Abstract
Type 2 diabetes mellitus is characterized by insulin resistance in the liver. Insulin is not only involved in carbohydrate metabolism, it also regulates protein synthesis. This work describes the expression of proteins in the liver of a diabetic mouse and identifies the metabolic pathways involved. Twenty-week-old diabetic db/db mice were hepatectomized, after which proteins were separated by 2D-Polyacrylamide Gel Electrophoresis (2D-PAGE). Spots varying in intensity were analyzed using mass spectrometry, and biological function was assigned by the Database for Annotation, Visualization and Integrated Discovery (DAVID) software. A differential expression of 26 proteins was identified; among these were arginase-1, pyruvate carboxylase, peroxiredoxin-1, regucalcin, and sorbitol dehydrogenase. Bioinformatics analysis indicated that many of these proteins are mitochondrial and participate in metabolic pathways, such as the citrate cycle, the fructose and mannose metabolism, and glycolysis or gluconeogenesis. In addition, these proteins are related to oxidation–reduction reactions and molecular function of vitamin binding and amino acid metabolism. In conclusion, the proteomic profile of the liver of diabetic mouse db/db exhibited mainly alterations in the metabolism of carbohydrates and nitrogen. These differences illustrate the heterogeneity of diabetes in its different stages and under different conditions and highlights the need to improve treatments for this disease.
1. Introduction
Type 2 Diabetes Mellitus (T2DM) is one of the fastest growing diseases, currently reaching pandemic levels. Its development is closely associated with obesity [1,2]. Physiological alterations resulting from high calorie intake do not only affect just a few key proteins or other functionally important biomolecules, but homeostasis is also guaranteed by the robustness of molecular networks [3,4]. It is thus conceivable that identification of deregulated pathways and networks is a fruitful approach as a starting point for the analyses of molecular mechanisms and validation of potential disease biomarkers.
The search for novel proteins or pathways linked to complex human diseases has been facilitated by proteomic technologies. Several proteomic studies have provided global proteome profiles of tissue and organs from T2DM patients or mouse (Mus musculus) models, identifying protein profiles associated with the pathogenesis of T2DM and its complications in tissues, such as in the pancreas [5], the adipose tissue [6], in muscles [7] and in the liver [8,9,10,11,12,13,14]. The liver is involved in the natural history of the ongoing epidemics of T2DM due to its participation in increased glucose production and dysregulated lipoprotein metabolism [15]. Additionally, several proteins that are exclusively or predominantly secreted from the liver are now known to directly affect glucose and lipid metabolism. Impaired insulin signaling in the liver and adipose tissue results in increased endogenous glucose production, which leads to hyperglycemia [15]. However, the mechanisms involved in the pathophysiology of this disease have not yet been clarified, and there could be other proteins involved in this process.
Due to the ethical and technical drawbacks of conducting studies in humans, testing in animal models for biomedical research is a good option. For the realization of this work, we use the db/db mouse, which is characterized by a mutation in the leptin receptor. This alteration results in hyperphagia, obesity (at about the age of 4 weeks), hyperinsulinemia (at about the age of 2 weeks), and insulin resistance for a later development of hyperglycemia (4–8 weeks); usually these mice do not live beyond 32–40 weeks [16].
Furthermore, age is associated with immunosenescence and by a chronic inflammatory state which contributes to multiple diseases [17]. Specifically, over time, all tissues, in particular the adipose and liver, exhibit important alterations in global gene expression profiles, accompanied by changes in the inflammatory state, cellular stress, fibrosis, altered capacity for apoptosis, xenobiotic metabolism, normal cell-cycle control, and DNA replication. Therefore, these changes influence the development of many diseases, including diabetes [18,19]. In a previous study, our research team reported alterations in proteins and metabolic pathways related to carbohydrates and lipids in 10-week-old mice [14]. Given these evidences, we anticipated that high blood glucose in db/db mice would be sufficient to predict a change in expression profile of hepatic proteins, which are related to diabetes complications. Therefore, the aim of this study was to analyze the changes in protein expression of the 20-week-old diabetic mouse liver.
2. Results
To confirm the conditions of diabetic and control mice, blood glucose was determined. At the time of sacrifice, blood glucose levels in db/db mice were higher than in the wild-type mice (488.2 ± 147 vs. 152 ± 22.5 mg/mL, p < 0.01).
