesicatoria strains expressing the AvrBs3 gene trigger Bs3-mediated resistance [116]. AvrBs3 encodes the AvrBs3 family type member, a large family of bacterial effectors that have a sequence identity of 80–99%. In their C-terminus, the AvrBs3-like proteins contain nuclear localization signals (NLSs) and a transcriptional activation domain (AD) that are critical to their virulence [131]. Two recessive genes in chilli, Bs5 andBs6, conferred resistance to all races of Bacterial Leaf Spot of chilli. These recessive genes were reported as more resilient than the dominant resistance genes, as they do not require complex gene-for-gene interactions [132]. The impact of two recessive genes, Bs5 andBs6 genes, showed thatBs5 confers a higher resistance level thanBs6 at 25 °C, but they confer higher resistance to P6 (X. euvesicatoria pv. euvesicatoria chilli strain XV157 of race 6) in combination, suggesting at least the action of the additive gene [133].
9. Genes Involved in Plant Defenses
Several defensives signaling mechanisms have been formed by plants to protect them from the adverse conditions of environment and pathogen attack. The effectiveness of phenylalanine ammonia-lyase (PAL) extracted from pepper (CaPAL1) in defense responses to pathogens is involved in salicylic acid (SA) biosynthesis, an important signal involved in plant systemic resistance [134]. The chilli leaf was induced with CaPAL1 gene by avirulent X. euvesicatoria pv. euvesicatoria infection. There was an increased vulnerability to virulent and avirulent X. euvesicatoria pv. euvesicatoria infection in CaPAL1-silenced chilli plants.
The CaCYP1 gene expression (cytochrome P450 from Capsicum annuum L. Bukang) was found after leaf hypersensitive reaction by infection of chilli plants with the non-host pathogens X. axonopodis pv. glycines [135]. The results suggested that in plant defense response pathways involving salicylic acid and abscisic acid signaling pathways, CaCYP1, a new cytochrome P450 in chilli plants, may play a role. In the plant defense response against pathogens, SA and ethylene are essential secondary signals. A cytoplasmic protein kinase (RLCK), receptor-like chilli (Capsicum annuum) gene (CaPIK1) was identified and transcriptionally activated by infection with X. euvesicatoria pv. euvesicatoria [136]. In chilli plants, silencing of CaPIK1 confers increased susceptibility to infections with X. euvesicatoria pv. euvesicatoria. The results indicate that CaPIK1 modulates the signalling needed for the defense response to pathogen infection based on salicylic acid. In chilli, the CaPIK1 gene was transcriptionally induced by virulent and avirulent X. euvesicatoria pv. euvesicatoria infection, leading at an early stage of infection to a high level of gene expression. These results support the possibility that CaPIK1 gene can confer as a signal transduction mediator in chilli plants’ defense responses to pathogen invasion.
10. Conclusions and Prospects
The challenges in BLS of chilli are to further develop detection platforms, determine the factors which account for the pathogenesis in specific hosts or strains, and include resistance in the breeding program. Further mapping of the prevalent strains is essential to improve management responses. The application of functional genomics and proteomics will help to identify the genes and proteins involved in the infection lifecycle of the different Xanthomonas species and strains. Mapping the genes that govern host–pathogen interactions will provide targets for future application of gene-editing technologies. These actions need to be integrated across the value chain from researchers through to farming stakeholders to meet the demand for this iconic fruit.
Author Contributions
D.U. initiated and designed the overall concept and wrote 90% of the manuscript. S.J.M. and A.J.Y. revised the manuscript, approved the final version, and approved it for publication. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by ACIAR Project No. SLAM/2018/145 Crop health and nutrient management of shallot-chilli-rice cropping systems in coastal Indonesia.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bosland, P.W.; Votava, E.J. Peppers: Vegetable and Spice Capsicums; CABI: Oxford, UK, 2012. [Google Scholar]
- Pickersgill, B. Relationships between weedy and cultivated forms in some species of chilli peppers (Genus Capsicum). Evolution 1971, 25, 683–691. [Google Scholar]
- Ramchiary, N.; Kole, C. The Capsicum Genome; Springer: New Delhi, India, 2019. [Google Scholar]
- Nunn, N.; Qian, N. The Columbian Exchange: A History of Disease, Food, and Ideas. J. Econ. Perspect. 2010, 24, 163–188. [Google Scholar] [CrossRef]
- Shaffer, M. Pepper: A History of the World’s Most Influential Spice; St. Martin’s Press: New York, NY, USA, 2013. [Google Scholar]
- Naves, E.R.; Silva, L.A.; Sulpice, R.; Araujo, W.L.; Nunes-Nesi, A.; Peres, L.E.P.; Zsogon, A. Capsaicinoids: Pungency beyond Capsicum. Trends Plant Sci. 2019, 24, 109–120. [Google Scholar] [CrossRef]
- Barbero, G.F.; Liazid, A.; Azaroual, L.; Palma, M.; Barroso, C.G. Capsaicinoid contents in peppers and pepper-related spicy foods. Int. J. Food Prop. 2016, 19, 485–493. [Google Scholar] [CrossRef]
- Tewksbury, J.J.; Nabhan, G.P. Directed deterrence by capsaicin in chillies. Nature 2001, 412, 403–404. [Google Scholar] [CrossRef]
- Mankowski, C.; Poole, C.D.; Ernault, E.; Thomas, R.; Berni, E.; Currie, C.J.; Treadwell, C.; Calvo, J.I.; Plastira, C.; Zafeiropoulou, E.; et al. Effectiveness of the capsaicin 8% patch in the management of peripheral neuropathic pain in European clinical practice: The ASCEND study. BMC Neurol. 2017, 17, 80. [Google Scholar] [CrossRef]
- Cserhati, T.; Forgacs, E.; Morais, M.H.; Mota, T.; Ramos, A. Separation and quantitation of colour pigments of chili powder (Capsicum frutescens) by high-performance liquid chromatography–diode array detection. J. Chromatogr. A 2000, 896, 69–73. [Google Scholar] [CrossRef]
- Li, B.; Yang, M.; Shi, R.; Ye, M. Insecticidal activity of natural capsaicinoids against several agricultural insects. Nat. Prod. Commun. 2019, 14, 1–7. [Google Scholar] [CrossRef]
- Kantar, M.B.; Anderson, J.E.; Lucht, S.A.; Mercer, K.; Bernau, V.; Case, K.A.; Le, N.C.; Frederiksen, M.K.; DeKeyser, H.C.; Wong, Z.; et al. Vitamin variation in Capsicum spp. provides opportunities to improve nutritional value of human diets. PLoS ONE 2016, 11, e0161464. [Google Scholar] [CrossRef]
- Olatunji, T.L.; Afolayan, A.J. The suitability of chili pepper (Capsicum annuum L.) for alleviating human micronutrient dietary deficiencies: A review. Food Sci. Nutr. 2018, 6, 2239–2251. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. Crops and Livestock Products. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 13 July 2022).
