Abstract
Aluminum (Al) toxicity is one of the environmental stress factors that affects crop growth, development, and productivity. MYB transcription factors play crucial roles in responding to biotic or abiotic stresses. However, the roles of MYB transcription factors in Al tolerance have not been clearly elucidated. Here, we found that GmMYB183, a gene encoding a R2R3 MYB transcription factor, is involved in Al tolerance. Subcellular localization studies revealed that GmMYB183 protein is located in the nucleus, cytoplasm and cell membrane. Overexpression of GmMYB183 in Arabidopsis and soybean hairy roots enhanced plant tolerance towards Al stress compared to the wild type, with higher citrate secretion and less Al accumulation. Furthermore, we showed that GmMYB183 binds the GmMATE75 gene promoter encoding for a plasma-membrane-localized citrate transporter. Through a dual-luciferase reporter system and yeast one hybrid, the GmMYB183 protein was shown to directly activate the transcription of GmMATE75. Furthermore, the expression of GmMATE75 may depend on phosphorylation of Ser36 residues in GmMYB183 and two MYB sites in P3 segment of the GmMATE75 promoter. In conclusion, GmMYB183 conferred Al tolerance by promoting the secretion of citrate, which provides a scientific basis for further elucidating the mechanism of plant Al resistance.
1. Introduction
Aluminum (Al), the most abundant metallic element in the Earth’s crust, is solubilized into a toxic trivalent cation (Al3+) in acidic soils with a pH value of less than 5.0. This solubilization inhibits plant root growth, blocking nutrients and water intake, resulting in severe losses in crop production [1,2]. Approximately 50% of the world’s potential arable land is acidic, and soil acidification is increasing due to industrial pollution and modern farming practices [3,4]. Therefore, Al toxicity presents a huge threat to agricultural production and productivity.
Plants employ a variety of strategies to mitigate the harmful effects of Al stress. One such strategy is the secretion of organic acids induced by Al, which plays a crucial role in most plant species [5,6,7]. Several plant species, including barley (Hordeum vulgare) [8], cabbage (Brassica oleracea) [9], camelina (Camelina sativa) [10], rape (Brassica napus) [11,12], rubber tree (Hevea brasiliensis) [13], and wheat (Triticum aestivum) [14], have been found to secrete malate, which forms chelates with Al and detoxifies it, whereas eucalyptus (Eucalyptus camaldulensis) [15], maize (Zea mays) [16], populous (Populous trichocarpa) [17], rice bean (Vigna umbellata) [18,19,20], Psychotria (Psychotria rubra) [21], sorghum (Sorghum bicolor) [22], soybean (Glycine max) [23,24,25], and wild soybean (Glycine soja) [26] have been shown to secrete citrate, which aids in the detoxification of Al. Additionally, the upregulation of transporters responsible for the secretion of citrate or malate enhances a plant’s tolerance to Al [1,6].
Transcription factors such as zinc finger proteins, WRKY, HD-ZIP, or NAC are known to play crucial roles in Al tolerance. One example is STOP1/ART1, a zinc finger transcription factor, which regulates the expression of various genes related to Al tolerance in different crop species, such as Arabidopsis (Arabidopsis thaliana) [27], cotton (Gossypium hirsutum) [28], pigeonpea (Cajanus caja) [29], rice (Oryza sativa) [30], sorghum, soybean [31] or tobacco (Nicotiana tabacum) [32]. Although the transcription of STOP1 is unaffected by Al stress, its activities at the post-transcriptional or post-translational levels are regulated by Al stress [33,34]. The SUMOylation of STOP1 has been found to be involved in modulating Al tolerance, suggesting that post-translational modifications of transcription factors are crucial in Al stress responses [35]. Similarly, WRKY22 has been shown to enhance Al tolerance by increasing the expression of OsFRDL4 and promoting citrate secretion in rice [36]. Another transcription factor, HvHOX9, a member of the HD-ZIP family, mediates Al resistance in barley roots by inhibiting Al binding to the root cell wall and increasing the apoplastic pH [37]. Furthermore, VuNAR1, a NAC transcription factor, promotes Al tolerance by activating WAK1 expression and regulating cell wall pectin metabolism [38]. However, the exact roles of the MYB family of genes in Al tolerance have not been fully elucidated.
In plants, the MYB family of transcription factors is considered one of the largest families and plays significant roles in regulating gene transcription networks involved in various developmental and stress-response mechanisms [39]. A study conducted by Wei et al. demonstrated that the overexpression of TaMYB344 in tobacco confers tolerance to heat, drought, and salt-induced stress [40]. Similarly, Shin et al. found that StMYB1R-1 activates genes associated with drought tolerance, leading to improved drought resistance in potatoes [41]. Additionally, the gene ARS1, which encodes the R1-MYB type transcription factor, is induced by salinity and contributes to salt tolerance in tomato leaves [42]. Similarly, GsMYB7 has been identified as a potential regulator of soybean’s tolerance to acidic aluminum stress through the regulation of downstream genes [43]. Hence, MYB transcription factors are believed to play crucial roles in plants’ response to Al stress.