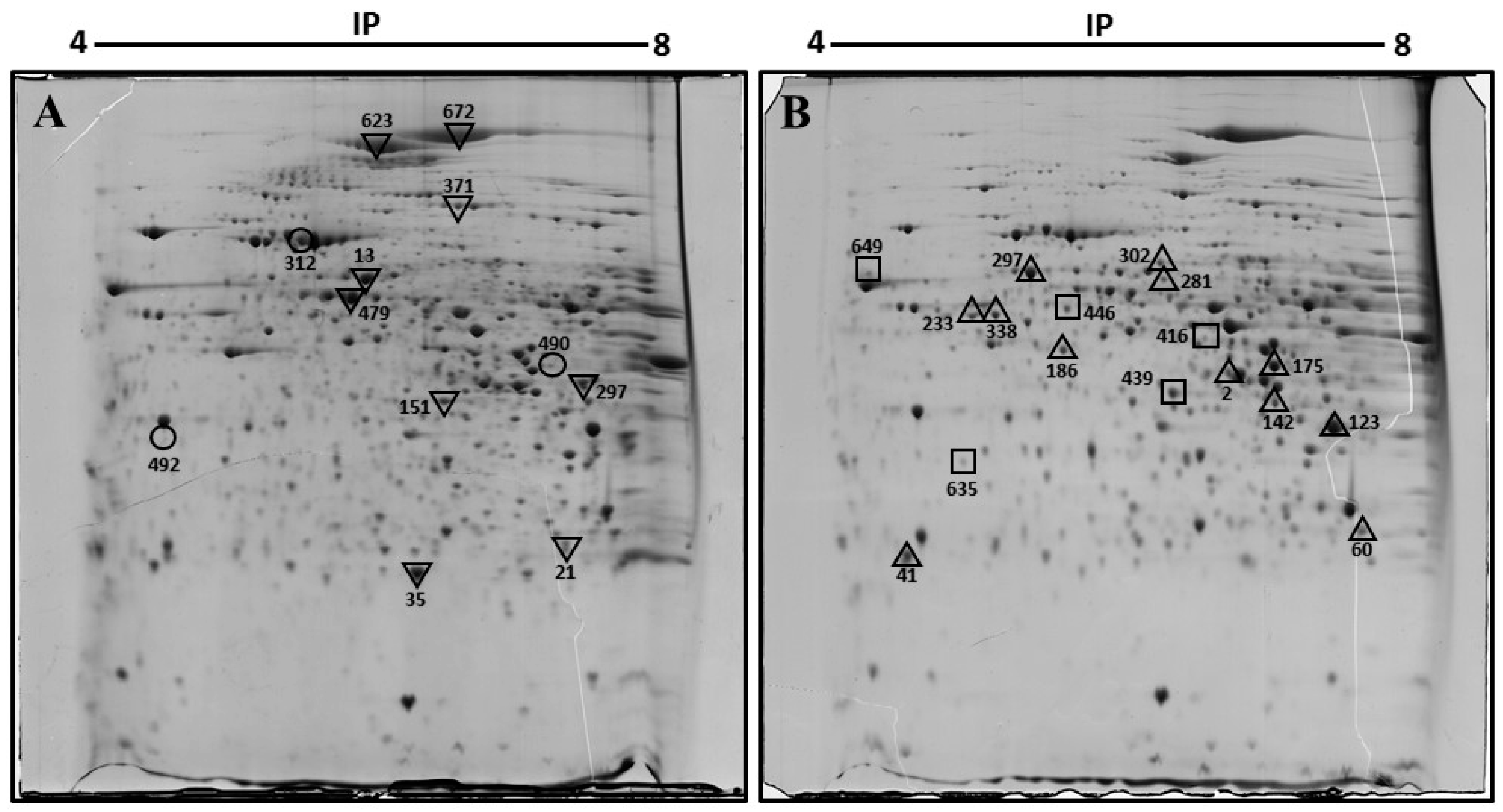
To identify the changes in protein expression associated with diabetes and obesity, we compared the expression profile of proteins in the liver of diabetic obese db/db mice versus non-diabetic mice using two-dimensional gel electrophoresis (2-DE). Three samples from liver homogenates obtained from each group were included for proteome analysis. Figure 1 displays representative 2D gel maps of liver proteins obtained from two groups. A total of approximately 600 spots were resolved on 2D gels. Spot volumes (area x intensity) were determined as a measure of protein expression using the ImageMaster 2D Platinum software. Spots that showed a 1.5-fold increase or decrease were included in the analysis, and proteins that are present in appreciable amounts were picked for identification by mass spectrometry (Figure 1).
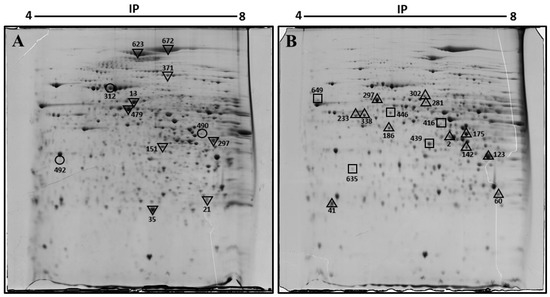
Figure 1.
Representative two-dimensional electrophoresis image of the liver of control (A) and diabetic obese db/db (B) mouse. Proteins were separated by isoelectric focusing (IEF) in the first dimension, pH 4–8 Linear, then by size in the second dimension. Spots found with differential expression are marked in the gels. In (A): ▽, diminished proteins in db/db mice. ○, disappeared proteins in db/db mice. In (B): △, increased proteins in db/db mice. □, appeared proteins in db/db mice liver. Spots 233 and 338 were the same protein, keratin, type I cytoskeletal 18.
The spots were excised from the gel, digested with trypsin, and identified by mass spectrometry. The results are shown in Table 1. Comprehensive analysis revealed that 29 spots registered a significant difference in diabetic hepatic tissue, 26 proteins were identified. Note that two spots were identified as the same protein, and that the identity of 3 spots could not be obtained. Of the 26 proteins identified, 12 protein spots were up-regulated and 8 were down-regulated in the liver samples of db/db mice compared with those of healthy mice. In addition, three spots appeared de novo, and in three other spots, expression was no longer detected in db/db mice.

Table 1.
Proteins differentially expressed in the db/db mouse liver.