- Food and Agriculture Organization of the United Nations. FAOSTAT. Available online: http://faostat.fao.org/site/567/default.aspx#ancor (accessed on 2 January 2022).
- Abewoy, D. Review on impacts of climate change on vegetable production and its management practices. Adv. Crop Sci. 2018, 6, 330. [Google Scholar] [CrossRef]
- Cook, A.A.; Stall, R.E. Differentiation of pathotypes among isolates of Xanthomonas vesicatoria. Plant Dis. Rep. 1969, 53, 617–622. [Google Scholar]
- Goode, M.; Sasser, M. Prevention-The key to controlling bacterial spot and bacterial speck of tomato. Plant Dis. 1980, 64, 831–834. [Google Scholar] [CrossRef]
- Pajcin, I.S.; Vlaijkov, V.R.; Cvetkovic, D.D.; Ignjatov, M.V.; Grahovac, M.S.; Vucurovic, D.G.; Grahovac, J.A. Selection of antagonists for biocontrol of Xanthomonas euvesicatoria. Acta Period. Technol. 2020, 51, 181–189. [Google Scholar] [CrossRef]
- Roach, R.; Mann, R.; Gambley, C.G.; Shivas, R.G.; Rodoni, B. Identification of Xanthomonas species associated with bacterial leaf spot of tomato, capsicum, and chilli crops in eastern Australia. Eur. J. Plant Pathol. 2018, 150, 595–608. [Google Scholar] [CrossRef]
- Schwartz, A.R.; Potnis, N.; Timilsina, S.; Wilson, M.; Patane, J. Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front. Microbiol. 2015, 6, 535. [Google Scholar] [CrossRef]
- Noble, T.J.; Young, A.J.; Barrerro, R.; Douglas, C.A.; Kelly, L.A.; Williams, B.; Mundree, S. Characterisation of the Pseudomonas savastanoi pv. phaseolicola population found in Eastern Australia associated with halo blight disease in Vigna radiata. Australas. Plant Pathol. 2020, 49, 515–524. [Google Scholar] [CrossRef]
- Doidge, E.M. A tomato canker. Ann. Appl. Biol. 1921, 7, 407–430. [Google Scholar] [CrossRef]
- Gardner, M.W.; Kendrick, J.B. Bacterial spot of tomato. J. Agric. Res. 1921, 21, 123–156. [Google Scholar]
- Dowson, W.J. On the systematic position and generic names of the gram-negative bacterial plant pathogens. Zent. Fur Bakteriol. Parasitenkd. Und Infekt. 1939, 2, 177–193. [Google Scholar]
- Young, J.M.; Dye, D.W.; Bradbury, J.F.; Panagopoulos, C.G.; Robbs, C.F. A proposed nomenclature and classification for plant pathogenic bacteria. N. Z. J. Agric. Res. 1978, 21, 153–177. [Google Scholar] [CrossRef]
- Vauterin, L.; Hoste, B.; Kersters, K.; Swings, J. Reclassification of Xanthomonas. Int. Syst. Ecol. Microbiol. 1995, 45, 472–489. [Google Scholar] [CrossRef]
- Šutic, D. Bakterioze crvenog patlidzana (Tomato Bacteriosis); Posebna Izd Inst Zastitu Bilja Beograd (Special Edition); Institut za zaštitu bilja: Beograd, Serbia, 1957; Volume 6, pp. 1–65. [Google Scholar]
- Jones, J.B.; Lacy, G.H.; Bouzar, H.; Stall, R.E.; Shcaad, N.W. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 2004, 27, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Timilsina, S.; Kara, S.; Jacques, M.A.; Potnis, N.; Minsavage, G.V.; Vallad, G.E.; Jones, J.B.; Saux, M.F. Reclassification of Xanthomonas gardneri (ex Šutič 1957) Jones et al. 2006 as a later heterotypic synonym of Xanthomonas cynarae Trébaol et al. 2000 and description of X. cynarae pv. cynarae and X. cynarae pv. gardneri based on whole genome analyses. Microbiol. Soc. 2018, 69, 792–806. [Google Scholar] [CrossRef]
- Moriniere, L.; Burlet, A.; Rosenthal, E.R.; Nesme, X.; Portier, P.; Bull, C.T.; Lavire, C.; Saux, M.F.; Bertolla, F. Clarifying the taxonomy of the causal agent of bacterial leaf spot of lettuce through a polyphasic approach reveals that Xanthomonas cynarae Trébaol et al. 2000 emend. Timilsina et al. 2019 is a later heterotypic synonym of Xanthomonas hortorum Vauterin et al. 1995. Syst. Appl. Microbiol. 2020, 43, 126087. [Google Scholar] [CrossRef] [PubMed]
- Stall, R.E.; Jones, J.B.; Minsavage, G.V. Durability of resistance in tomato and pepper to Xanthomonads causing bacterial spot. Ann. Rev. Phytopathol. 2009, 47, 265–284. [Google Scholar] [CrossRef]