Tamba black soybean (TBS) is an Al-tolerant genotype with significant potential for Al tolerance via the secretion of citrate under Al stress (Figure S1). Previously, we studied the effect of GmMYB183 on the Al stress response in TBS. We observed significant phosphorylation shifts, including upregulated phosphorylation of a serine residue (Ser36) [44]. In this study, our objective was to further investigate the function of GmMYB183 in the Al stress response. Our findings revealed that overexpression of GmMYB183 leads to increased tolerance to Al in transgenic Arabidopsis and soybean hairy roots. Additionally, we discovered that GmMYB183 directly binds to the promoter of GmMATE75, resulting in enhanced expression of this gene. In addition, GmMYB183 that binds to the MYB sites in the P3 segment of the GmMATE75 promoter may depend on phosphorylation of Ser36 residues in GmMYB183.
2. Materials and Methods
2.1. Plant Culture and Treatment
TBS seedlings were cultured as previously described [44]. Briefly, pre-treated roots were transferred to a 50 µmol/L AlCl3 solution (containing 0.5 mmol/L CaCl2, pH 4.5) for 3 h, 6 h, 9 h, 12 h, 24 h, 48 h or 96 h. Arabidopsis seeds (ecotype Columbia, Col-0) were directly germinated on mixed soil (substrate: vermiculite = 1:1) and grown in a controlled environment at day/night temperatures of 25/22 °C, with 16 h of light (120 µmol photons m−2·s−1).
2.2. GmMYB183 Gene Isolation and Sequence Analysis
Based on the previous analysis of the quantitative phosphoproteomics of TBS, we observed that, under acidic aluminum exposure, GmMYB183 was hyperphosphorylated at Ser36. We then obtained the GmMYB183 gene sequence data from Phytozome 12 database, using a gene ID, Glyma.06G187600. The specific primers of GmMYB183 amplification were designed from the full-length cDNA, which were used to clone the gene from TBS roots using RT-PCR. The primers in this study are shown in Table S1.
We used the ExPaSy platform (https://www.expasy.org/, accessed on 24 February 2020) to predict physical and chemical properties of the protein. Localization of the GmMYB183 gene was predicted using NetNES 1.1, and then the BLAST tool was used to search for GmMYB183 homologous proteins in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 24 February 2020). Conserved domains and the three-dimensional structure of the GmMYB183 protein were predicted using the Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 24 February 2020) and PDB software (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index, accessed on 24 February 2020). In addition, DNAMAN was used to perform homology studies of the GmMYB183 protein. Then, the MEGA 7.0 software was used to construct phylogenetic trees through the neighbor-joining method with 1000 bootstrap replications.
2.3. RNA Extraction and Quantitative Real-Time PCR (qRT-qPCR)
Total RNA was extracted using the RNAiso Plus kit (Takara, Shiga, Japan) according to the manufacturer’s instructions. First-strand cDNA was synthesized using Prime ScriptTM reagent Kit with a gDNA Eraser kit (Takara, Shiga, Japan). Quantitative real-time PCR (RT-qPCR) was performed according to our previous study with 40SrRNA gene (GenBank: XM_003549836.4) as an internal control [44]. Primers used in this study are presented in Table S1.
2.4. Vector Construction and Transformation of GmMYB183 into Arabidopsis and Soybean Hairy Roots
Encoding region of GmMYB183 cDNA was ligated into a pMD™ 19-T vector (Takara, Shiga, Japan). Fragments encoding GmMYB183 were amplified from the pMD™ 19-T-GmMYB183 vector using a pair of specific primers with terminal Xba I and Sma I restriction sites (Table S1). Moreover, the coding region of the GmMYB183 gene was mutated using Mut Express® II Fast Mutagenesis Kit V2 (Vazyme, Nanjing, China) according to the manufacturer’s instructions. Expression vectors, pBI121-GmMYB183-eGFP (OE) and pBI121-GmMYB183-S36A-eGFP (OE-m), were created by inserting the coding region of the GmMYB183 gene into the same restriction sites under the control of the CaMV 35S promoter and NOS terminator of the expression box. Then, pBI121-eGFP, pBI121-GmMYB183-eGFP and pBI121-GmMYB183-S36A-eGFP were introduced into the Agrobacterium tumefaciens strain GV3101 through the heat shock method. Full-flowering Arabidopsis plants with fruit pods and flowers were used for genetic transformation through the floral dip method. The surface of Arabidopsis seeds was disinfected with 7% sodium hypochlorite for 10 min, washed with sterile water for three times, then spread on the medium of 1/2 MS (containing 500 mg/L Cefixime). After germination, the Arabidopsis seedlings with green fluorescence were screened out using an LUYOR-3415RG hand-held fluorescent protein excitation light source (as indicated by the arrow in Figure S2). The expression of GmMYB183 was further detected by RT-qPCR. The seeds of T0 generation Arabidopsis were disinfected and cultured on 1/2 MS medium. The single-copy T1 generation of transgenic Arabidopsis thaliana was screened according to the ratio of transgenic/wild type = 3:1. Among them, three single-copy lines of OE-1, OE-3 and OE-7 were screened in GmMYB183 transgenic lines, and three single-copy lines of OE-m2, OE-m5 and OE-m6 were screened in GmMYB183-S36A transgenic lines. Homozygous T2 and T3 generations were further screened for subsequent studies.