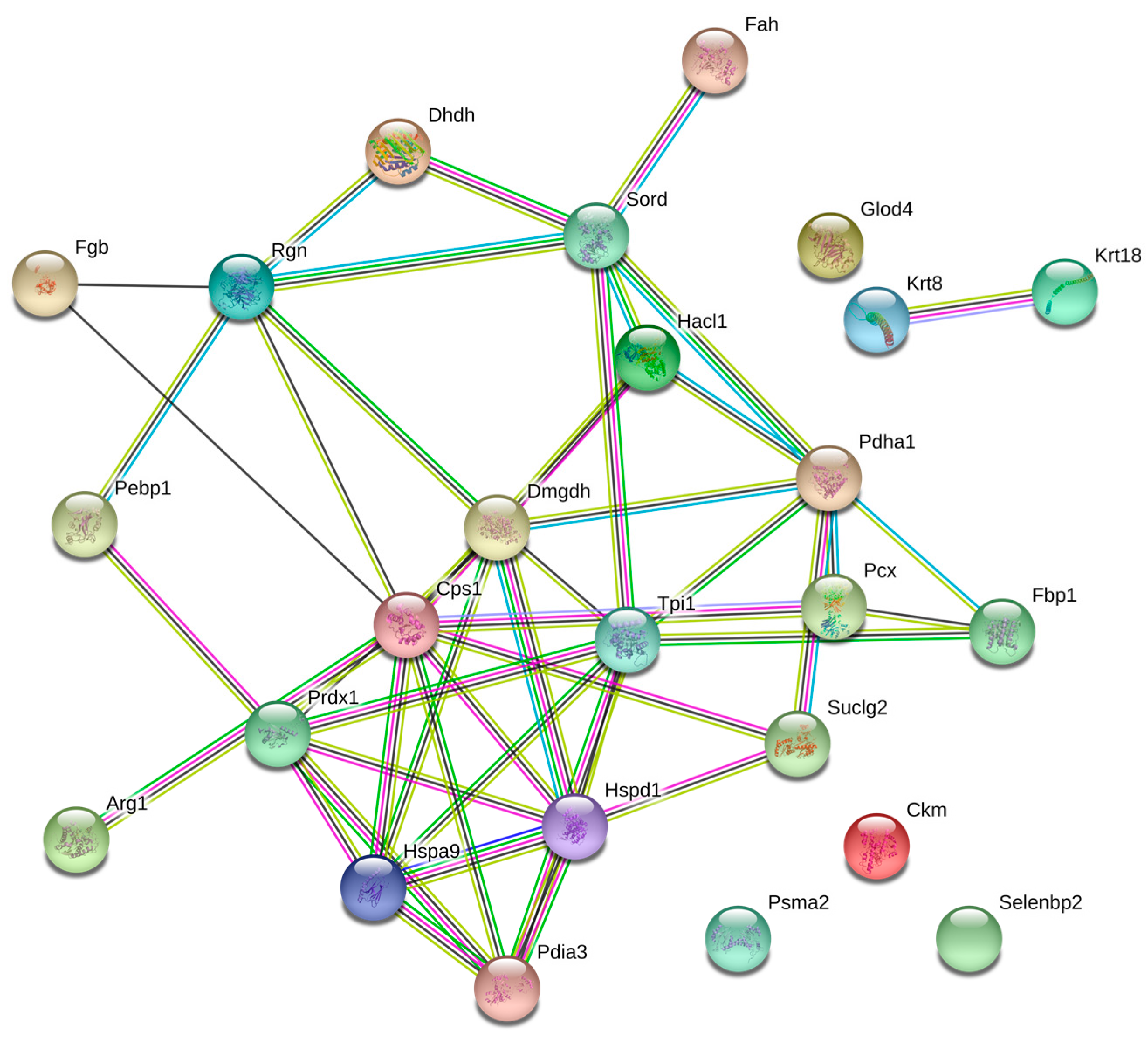
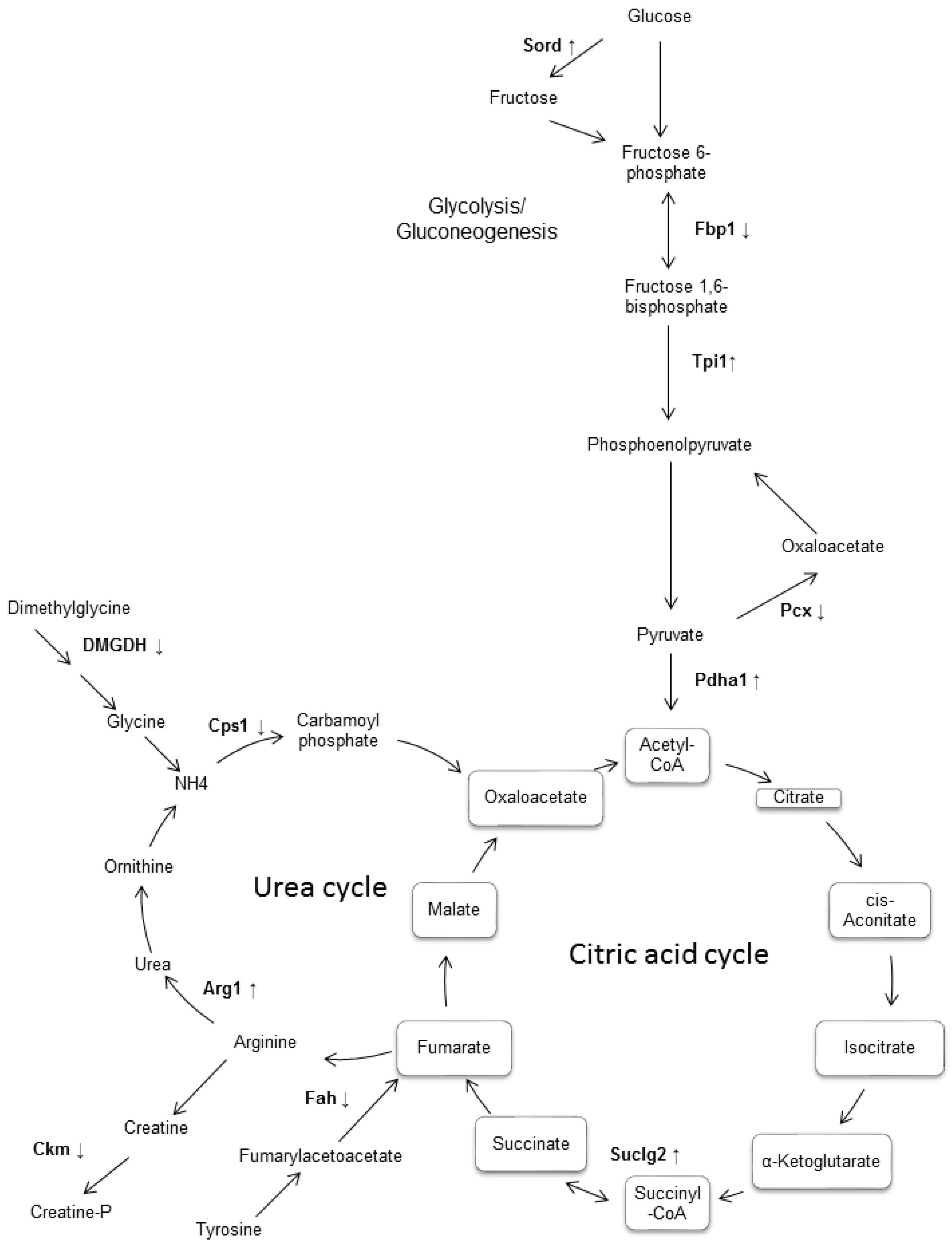
To achieve a better understanding of the involvement of these proteins in the biological processes related to the pathophysiology of diabetes, these molecules were uploaded in the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, with the purpose of inferring a network from these proteins, which is shown in Figure 2. Likewise, the altered proteins were classified using Gene Ontology (GO) database and Kyoto Encyclopedia of Genes and Genomes Pathway (KEEG) pathway. For this purpose, the Database for Annotation, Visualization and Integrated Discovery (DAVID) software was used. Classification of proteins by biological process indicated that most of proteins were associated with glucose metabolism, cellular amide metabolism, glutamine family amino acid metabolism, carboxylic acid catabolism, and oxidation-reduction processes (Figure 3).