- Keen, N.; Kobayashi, D.; Tamaki, S.; Shen, H.; Stayton, M.; Lawrence, D.; Sharma, A.; Midland, S.; Smith, M.; Sims, J. Avirulence gene D from Pseudomonas syringae pv. tomato and its interaction with resistance gene Rpg4 in soybean. In Current Plant Science and Biotechnology in Agriculture; Hennecke, H., Verma, D.P., Eds.; Springer-Science: Davis, CA, USA, 1991; pp. 37–44. [Google Scholar]
- Cook, A.A.; Guevara, Y.G. Hypersensitivity in Capsicum chacoense to race 1 or the bacterial spot pathogen of pepper. Plant Dis. 1984, 68, 329–330. [Google Scholar] [CrossRef]
- Sahin, F.; Miller, S.A. Resistance in Capsicum pubescens to Xanthomonas campestris pv. vesicatoria pepper race 6. Plant Dis. 1998, 82, 794–799. [Google Scholar] [CrossRef]
- Kousik, C.S.; Ritchie, D.F. Isolation of pepper races 4 and 5 of Xanthomonas campestris pv. vesicatoria from diseased pepper in southeastern U.S fields. Plant Dis. 1995, 79, 540. [Google Scholar] [CrossRef]
- Gambley, C. Management and detection of bacterial leaf spot in capsicum and chili crops. Hort Innovation. Available online: https://www.horticulture.com.au/globalassets/laserfiche/assets/project-reports/vg14010/vg14010---final-report-complete.pdf (accessed on 4 April 2022).
- Ivey, M.L.L.; Strayer, A.; Sidhu, J.K.; Minsavage, G.V. Bacterial leaf spot of bell pepper (Capsicum annuum) in Louisiana is caused by Xanthomonas euvesicatoria Pepper Races 1 and 3. Dis. Not. 2016, 100, 853. [Google Scholar] [CrossRef]
- Burlakoti, R.R.; Hsu, C.; Chen, J.; Wang, J. Population dynamics of Xanthomonads associated with bacterial spot of tomato and pepper during 27 years across Taiwan. Plant Dis. 2018, 102, 1348–1356. [Google Scholar] [CrossRef] [PubMed]
- Garton, J.E. Evaluation of Race and Copper Tolerant Strains of Xanthomonas axonopodis pv. vesicatoria, Causal Agent of Bacterial Leaf Spot of Bell Pepper in Georgia. Master’s Thesis, University of Georgia, Athens, GA, USA, 2009. [Google Scholar]
- An, S.; Potnis, N.; Dow, M.; Vorholter, F.; He, Y.; Becker, A.; Teper, D.; Li, Y.; Wang, N.; Bleris, L.; et al. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 2020, 44, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Hugouvieux, V.; Barber, C.E.; Daniels, M.J. Entry of Xanthomonas campestris pv. Campestris into Hydathodes of Arabidopsis thaliana Leaves: A System for Studying Early Infection Events in Bacterial Pathogenesis. Mol. Plant Microbe Interact. 1998, 11, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Almeida, R.P.P.; Lindow, S. Living in two worlds: The plant and insect lifestyles of Xylella fastidiosa. Annu. Rev. Phytopathol. 2008, 46, 243–271. [Google Scholar] [CrossRef]
- Ryan, R.P.; Vorholter, F.; Potnis, N.; Jones, J.B.; Sluys, M.V.; Bogdanove, A.J.; Dow, J.M. Pathogenomics of Xanthomonas: Understanding bacterium-plant interactions. Nat. Rev. Microbiol. 2011, 9, 344–355. [Google Scholar] [CrossRef]
- Petrocelli, S.; Tondo, M.L.; Daurelio, L.D.; Orellano, E.G. Modifications of Xanthomonas axonopodis pv. citri lipopolysaccharide affect the basal response and the virulence process during citrus canker. PLoS ONE 2012, 7, e40051. [Google Scholar] [CrossRef]
- Silipo, A.; Molinaro, A.; Lanzetta, R.; Parrilli, M. The structures of the lipid A moieties from the lipopolysaccharides of two phytopathogenic bacteria, Xanthomonas campestris pv. pruni and Xanthomonas fragariae. Eur. J. Org. Chem. 2004, 6, 1336–1343. [Google Scholar] [CrossRef]
- Castiblanco, L.F.; Sundin, G.W. New insights on molecular regulation of biofilm formation in plant-associated bacteria. J. Integr. Plant Biol. 2016, 58, 362–372. [Google Scholar] [CrossRef]
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Vojnov, A.A.; Slater, H.; Daniels, M.J.; Dow, J.W. Expression of the gum operon directing xanthan biosynthesis in Xanthomonas campestris and its regulation in planta. Mol. Plant Microbe Interact. 2001, 14, 768–774. [Google Scholar] [CrossRef]
- Katzen, F.; Becker, A.; Zorreguieta, A.; Puhler, A.; Lelpi, L. Promoter analysis of the Xanthomonas campestris pv. campestris gum operon directing biosynthesis of the xanthan polysaccharide. J. Bacteriol. 1996, 178, 4313–4318. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; Vik, A.; Friedman, L.; Kolter, R. Biofilms: The matrix revisited. Trends Microbiol. 2005, 13, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Moreira, L.M.; Facincani, A.P.; Ferreira, C.B.; Ferreira, R.M.; Ferro, M.I.T.; Gozzo, F.C.; de Oliveira, J.C.F.; Ferro, J.A.; Soares, M.R. Chemotactic signal transduction and phosphate metabolism as adaptive strategies during citrus canker induction by Xanthomonas citri. Funct. Integr. Genom. 2015, 2, 197–210. [Google Scholar] [CrossRef]
- Bashan, Y.; Diab, S.; Okon, Y. Survival of Xanthomonas campestris pv. vesicatoria in pepper seeds and roots in symptomless and dry leaves in non-host plants and in the soil. Plant Soil. 1982, 68, 161–170. [Google Scholar]
- Osdaghi, E.; Jones, J.B.; Sharma, A.; Goss, E.M.; Abrahamian, P.; Newberry, E.A.; Potnis, N.; Carvalho, R.; Choudhary, M.; Paret, M.L.; et al. A centenary for bacterial spot of tomato and pepper. Mol. Plant Pathol. 2021, 22, 1500–1519. [Google Scholar] [PubMed]
- European and Mediterranean Plant Protection Organization. PM 7/110 (1) Xanthomonas spp. (Xanthomonas euvesicatoria, Xanthomonas gardneri, Xanthomonas perforans, Xanthomonas vesicatoria) causing bacterial spot of tomato and sweet pepper. Bull. OEPP/EPPO Bull. 2013, 43, 7–20. [CrossRef]
- Young, A.J.; Marney, T.S.; Herrington, M.; Hutton, D.; Gomez, A.O.; Villiers, A.; Campbell, P.R.; Geering, A.D.W. Outbreak of angular leaf spot in an Australian strawberry germplasm collection. Australas. Plant Pathol. 2011, 40, 286–292. [Google Scholar]
- Bouzar, H.; Jones, J.B.; Stall, R.E.; Somodi, G.C.; Kelly, R.O.; Daouzli, N. Phenotypic characterization of Xanthomonas campestris pv. vesicatoria strains from the Caribbean and Central America. Phytopathology 1994, 84, 1069. [Google Scholar]
- Jones, J.B.; Somodi, G.C.; Scott, J.W. Increased ELISA sensitivity using a modified extraction buffer for detection of Xanthomonas campestris pv. vesicatoria in leaf tissue. J. Appl. Microbiol. 1997, 83, 397–401. [Google Scholar] [CrossRef]
- Massomo, S.M.S.; Nielsen, H.; Mabagala, R.B.; Mansfeld-Giese, K.; Hockenhull, J.; Mortensen, C.N. Identification and characterisation of Xanthomonas campestris pv. campestris strains from Tanzania by pathogenicity tests, Biolog, rep-PCR and fatty acid methyl ester analysis. Eur. J. Plant Pathol. 2003, 109, 775–789. [Google Scholar] [CrossRef]
- Bouzar, H.; Jones, J.B.; Minsavage, G.V.; Stall, R.E.; Scoot, J.W. Proteins unique to phenotypically distinct groups of Xanthomonas campestris pv. vesicatoria revealed by silver staining. Phytopathology 1994, 84, 39–44. [Google Scholar] [CrossRef]
- Jones, J.B.; Bouzar, H.; Stall, R.E.; Almira, E.C.; Roberts, P.D.; Bowen, B.W.; Sudberry, J.; Strickler, P.M.; Chun, J. Systematic analysis of xanthomonads (Xanthomonas spp.) associated with pepper and tomato lesions. Int. J. Syst. Evol. 2000, 50, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Leite, R.P.; Minsavage, G.V.; Bonas, U.; Stall, R.E. Detection and identification of phytopathogenic Xanthomonas strains by amplification of DNA sequences related to the hrp genes of Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 1994, 60, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Moretti, C.; Amatulli, M.T.; Buonaurio, R. PCR-based assay for the detection of Xanthomonas euvesicatoria causing pepper and tomato bacterial spot. Lett. Appl. Microbiol. 2009, 49, 466–471. [Google Scholar] [CrossRef]
- Park, D.S.; Shim, J.K.; Kim, J.S.; Lim, C.K.; Shrestha, R.; Hahn, J.H.; Kim, H.G. Sensitive and specific detection of Xanthomonas campestris pv. vesicatoria by PCR using pathovar-specific primers based on rhs family gene sequences. Microbiol. Res. 2009, 164, 36–42. [Google Scholar] [CrossRef]
- Koenraadt, H.; Betteray, B.; Germain, R.; Hiddink, G.; Jones, J.B.; Oosterhof, J.; Rijlaarsdam, A.; Roorda, P.; Woudt, B. Development of specific primers for the molecular detection of bacterial spot of pepper and tomato. Acta Hortic. 2009, 808, 99–102. [Google Scholar] [CrossRef]