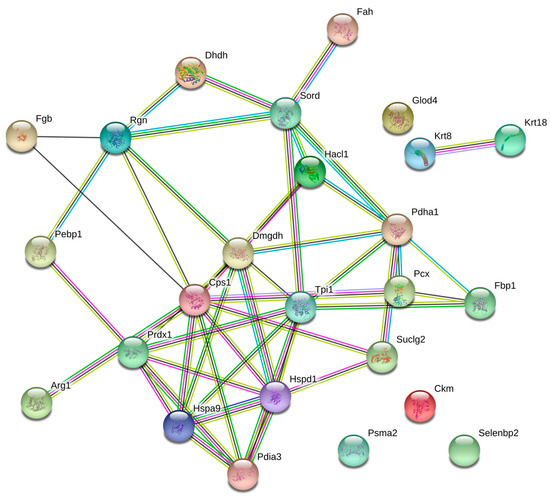
Figure 2.
Network of differentially expressed proteins. The nodes stand for differentially expressed genes, and the lines stand for the interactions between two proteins.
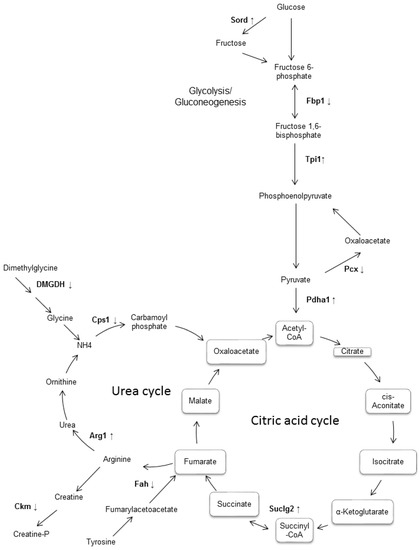
Figure 3.
Representative diagram of the main metabolic pathways which involved some of differentially expressed proteins in the liver of db/db diabetic mouse. It was observed that most of the differentially expressed enzymes were found to participate in the metabolic pathways of gluconeogenesis/glycolysis, as well as in the metabolism of fructose and in the citric acid cycle. Although no enrichment of the urea cycle was observed, this was included because it binds both to the citric acid cycle and to the metabolic pathways of the enriched amino acids found. The enzymes that were found differentially expressed in this work are shown in bold, with an arrow indicating if it was increased or decreased.
Protein classification by molecular function revealed that most proteins were implicated in purine nucleoside binding and vitamin binding. Classification of these proteins using GO database showed that differentially expressed proteins comprised mitochondrial proteins, especially matrix. Finally, to investigate pathways which were differentially expressed in diabetic mice livers, the KEEG pathway was used to sort and showed that most of these molecules are involved in the tricarboxylic acid cycle, the fructose and mannose metabolism, the arginine and proline metabolism, and glycolysis or gluconeogenesis (Table 2).

Table 2.
Enrichment analysis of metabolic pathways and biological function of differentially expressed proteins.
3. Discussion
This paper describes alterations found in the expression of 26 proteins identified in the liver of the diabetic mouse (db/db), which are associated with the metabolism of carbohydrates and amino acids, in addition to oxidation–reduction reactions. The findings in diabetic mice suggest that glucose metabolism is impaired in the liver of the 20 weeks-old db/db mice, more specifically in glycolysis and gluconeogenesis, the fructose and mannose metabolism, and the citrate cycle (Figure 3).
Regarding metabolic pathways of glycolysis and gluconeogenesis, we found three enzymes altered: fructose-1,6-bisphosphatase 1, pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial and triose phosphate isomerase 1, as well as pyruvate carboxylase mitochondrial, which together with the three previous enzymes are involved in the metabolic processes of glucose (GO:0006006). Previously, it has been reported that in diabetes and obesity gluconeogenesis is damaged [15,20]. Specifically, the proteins identified as altered in the present study have also been found to be associated with damage related to diabetes in previous studies. Indeed, fructose-1,6-bisphosphatase 1 has been associated with the regulation of appetite and adiposity [21]. Moreover, the finding of decreased expression of the pyruvate carboxylase enzyme may be because the model of diabetes we used begins to lower insulin levels after 16 weeks of age [22]. Thus, the mice used in this work would be in the process of losing weight. Contrary to our results, the enzyme triose phosphate isomerase 1 was found increased in a human cell line exposed to high concentrations of glucose, but interestingly, the expression of this enzyme decreased when the glucose concentration was increased even more [10], which would be consistent with our results. Moreover, the enzyme sorbitol dehydrogenase, which is responsible for forming sorbitol from glucose or fructose, was found to have increased. This result supports the reports claiming that pericytes cultured under high glucose levels overexpress this enzyme, producing reactive oxygen species [23], which is a determining factor in the process of insulin resistance. In addition, another research group reported that sorbitol dehydrogenase is decreased in the liver of mice with caloric restriction [24], which would support our results. Also, high levels of fructose lead to insulin resistance [25]. Another enzyme found differentially expressed was pyruvate dehydrogenase E1 component subunit alpha, somatic form, a mitochondrial enzyme responsible for transforming pyruvate into acetyl-CoA. The increase in expression matches earlier reports [13]. Succinate-CoA ligase [GDP-forming] subunit beta is mitochondrial and is involved in the citric acid cycle where it converts succinate to succinyl-CoA [13], an event that may be linked to the differential expression of other proteins found in this work, and which are part of the mitochondria (GO:0005739). This result confirms the fact that mitochondria have an important role in the pathophysiology of diabetes [26].