- Strayer-Scherer, A.; Jones, J.B.; Paret, M.L. Recombinase polymerase amplification assay for field detection of tomato bacterial spot pathogens. Phytopathology 2019, 109, 690–700. [Google Scholar] [CrossRef]
- Larrea-Sarmiento, A.; Dhakal, U.; Boluk, G.; Fatdal, L.; Alvarez, A.; Strayer-Scherer, A.; Paret, M.; Jones, J.; Jenkins, D.; Arif, M. Development of a genome-informed loop-mediated isothermal amplification assay for rapid and specific detection of Xanthomonas euvesicatoria. Sci. Rep. 2018, 8, 14298. [Google Scholar] [CrossRef]
- Stehlikova, D.; Beran, P.; Cohen, S.P.; Curn, V. Development of real-time and colorimetric loop mediated isothermal amplification assay for detection of Xanthomonas gardneri. Microorganisms 2020, 8, 1301. [Google Scholar] [CrossRef]
- Pecenka, J.; Kacanova, M.; Baranek, M.; Gazdik, F.; Ragasova, L.; Penazova, E.; Cechova, J.; Beran, P.; Eichmeier, A. Species-specific PCR primers for the detection of poorly distinguishable Xanthomonas euvesicatoria. Crop Prot. 2020, 127, 104978. [Google Scholar] [CrossRef]
- Liu, H.; Dong, C.; Zhao, T.; Han, J.; Wang, T.; Wen, X.; Huang, Q. Functional analysis of the ferric uptake regulator gene fur in Xanthomonas vesicatoria. PLoS ONE 2016, 11, e0149280. [Google Scholar] [CrossRef][Green Version]
- Beran, P.; Mraz, I. Species-specific PCR primers for detection of Xanthomonas vesicatoria. Crop Prot. 2012, 43, 213–215. [Google Scholar] [CrossRef]
- Albuquerque, P.; Caridade, C.M.R.; Rodrigues, A.S.; Marcal, A.R.S.; Cruz, J.; Cruz, L.; Santos, C.L.; Mendes, M.V.; Tavares, F. Evolutionary and experimental assessment of novel markers for detection of Xanthomonas euvesicatoria in plant samples. PLoS ONE 2012, 7, e37836. [Google Scholar] [CrossRef]
- Boudon, S.; Manceau, C.; Notteghem, J. Structure and origin of Xanthomonas arboricola pv. pruni populations causing bacterial spot of stone fruit trees in Western Europe. APS 2005, 95, 1081–1088. [Google Scholar] [CrossRef]
- Hamza, A.A.; Robene-Soustrade, I.; Jouen, E.; Lefeuvre, P.; Chiroleu, F.; Fisher-Le Saux, M.; Gagnevin, L.; Provust, O. Multilocus sequence analysis and amplified fragment length polymorphism-based characterization of xanthomonads associated with bacterial spot of tomato and pepper and their relatedness to Xanthomonas species. Syst. Appl. Microbiol. 2012, 35, 183–190. [Google Scholar] [CrossRef]
- Dutta, B.; Gitaitis, R.; Sanders, H.; Booth, C.; Smith, S.; Langston, D.B., Jr. Role of blossom colonization in pepper seed infestation by Xanthomonas euvesicatoria. Phytopathology 2014, 104, 232–239. [Google Scholar] [CrossRef]
- Dutta, B.; Langston, D.B.; Luo, X.; Carlson, S.; Kichler, J.; Gitaitis, R. A risk assessment model for bacterial leaf spot of pepper (Capsicum annuum), caused by Xanthomonas euvesicatoria, based on concentrations of macronutrients, micronutrients, and micronutrient ratios. Phytopathology 2017, 107, 1331–1338. [Google Scholar] [CrossRef]
- Johnson, G.F. The early history of copper fungicides. Agric. Hist. 1935, 9, 67–79. [Google Scholar]
- Conover, R.A.; Gerhold, N.R. Mixtures of copper and maneb or mancozeb for control of bacterial spot of tomato and their compatibility for control fungus diseases. Proc. Annu. Meet. Fla. State Hort. Soc. 1981, 94, 154–156. [Google Scholar]
- Marco, G.M.; Stall, R.E. Control of bacterial spot of pepper initiated by strains of Xanthomonas campestris pv. vesicatoria that differ in sensibility to copper. Plant Dis. 1983, 67, 779–781. [Google Scholar] [CrossRef]
- Li, J.; Wang, N. Foliar application of biofilm formation-inhibiting compounds enhances control of citrus canker caused by Xanthomonas citri subsp. citri. Phytopathology 2014, 104, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, S.; Huang, Y.; Jones, J. Evaluation of a small molecule compound 3-indolylacetonitrile for control of bacterial spot-on tomato. Crop Prot. 2019, 120, 7–12. [Google Scholar] [CrossRef]
- Qiao, K.; Liu, Q.; Huang, Y.; Xia, Y.; Zhang, S. Management of bacterial spot of tomato caused by copper-resistant Xanthomonas perforans using a small molecule compound carvacrol. Crop Prot. 2020, 132, 105114. [Google Scholar] [CrossRef]
- Abbasi, P.A.; Soltani, N.; Cuppels, D.A.; Lazarovits, G. Reduction of bacterial spot disease severity on tomato and pepper plants with foliar applications of ammonium lignosulfonate and potassium phosphate. Plant Dis. 2002, 86, 1232–1236. [Google Scholar]
- Carvalho, R.; Duman, K.; Jones, J.B.; Paret, M.L. Bactericidal activity of copper-ziinc hybrid nanoparticles on copper-tolerant Xanthomonas perforans. Sci. Rep. 2019, 9, 20124. [Google Scholar] [CrossRef]
- Srivastava, V.; Deblais, L.; Kathayat, D.; Rotondo, F.; Helmy, Y.A.; Miller, S.A.; Rajashekara, G. Novel small molecule growth inhibitors of Xanthomonas spp. causing bacterial spot of tomato. Phytopathology 2021, 111, 940–953. [Google Scholar] [CrossRef]
- He, D.C.; He, M.H.; Amalin, D.M.; Liu, W.; Alvindia, D.G.; Zhan, J. Biological control of plant diseases: An evolutionary and eco-economic consideration. Pathogens 2021, 10, 1311. [Google Scholar] [CrossRef]
- Pajcin, I.; Vlajkov, V.; Frohme, M.; Grebinyk, S.; Grahovac, M.; Mojicevic, M.; Grahovac, J. Pepper bacterial spot control by Bacillus velezensis: Bioprocess solution. Microorganisms 2020, 8, 1463. [Google Scholar] [CrossRef]
- Yi, H.; Yang, J.; Ryu, C. ISR meets SAR outside: Additive action of the endophyte Bacillus pumilus INR7 and the chemical inducer, benzothiadiazole, on induced resistance against bacterial spot in field-grown pepper. Front. Plant Sci. 2013, 14, 122. [Google Scholar] [CrossRef]
- Da Silva, A.P.S.; Olivares, F.L.; Sudre, C.P.; Peres, L.E.P.; Canellas, N.A.; da Silva, R.M.; Cozzolino, V.; Canellas, L.P. Attenuations of bacterial spot disease Xanthomonas euvesicatoria on tomato plants treated with biostimulants. Chem. Biol. Technol. 2021, 8, 42. [Google Scholar] [CrossRef]
- Le, K.D.; Kim, J.; Yu, N.H.; Kim, B.; Lee, C.W.; Kim, J. Biological control of tomato bacterial wilt, kimchi cabbage soft rot, and red pepper bacterial leaf spot using Paenibacillus elgii JCK-5075. Front. Plant Sci. 2020, 11, 775. [Google Scholar] [CrossRef] [PubMed]
- Moss, W.P.; Byrne, J.M.; Campbell, H.L. Biological control of bacterial spot of tomato using hrp mutants of Xanthomonas campestris pv. vesicatoria. Biol. Control. 2007, 41, 199–206. [Google Scholar] [CrossRef]
- Moineau, S. Bacteriophage. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 280–283. [Google Scholar]
- Flaherty, J.E.; Jones, J.B.; Harbaugh, B.K.; Somodi, G.C.; Jackson, L.E. Control of bacterial spot on tomato in the greenhouse and field with H-mutant bacteriophages. J. Am. Soc. Hortic. Sci. 2000, 35, 882–884. [Google Scholar] [CrossRef]
- Gasic, K.; Ivanovic, M.M.; Ignjatov, M.; Calic, A.; Obradovic, A. Isolation and characterization of Xanthomonas euvesicatoria bacteriophages. Plant Pathol. J. 2011, 93, 415–423. [Google Scholar]
- Gasic, K.; Kuzmanovic, N.; Ivanovic, M.; Prokic, A.; Sevic, M.; Obradovic, A. Complete genom of the Xanthomonas euvesicatoria specific bacteriophage KФ1, its survival and potential in control of pepper bacterial spot. Front. Microbiol. 2018, 9, 2021. [Google Scholar] [CrossRef]
- Solis-Sanchez, G.A.; Quinones-Aguilar, E.E.; Fraire-Velasquez, S.; Vega-Arreguin, J.; Rincon-Enriquez, G. Complete genome sequence of XaF13, a novel bacteriophage of Xanthomonas vesicatoria from Mexico. Microbiol. Resour. Announc. 2020, 9, e01371-19. [Google Scholar] [CrossRef]
- Rios-Sandoval, M.; Quinones-Aguilar, E.E.; Solis-Sanchez, G.A.; Enriquez-Vara, J.N.; Rincon-Enriquez, G. Complete genome sequence of Xanthomonas vesicatoria bacteriophage ФXaF18, a contribution to the biocontrol of bacterial spot of pepper in Mexico. Microbiol. Resour. Announc. 2020, 9, e00213-20. [Google Scholar] [CrossRef]
- Omnilytics. Find a Natural, Safe, organic, Eco-Friendly, and Effective Pesticide. Available online: https://www.agriphage.com/product-info/ (accessed on 10 January 2022).