On the other hand, proteins that participate in the metabolism of nitrogen compounds were differentially expressed. Specifically, we found arginine and proline metabolism (mmu00330), as well as glutamine family amino acid metabolic processes (GO:0009064). These are also related to the urea cycle (Figure 3). Diabetes is traditionally considered a carbohydrate disease and to a lesser extent a lipidic one. Considering that insulin is the key anabolic regulator for peripheral protein metabolism, it stands to reason that blood amino acid availability will be altered in diabetic patients, as previously reported [27,28]. With respect to the metabolic pathway of arginine and proline metabolism, three enzymes involved were discovered: carbamoyl-phosphate synthase [ammonia], mitochondrial, creatine kinase and arginase-1. This finding reinforces an earlier report released in Akt1 +/− and Akt2 −/− mice [12]. The enzyme fumaryl-acetoacetase, in addition to participating in the glutamine family amino acid metabolic process, is also involved in apoptotic mechanisms in the liver, a phenomenon widely reported in diabetes [29,30].
Oxidation–reduction processes and diabetes have been extensively studied [31]. In this case, five altered proteins were identified: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase, dimethylglycine dehydrogenase, mitochondrial; peroxiredoxin-1, pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial and sorbitol dehydrogenase. Controversially, in a previous study peroxiredoxin-1 overexpression was found in the same model [32]. Such effect may be due to the age difference between the two models used in the experiments. In addition to redox processes, dimethylglycine dehydrogenase transforms dimethylglycine to sarcosine, which then becomes glycine and enters the urea cycle. Interestingly, low plasma levels of dimethylglycine were significantly associated with higher blood glucose levels [33]. Therefore, our finding about down-expression of dimethylglycine dehydrogenase supports the involvement of this enzyme in the development of T2DM.
Moreover, in this study it was found that vitamin binding is altered in the diabetic mouse. This molecular function was altered in the diabetic mouse liver through the participation of proteins 2-hydroxyacyl-CoA lyase 1, dimethylglycine dehydrogenase, and pyruvate carboxylase mitochondrial. Therefore, it is suggested that to prevent deficiencies and maintain health, the majority of diabetic individuals should receive daily vitamins at recommended doses from consumption of natural food sources and fortified foods [34]. Either way, 2-hydroxyacyl-CoA lyase 1, along with glyoxalase domain-containing protein 4 have been superficially studied and so far, their relationship with diabetes or obesity is not clear.
Another process closely related to diabetes is inflammation, which is also associated with oxidative stress [35,36]. An important molecule linking the latter two processes is the mitochondrial 60 kDa heat shock protein, also called HSP60, which is overexpressed during the oxidative stress, present in diabetes [35]. This protein binds toll-like receptors of immune cells and induces the inflammatory process through NF-κB [35], a molecule that although in this work was found not differentially expressed, was indeed found increased in a previous study in this type of mouse [37].
It has been proposed that overexpression of regucalcin blocks the breakdown of both glycogen and this protein, which together with selenium-binding protein 2 are down-expressed in ten week old mouse liver db/db [14]: this can reinforce the results presented in this paper. Finally, phosphatidylethanolamine binding protein 1, also known as RKIP1 appears to have an important role in the pathophysiology of diabetes. Based on previous report which demonstrated that inactivation of this protein results in enhanced beta cell and pancreatic growth [38].
Given that one of the main characteristics of diabetes is the presence of insulin resistance [1], it is to be expected that the proteins identified in this work show some association with this characteristic. In this context, it has been reported that three of the molecules identified with differential expression, carbamoyl-phosphate synthase; dimethylglycine dehydrogenase; and CPS1, fibrinogen beta chain. Previously, they have been associated with insulin resistance, specifically simple nucleotide polymorphisms [33,39,40]. Whereas in a murine model knockout of pyruvate dehydrogenase E1 component subunit alpha, a protein found to be increased in our study, cells were found to be more sensitive to insulin [41]. As well as other proteins, such as 60 kDa heat shock protein, peroxiredoxin-1, pyruvate carboxylase and regucalcin; they have been found increased in different models of insulin resistance [32,42,43,44].
There are some previous studies that address from a proteomics approach the effect of diabetes on the liver [8,9,10,11,12,13,14]. However, there is little similarity with the observed differential protein expression in these studies. This may be due to several factors, including organisms used: human [9] and cell lines [10]; also, the study of a specific organelle such as mitochondria [8], finding specific post translational modifications: protein lysine malonylation [11] and acetylation [13], as well as different organism genetically modified [12] or different age [14]. This variety of results can be interpreted as the effect of the heterogeneity and complexity of diabetes. We must also be cautious when interpreting these results, because it cannot be excluded that the alteration of some proteins may be due to the effect of time, and not exclusively to diabetes. Nonetheless, it should be noted that the main limitation of the present study is the lack of confirmation of differentially expressed proteins directly related to diabetes and obesity, by some other technique, such as western blotting, immunohistochemistry, or enzyme-linked immunosorbent assays. In addition, it would be advisable to use of mice of different age groups. Consequently, if we take into account these considerations, the realization of these studies in the future will allow us to better understand the pathophysiology of diabetes.
Another research group conducted a bioinformatics analysis of differentially expressed genes in the liver of patients with diabetes [45]. However, the metabolic pathways found in the study and those found by our team do not match. Differences may be due to the fact that Kutmon et al. analyzed gene expression, while we analyzed protein expression, as well as different species: humans and mice, respectively [45].
4. Materials and Methods
4.1. Animals
Heterozygote non-diabetic db/+ mice (BKS.Cg-m +/+ Leprdb/OlaHsd:), all black and lean, were purchased from Harlan Laboratories (Mexico City, Mexico); they were crossbred to get the db/db mice. All animal procedures were performed in accordance with Mexican legislation, NOM-062-ZOO-1999, and with the Guide for the care and use of laboratory animals of the National Institutes of Health (NIH, Bethesda, MD, USA). Mice were kept in the animal facility of the University of Guanajuato in Leon, Mexico under standard housing conditions with water and food ad libitum and with a 12 h:12 h light–dark cycle. Experimental procedures were approved by the Medical Science Department Ethics Committee at the University of Guanajuato, Mexico.
4.2. Sample Preparation and 2D-Polyacrylamide Gel Electrophoresis
Twenty-week-old males were selected and grouped in db/db and wild type mice (n = 3). Mice were sacrificed by cervical dislocation and then blood levels of glucose were determined using a commercial kit (Accutrend® GCT, Roche, San Francisco, CA, USA). The liver of mice was extracted and stored in: Tris/HCl 20 mM, 10 mM EDTA, 2 mM DTT, pH 7.8 and a protease inhibitor (20 mM PMSF), then stored at −70 °C until the sample was processed. Cellular proteins were obtained by a sample grinding kit (GE Healthcare, Deutsch, Switzerland) and resuspended in 200 μL of buffer (7 M urea, 4% CHAPS, 60 mM DTT, 2 mM TBP, 2 mM thiourea, 2% IPG). Protein concentrations were determined by 2D Quant Kit (GE Healthcare). Two-dimensional gel electrophoresis was performed as previously described [5]. The protein separation pattern was visualized in 2-D gels by colloidal Coomassie Brilliant Blue staining. The gels were scanned and digital images were analyzed and compared, in a GS-800 densitometer (Bio-Rad, Hercules, CA, USA) and the ImageMaster 2D Platinum 7.0 software (GE Healthcare) as previously described [14]. Spots were quantitatively compared between diabetic and normal control groups, using the liver protein samples from three diabetic mice and three control mice. Data are presented as fold change (increase or decrease) versus matched controls. Each experiment was performed in triplicate. Statistical differences between groups were calculated using ANOVA and fold changes ≥1.5 with p values ≤0.05 considered significant.
4.3. Mass Spectrometry
The spots of interest were cut, reduced, alkylated, digested, and automatically transferred to a Matrix-assisted laser desorption/ionization (MALDI) analysis target by a Proteineer SP II and DP robot using the SPcontrol 3.1.48.0 v software (Bruker Daltonics, Bremen, Germany), with the aid of a DP Chemicals 96 gel digestion kit (Bruker Daltonics, Germany) and processed in a MALDI-TOF Autoflex (Bruker Daltonics, Germany) to obtain a mass fingerprint. We performed 100 satisfactory shots in 20 shotsteps, the peak resolution threshold was set at 1500, the signal/noise ratio of tolerance was 6, and contaminants were not excluded [5]. The spectrum was annotated by the Flex Analysis 1.2 v SD1 Patch 2 (Bruker Daltonics, Germany). The search engine MASCOT was used to compare the fingerprints against the UNIPROT [46] with the following parameters: Taxon-mouse, mass tolerance of up to 200 ppm, one miss-cleavage allowed, and as the fixed modification carbamidomethyl cysteine and oxidation of methionine as the variable modification.
4.4. Bioinformatics Analysis
The network of proteins was constructed by Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, which provides a critical assessment and integration of protein-protein interactions, including direct (physical) as well as indirect (functional) associations [47]. Interactions among proteins in STRING were provided with a confidence score and the confidence score more than 0.4 was defined as the cut-off criterion. To understand the relationship among differentially expressed proteins in liver of db/db mice and their interaction with other proteins, differentially expressed proteins by Gene Ontology (GO) assignment was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v. 6.7 [48]. Proteins were uploaded into the DAVID functional annotation tool and compared to the mouse proteome background. Only enriched pathways and GO in functional annotation tool terms with a minimum of 3-fold enrichment and a Fisher’s Exact test p-value < 0.05 were defined as statistically significant.
5. Conclusions
In this study, twenty-six differentially expressed proteins were identified in the liver of db/db diabetic mice, which are involved in various metabolic processes related to carbohydrate and nitrogen metabolism. Results that do not agree entirely with those reported previously. However, they do reflect the importance of studying each of the variants of the disease to better understand the pathophysiology and etiology to design new pharmaceutical, nutritional, and physical activity treatments to reduce their prevalence.
Author Contributions
E.C.F.-P., S.E.-G. and V.P.-V. conceived of the study and participated in its design and coordination and helped to draft the manuscript. J.R.-E., K.V.-O. and J.M.G.-F. participated in the design of the study and collected samples. E.C.F.-P. and M.H.-O. designed the study and performed the statistical analysis. All authors read and approved the final manuscript.