- Sevic, M.; Gasic, K.; Ignjatov, M.; Mijatovic, M.; Prokic, A.; Obradovic, A. Integration of biological and conventional treatments in control of pepper bacterial spot. Crop Prot. 2019, 119, 46–51. [Google Scholar] [CrossRef]
- Restrepo, M.; Babu, B.L.; Reyes, L.F.; Chalmers, J.D.; Soni, N.J.; Sibila, O.; Faverio, P.; Cilloniz, C.; Rodriguez-Cintron, W.; Aliberti, S. Burden and risk factors for Pseudomonas aeruginosa community-acquired pneumonia: A multinational point prevalence study of hospitalised patients. Eur. Respir. J. 2018, 52, 1701190. [Google Scholar] [CrossRef]
- Keswani, C.; Prakash, O.; Bharti, N.; Vílchez, J.I.; Sansinenea, E.; Lally, R.D.; Borriss, R.; Singh, S.P.; Gupta, V.K.; Fraceto, L.F.; et al. Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci. Total Environ. 2019, 690, 841–852. [Google Scholar] [CrossRef]
- Vilchez, J.I.; Navas, A.; Gonzales-Lopez, J.; Arcos, S.C.; Manzanera, M. Biosafety test for plant growth-promoting bacteria: Proposed environmental and human safety index (EHSI) protocol. Front. Microbiol. 2016, 6, 1514. [Google Scholar] [CrossRef] [PubMed]
- Pena, R.T.; Blasco, L.; Ambroa, A.; González-Pedrajo, B.; Fernández-García, L.; López, M.; Bleriot, I.; Bou, G.; García-Contreras, R.; Wood, T.K.; et al. Relationship between quorum sensing and secretion systems. Front. Microbiol. 2019, 10, 1100. [Google Scholar] [CrossRef] [PubMed]
- Timilsina, S.; Potnis, N.; Newberry, E.A.; Liyanapathiranage, P.; Iruegas-Bocardo, F.; White, F.F.; Goss, E.M.; Jones, J.B. Xanthomonas diversity, virulence and plant–pathogen interactions. Nat. Rev. Microbiol. 2020, 18, 415–427. [Google Scholar] [CrossRef]
- Abrahamian, P.; Timilsina, S.; Minsavage, G.V.; Suhmita, K.C.; Goss, E.M.; Jones, J.B.; Vallad, G.E. The type III effector AvrBst enhance Xanthomonas perforans fitness in field-grown tomato. Phytopathology 2018, 108, 1355–1362. [Google Scholar] [CrossRef] [PubMed]
- Potnis, N.; Krasileva, K.; Chow, V.; Almeida, N.F.; Patil, P.B.; Ryan, R.P.; Sharlach, M.; Berhlau, F.; Dow, J.M.; Momol, M.T.; et al. Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genom. 2011, 12, 146. [Google Scholar] [CrossRef]
- Liyanapathiranage, P.; Wagner, N.; Avram, O.; Pupko, T.; Potnis, N. Phylogenetic distribution and evolution of type VI secretion system in the Genus Xanthomonas. Front. Microbiol. 2022, 13, 840308. [Google Scholar] [CrossRef]
- Sole, M.; Scheibner, F.; Hoffmeister, A.; Hartman, N.; Hause, G.; Rother, A.; Jordan, M.; Lautier, M.; Arlat, M.; Buttner, D. Xanthomonas campestris pv. vesicatoria secretes proteases and xylanases via the Xps type II secretion system and outer membrane vesicles. J. Bacteriol. 2015, 197, 2879–2893. [Google Scholar] [CrossRef]
- Szczesny, R.; Jordan, M.; Schramm, C.; Schulz, S.; Cogez, V.; Bonas, U.; Buttner, D. Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria. New Pathologist. 2010, 187, 983–1002. [Google Scholar] [CrossRef]
- Barak, J.D.; Vancheva, T.; Lefeuvre, P.; Jones, J.B.; Timilsina, S.; Minsavage, G.V.; Vallad, G.E.; Koebnik, E. Whole-genome sequences of Xanthomonas euvesicatoria strains clarify taxonomy and reveal a stepwise erosion of Type 3 Effectors. Front. Plant Sci. 2016, 7, 1805. [Google Scholar] [CrossRef]
- Buttner, D.; Bonas, U. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol.-Microbiol. Rev. 2010, 34, 107–133. [Google Scholar] [CrossRef]
- Lu, H.; Patil, P.; Sluys, M.V.; White, F.F.; Ryan, R.P.; Dow, J.M.; Rabinowicz, P.; Salzberg, S.L.; Leach, J.E.; Sonti, R.; et al. Acquisition and evolution of plant pathogenesis-associated gene clusters and candidate determinants of tissue-specificity in Xanthomonas. PLoS ONE 2008, 3, e3828. [Google Scholar] [CrossRef]
- Dow, J.M.; Feng, J.; Barber, C.E.; Tang, J.; Daniels, M.J. Novel genes involved in the regulation of pathogenicity factor production within the rpf gene cluster of Xanthomonas campestris. Microbiology 2000, 146, 885–891. [Google Scholar] [CrossRef][Green Version]
- Teper, D.; Salomon, D.; Sunitha, S.; Kim, J.; Mudgett, M.B.; Sessa, G. Xanthomonas euvesicatoria type III effector XopQ interacts with tomato and pepper 13-4-4 isoforms to suppress effector-triggered immunity. Plant J. 2014, 77, 297–309. [Google Scholar] [CrossRef]
- Marois, E.; den Ackerveken, G.V.; Bonas, U. The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol. Plant Microbe Interact. 2002, 15, 637–646. [Google Scholar] [CrossRef]
- Szurek, B.; Rossier, O.; Hause, G.; Bonas, U. Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell. Mol. Microbiol. 2002, 46, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Figueiredo, F.; Jones, J.; Wang, N. HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv.citri. Mol. Plant Microbe Interact. 2011, 24, 649–661. [Google Scholar] [CrossRef]