Acknowledgments
We are grateful to Gabriel Martínez-Batallar for its technical assistance. The authors acknowledge to Salvador Uribe (IFC-UNAM) for the editing of the English-language version and critically read the paper. This work was partially supported by a grant from Consejo de Ciencia y Tecnología del Estado de Guanajuato (09-16-K662-079 A02), and Promep (UGTO-PTC-120) to VPV.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| T2DM | Type 2 diabetes mellitus |
| IEF | Isoelectric focusing |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| DAVID | Database for Annotation, Visualization and Integrated Discovery |
| GO | Gene Ontology |
| BP | Biological Process |
| CC | Cellular Component |
| MF | Molecular Function |
| HCl | Hydrochloric acid |
| EDTA | Ethylenediaminetetraacetic acid |
| DTT | 1,4-Dithiothreitol |
| KEEG | Kyoto Encyclopedia of Genes and Genomes Pathway |
| PMSF | Phenylmethylsulfonyl fluoride |
| CHAPS | 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate hydrate |
| TBP | tributyl phosphine |
| IPG | Immobilized pH gradient |
| ANOVA | Analysis of variance |
| MALDI-TOF | matrix-assisted laser desorption/ionization Time-Of-Flight |
| UNIPROT | Universal Protein |
References
- Zimmet, P.; Alberti, K.G.; Magliano, D.J.; Bennett, P.H. Diabetes mellitus statistics on prevalence and mortality: Facts and fallacies. Nat. Rev. Endocrinol. 2016, 12, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Alcalde-Rabanal, J.E.; Orozco-Nunez, E.; Espinosa-Henao, O.E.; Arredondo-Lopez, A.; Alcayde-Barranco, L. The complex scenario of obesity, diabetes and hypertension in the area of influence of primary healthcare facilities in mexico. PLoS ONE 2018, 13, e0187028. [Google Scholar] [CrossRef] [PubMed]
- Keith, B.P.; Robertson, D.L.; Hentges, K.E. Locus heterogeneity disease genes encode proteins with high interconnectivity in the human protein interaction network. Front. Genet. 2014, 5, 434. [Google Scholar] [CrossRef] [PubMed]
- Kontou, P.I.; Pavlopoulou, A.; Dimou, N.L.; Pavlopoulos, G.A.; Bagos, P.G. Network analysis of genes and their association with diseases. Gene 2016, 590, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Perez-Vazquez, V.; Guzman-Flores, J.M.; Mares-Alvarez, D.; Hernandez-Ortiz, M.; Macias-Cervantes, M.H.; Ramirez-Emiliano, J.; Encarnacion-Guevara, S. Differential proteomic analysis of the pancreas of diabetic db/db mice reveals the proteins involved in the development of complications of diabetes mellitus. Int. J. Mol. Sci. 2014, 15, 9579–9593. [Google Scholar] [CrossRef] [PubMed]
- Ke, M.; Wu, H.; Zhu, Z.; Zhang, C.; Zhang, Y.; Deng, Y. Differential proteomic analysis of white adipose tissues from T2d KKAy mice by LC-ESI-QTOF. Proteomics 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Giebelstein, J.; Poschmann, G.; Hojlund, K.; Schechinger, W.; Dietrich, J.W.; Levin, K.; Beck-Nielsen, H.; Podwojski, K.; Stuhler, K.; Meyer, H.E.; et al. The proteomic signature of insulin-resistant human skeletal muscle reveals increased glycolytic and decreased mitochondrial enzymes. Diabetologia 2012, 55, 1114–1127. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Darshi, M.; Ma, Y.; Perkins, G.A.; Shen, Z.; Haushalter, K.J.; Saito, R.; Chen, A.; Lee, Y.S.; Patel, H.H.; et al. Quantitative proteomic and functional analysis of liver mitochondria from high fat diet (hfd) diabetic mice. Mol. Cell. Proteom. 2013, 12, 3744–3758. [Google Scholar] [CrossRef] [PubMed]
- Valle, A.; Catalan, V.; Rodriguez, A.; Rotellar, F.; Valenti, V.; Silva, C.; Salvador, J.; Fruhbeck, G.; Gomez-Ambrosi, J.; Roca, P.; et al. Identification of liver proteins altered by type 2 diabetes mellitus in obese subjects. Liver Int. 2012, 32, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Chou, H.C.; Chen, Y.H.; Chan, H.L. High glucose-induced proteome alterations in hepatocytes and its possible relevance to diabetic liver disease. J. Nutr. Biochem. 2013, 24, 1889–1910. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Cai, T.; Li, T.; Xue, P.; Zhou, B.; He, X.; Wei, P.; Liu, P.; Yang, F.; Wei, T. Lysine malonylation is elevated in type 2 diabetic mouse models and enriched in metabolic associated proteins. Mol. Cell. Proteom. 2015, 14, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.A.; Wang, W.; Taylor, J.F.; Khattab, O.S.; Chen, Y.H.; Edwards, R.A.; Yazdi, P.G.; Wang, P.H. Hepatic proteomic analysis revealed altered metabolic pathways in insulin resistant akt1(+/−)/akt2(−/−) mice. Metabolism 2015, 64, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Holper, S.; Nolte, H.; Bober, E.; Braun, T.; Kruger, M. Dissection of metabolic pathways in the db/db mouse model by integrative proteome and acetylome analysis. Mol. Biosyst. 2015, 11, 908–922. [Google Scholar] [CrossRef] [PubMed]
- M Guzmán-Flores, J.; C Flores-Pérez, E.; Hernández-Ortíz, M.; López-Briones, S.; Ramírez-Emiliano, J.; Encarnación-Guevara, S.; Pérez-Vázquez, V. Comparative proteomics of liver of the diabetic obese db/db and non-obese or diabetic mice. Curr. Proteom. 2016, 13, 231–236. [Google Scholar] [CrossRef]
- Sharabi, K.; Tavares, C.D.; Rines, A.K.; Puigserver, P. Molecular pathophysiology of hepatic glucose production. Mol. Asp. Med. 2015, 46, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Chatzigeorgiou, A.; Halapas, A.; Kalafatakis, K.; Kamper, E. The use of animal models in the study of diabetes mellitus. In Vivo 2009, 23, 245–258. [Google Scholar] [PubMed]
- Guarner, V.; Rubio-Ruiz, M.E. Low-grade systemic inflammation connects aging, metabolic syndrome and cardiovascular disease. Interdiscip. Top. Gerontol. 2015, 40, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Lebel, M.; de Souza-Pinto, N.C.; Bohr, V.A. Metabolism, genomics, and DNA repair in the mouse aging liver. Curr. Gerontol. Geriatr. Res. 2011, 2011, 859415. [Google Scholar] [CrossRef] [PubMed]