- Popov, G.; Fraiture, M.; Brunner, F.; Sessa, G. Multiple Xanthomonas euvesicatoria type III effectors inhibit flg22-triggered immunity. Mol. Plant-Microbe. Interact. 2016, 29, 651–660. [Google Scholar] [CrossRef]
- Raffeiner, M.; Ustun, S.; Guerra, T.; Spinti, D.; Fitzner, M.; Sonnewald, S.; Baldermann, S.; Bornke, F. The Xanthomonas type-III effector XopS stabilizes CaWRKY40a to regulate defense responses and stomatal immunity in pepper (Capsicum annuum). Plant. Cell 2022, 34, 1684–1708. [Google Scholar] [CrossRef] [PubMed]
- Schulze, S.; Kay, S.; Buttner, D.; Egler, M.; Eschen-Lippold, L.; Hause, G.; Kruger, A.; Lee, J.; Muller, O.; Scheel, D.; et al. Analysis of new type III effectors from Xanthomonas uncovers XopB and XopS as suppressors of plant immunity. New Phytol. 2012, 195, 894–911. [Google Scholar] [CrossRef] [PubMed]
- Szczesny, R.; Büttner, D.; Escolar, L.; Schulze, S.; Seiferth, A.; Bonas, U. Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytolog. 2010, 187, 1058–1074. [Google Scholar] [CrossRef]
- Zhao, B.; Dahlbeck, D.; Krasileva, K.V.; Fong, R.W.; Staskawicz, B.J. Computational and biochemical analysis of the Xanthomonas effector avrbs2 and its role in the modulation of Xanthomonas type three effector delivery. PLoS Pathog. 2011, 7, e1002408. [Google Scholar] [CrossRef][Green Version]
- Ki, J.; Stork, W.; Mudget, M.B. Xanthomonas type iii effector xopd desumoylates tomato transcription factor slerf4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 2013, 13, 143–154. [Google Scholar] [CrossRef]
- Teper, D.; Girija, A.M.; Bosis, E.; Popov, G.; Savidor, A.; Sessa, G. The Xanthomonas euvesicatoria type III effector XopAU is an active protein kinase that manipulates plant MAP kinase signaling. PLoS Pathog. 2018, 14, e1006880. [Google Scholar] [CrossRef] [PubMed]
- Priller, J.P.R.; Reid, S.; Konein, P.; Dietrich, P.; Sonnewald, S. The Xanthomonas campestris pv. vesicatoria Type-3 Effector XopB Inhibits plant defence responses by interfering with ROS production. PLoS ONE 2016, 11, e0159107. [Google Scholar] [CrossRef]
- Kim, J.; Li, X.; Roden, J.A.; Taylor, K.W.; Aakre, C.D.; Su, B.; Lalonde, S.; Kirik, A.; Chen, Y.; Baranage, G.; et al. Xanthomonas T3S effector Xopn suppresses pamp-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell. 2009, 21, pp. 1305–1323. Available online: www.plantcell.org/cgi/doi/10.1105/tpc.108.063123 (accessed on 1 April 2022).
- Bartetzko, V.; Sonnewald, S.; Vogel, F.; Hartner, K.; Stadler, R.; Hammes, U.Z.; Bornke, F. The Xanthomonas campestris pv. vesicatoriaType III Effector Protein XopJ Inhibits Protein Secretion: Evidence for Interference with Cell Wall–Associated Defense Responses. Mol. Plant Microbe Interact. 2009, 22, 655–664. [Google Scholar] [CrossRef]
- Kousik, C.S.; Ritchie, D.F. Response of bell pepper cultivars to bacterial spot pathogen races that individually overcome major resistance genes. Plant Dis. 1998, 82, 181–186. [Google Scholar] [CrossRef]
- Horvath, D.M.; Pauly, M.H.; Hutton, S.F.; Vallad, G.E.; Scott, J.W.; Jones, J.B.; Stall, R.E.; Dahlbeck, D.; Staskawicz, B.J.; Tricoli, D.; et al. The pepper BS2 gene confers effective field resistance to bacterial leaf spot and yield enhancement in Florida tomatoes. Acta Hortic. 2015, 1069, 47–51. [Google Scholar] [CrossRef]
- Tai, T.H.; Dahlbeck, D.; Clark, E.T.; Gajiwala, P.; Pasion, R.; Whalen, M.C.; Stall, R.E.; Staskawicz, B.J. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 1999, 96, 14153–14158. [Google Scholar] [CrossRef]
- Jordan, T.; Romer, P.; Meyer, A.; Szczesny, R.; Pierre, M.; Piffanelli, P.; Bendahmane, A.; Bonas, U.; Lahaye, T. Physical delimitation of the pepper Bs3 resistance gene specifying recognition of the AvrBs3 protein from Xanthomonas campestris pv. vesicatoria. Theor. Appl. Genet. 2006, 113, 896–905. [Google Scholar] [CrossRef]
- Adhikari, P.; Adhikari, T.B.; Louws, F.J.; Panthee, D.R. Advances and challenges in bacterial spot resistance breeding in tomato (Solanum lycopersicum L.). Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef]
- Vallejos, C.E.; Jones, V.; Stall, R.E.; Jones, J.B.; Minsavage, G.V.; Schultz, D.C.; Rodrigues, E.; Olsen, L.E.; Mazourek, M. Characterization of two recessive gene controlling resistance to all races of bacterial spot in peppers. Theor. Appl. Genet. 2010, 121, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Kim, S.Y.; Paek, K.H.; Choi, D.; Park, J.M. Suppression of a CaCYP1, a novel cytochrome P450 gene, compromises the basal pathogen defense response of pepper plants. Biochem. Biophys. Res. Commun. 2006, 345, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Hwang, B.K. The pepper receptor-like cytoplasmic protein kinase CaPIK1 is involved in plant signalling of defence and cell-death responses. Plant J. 2011, 66, 642–655. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).