- Fatima, F.; Nawaz, M. Long distance metabolic regulation through adipose-derived circulating exosomal mirnas: A trail for rna-based therapies? Front. Physiol. 2017, 8, 545. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.T.; Hsia, D.S.; Chacko, S.K.; Rodriguez, L.M.; Haymond, M.W. Increased gluconeogenesis in youth with newly diagnosed type 2 diabetes. Diabetologia 2015, 58, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Visinoni, S.; Khalid, N.F.; Joannides, C.N.; Shulkes, A.; Yim, M.; Whitehead, J.; Tiganis, T.; Lamont, B.J.; Favaloro, J.M.; Proietto, J.; et al. The role of liver fructose-1,6-bisphosphatase in regulating appetite and adiposity. Diabetes 2012, 61, 1122–1132. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, K.; Ramarao, P. Animal models in type 2 diabetes research: An overview. Indian J. Med. Res. 2007, 125, 451–472. [Google Scholar] [PubMed]
- Amano, S.; Yamagishi, S.; Kato, N.; Inagaki, Y.; Okamoto, T.; Makino, M.; Taniko, K.; Hirooka, H.; Jomori, T.; Takeuchi, M. Sorbitol dehydrogenase overexpression potentiates glucose toxicity to cultured retinal pericytes. Biochem. Biophys. Res. Commun. 2002, 299, 183–188. [Google Scholar] [CrossRef]
- Hagopian, K.; Ramsey, J.J.; Weindruch, R. Caloric restriction counteracts age-related changes in the activities of sorbitol metabolizing enzymes from mouse liver. Biogerontology 2009, 10, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Havel, P.J. Dietary fructose: Implications for dysregulation of energy homeostasis and lipid/carbohydrate metabolism. Nutr. Rev. 2005, 63, 133–157. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wei, S.; Yang, F. Mitochondria in the pathogenesis of diabetes: A proteomic view. Protein Cell 2012, 3, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Newsholme, P.; Abdulkader, F.; Rebelato, E.; Romanatto, T.; Pinheiro, C.H.; Vitzel, K.F.; Silva, E.P.; Bazotte, R.B.; Procopio, J.; Curi, R.; et al. Amino acids and diabetes: Implications for endocrine, metabolic and immune function. Front. Biosci. 2011, 16, 315–339. [Google Scholar] [CrossRef]
- Tessari, P.; Cecchet, D.; Cosma, A.; Puricelli, L.; Millioni, R.; Vedovato, M.; Tiengo, A. Insulin resistance of amino acid and protein metabolism in type 2 diabetes. Clin. Nutr. 2011, 30, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; van Den Berg, I.E.; Al-Dhalimy, M.; Groopman, J.; Ou, C.N.; Ryabinina, O.; Iordanov, M.S.; Finegold, M.; Grompe, M. Chronic liver disease in murine hereditary tyrosinemia type 1 induces resistance to cell death. Hepatology 2004, 39, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Schattenberg, J.M.; Schuchmann, M. Diabetes and apoptosis: Liver. Apoptosis 2009, 14, 1459–1471. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, T.; Araki, E. Impact of mitochondrial ros production in the pathogenesis of diabetes mellitus and its complications. Antioxid. Redox Signal. 2007, 9, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Xia, N.; Yuan, X.; Zhu, X.; Xu, G.; Cui, S.; Zhang, T.; Zhang, W.; Zhao, Y.; Wang, S.; et al. Prdx1 is involved in palmitate induced insulin resistance via regulating the activity of p38mapk in hepg2 cells. Biochem. Biophys. Res. Commun. 2015, 465, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, M.; Wang, T.J.; Clish, C.; Engstrom, G.; Nilsson, P.; Gerszten, R.E.; Melander, O. Dimethylglycine deficiency and the development of diabetes. Diabetes 2015, 64, 3010–3016. [Google Scholar] [CrossRef] [PubMed]
- Martini, L.A.; Catania, A.S.; Ferreira, S.R. Role of vitamins and minerals in prevention and management of type 2 diabetes mellitus. Nutr. Rev. 2010, 68, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Juwono, J.; Martinus, R.D. Does hsp60 provide a link between mitochondrial stress and inflammation in diabetes mellitus? J. Diabetes Res. 2016, 2016, 8017571. [Google Scholar] [CrossRef] [PubMed]
- Guzman-Flores, J.M.; Lopez-Briones, S. Cells of innate and adaptive immunity in type 2 diabetes and obesity. Gac. Med. Mex. 2012, 148, 381–389. [Google Scholar] [PubMed]
- Jimenez-Flores, L.M.; Lopez-Briones, S.; Macias-Cervantes, M.H.; Ramirez-Emiliano, J.; Perez-Vazquez, V. A ppargamma, nf-kappab and ampk-dependent mechanism may be involved in the beneficial effects of curcumin in the diabetic db/db mice liver. Molecules 2014, 19, 8289–8302. [Google Scholar] [CrossRef] [PubMed]
- Pardo, F.N.; Altirriba, J.; Pradas-Juni, M.; Garcia, A.; Ahlgren, U.; Barbera, A.; Slebe, J.C.; Yanez, A.J.; Gomis, R.; Gasa, R. The role of raf-1 kinase inhibitor protein in the regulation of pancreatic beta cell proliferation in mice. Diabetologia 2012, 55, 3331–3340. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Wood, A.R.; Lyssenko, V.; Weedon, M.N.; Knowles, J.W.; Alkayyali, S.; Assimes, T.L.; Quertermous, T.; Abbasi, F.; Paananen, J.; et al. Genetic variants associated with glycine metabolism and their role in insulin sensitivity and type 2 diabetes. Diabetes 2013, 62, 2141–2150. [Google Scholar] [CrossRef] [PubMed]
- Maumus, S.; Marie, B.; Vincent-Viry, M.; Siest, G.; Visvikis-Siest, S. Analysis of the effect of multiple genetic variants of cardiovascular disease risk on insulin concentration variability in healthy adults of the stanislas cohort. The role of fgb-455 g/a polymorphism. Atherosclerosis 2007, 191, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.S.; Ghoshal, P.; Srinivasan, M.; Kim, S.; Cline, G.; Patel, M.S. Liver-specific pyruvate dehydrogenase complex deficiency upregulates lipogenesis in adipose tissue and improves peripheral insulin sensitivity. Lipids 2010, 45, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Samuel, V.T.; Shulman, G.I. The pathogenesis of insulin resistance: Integrating signaling pathways and substrate flux. J. Clin. Investig. 2016, 126, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.S.; Wu, T.E.; Juan, C.C.; Lin, H.D. Myocardial heat shock protein 60 expression in insulin-resistant and diabetic rats. J. Endocrinol. 2009, 200, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Murata, T. Involvement of regucalcin in lipid metabolism and diabetes. Metabolism 2013, 62, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Kutmon, M.; Evelo, C.T.; Coort, S.L. A network biology workflow to study transcriptomics data of the diabetic liver. BMC Genom. 2014, 15, 971. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. Uniprot: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The string database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).