The full-length coding sequence of GmMYB183, amplified from the pMD™ 19-T-GmMYB183 vector, was sub-cloned into the pBin35S-Red3 vector between EcoR I and Xho I restriction sites, downstream of the 35S promoter to generate a GmMYB183 overexpression vector (OX). The empty pBin35S-Red3 vector was used as the negative control (EV). Site-directed mutations were established as above. The GmMYB183 overexpressing vector and the empty pBin35S-Red3 vector were introduced into the Agrobacterium rhizogenes strain K599 for transformation of black soybean hairy roots. Soybeans germinated in the soil for 4–5 days, and samples were taken before cotyledon was fully developed. The hypocotyls with cotyledon were cut 1 cm away from cotyledon and used as explants. The cotyledon was infected in the prepared infection solution for 1 h, and the positive hair roots were observed and screened after 14 days of moisturized culture.
2.5. Subcellular Localization of the GmMYB183 Protein
For subcellular localization, Arabidopsis roots were mounted on glass slides, covered and viewed using an inverted LSM800 laser scanning microscope (Carl Zeiss, Oberkochen Germany). The empty pBI121-eGFP vector was used as the negative control.
2.6. Phenotypic Identification of GmMYB183 Transgenic Lines
The Arabidopsis seeds were surface sterilized with 7% sodium hypochlorite for 15 min, washed three times in sterile water, and then incubated in 1/2 MS agar medium. For relative root growth, the seedlings, after germination for 3 d, were transplanted onto a 0, 50, 100, and 150 μmol/L AlCl3 solution (containing 0.5 mmol/L CaCl2, pH 4.5) for 7 d. The root relative growth was evaluated according to the procedures described by Min et al. [45]. We then determined citrate secretion and hematoxylin staining according to our previous work [46]. All treatments were performed in triplicates, and each replicate contained 3 plants.
2.7. Yeast Two-Hybrid and Yeast One-Hybrid Assays
To evaluate the interaction between the GmMYB183 and 14-3-3 protein (GmSGFa), yeast two-hybrid assays were carried out using AH109 yeast strain, according to the manufacturer’s instructions. The GmSGFa gene was cloned into the pGADT7 vector to generate the pGADT7-GmSGFa construct, while the GmMYB183 or GmMYB183-S36A was cloned into pGBKT7 vector to generate the pGBKT7-GmMYB183 or pGBKT7-GmMYB183-S36A construct. The AH109 yeast strain was co-transformed with different construct combinations and incubated in a medium lacking Trp (T), Leu (L), His (H), and Adenine (A), but with 10 mmol/L 3-AT.
On the other hand, to evaluate the binding of GmMYB183 to the GmMATE75 promoter, yeast one-hybrid assays were performed according to the procedures described by Yu et al. [47]. Briefly, the full-length GmMYB183 gene was ligated between the EcoRI and XhoI sites and fused in frame in yeast GAL4-AD effector plasmids. The four fragments of the GmMATE75 promoter were inserted into p178 vector to generate reporter plasmids. Subsequently, the different plasmid combinations were co-transformed into EGY48 yeast strain, and the interactions were tested on an SD medium lacking Trp and Ura, for 2~3 days, at 28 °C. Positive clones were incubated on X-gal plates with filter paper for 8 h at 28 °C.
2.8. Dual-LUC Assays
Dual-LUC assays were performed to quantify the binding affinity of the GmMYB183 protein to the GmMATE75 gene promoter. The ORFs of GmMYB183 were ligated into Xba I and Sma I restriction sites of pGreenII 62-SK vector to generate 35S-GmMYB183 effector plasmids. On the other hand, the GmMATE75 promoter was cloned into pGreen 0800-Luc at the Xho I and Hind III restriction sites to generate proGmMATE75-Luc reporter plasmids. The 35S-GmMYB183 vector was mutated by PCR to generate 35S-GmMYB183-S36A, while proGmMATE75-Luc vector was mutated by PCR to generate proGmMATE75-m1-Luc, proGmMATE75-m2-Luc and proGmMATE75-m1m2-Luc. The reporter and effector vectors were transformed into Agrobacterium tumefaciens strain GV3101 (pSoup). We then conducted infiltration in tobacco leaves and detection following the described protocols [48].
2.9. Statistical Analysis
Data for the relative expression, root growth, citrate secretion, and LUC/REN were presented as a mean ± the standard error of the mean (SEM). The data were processed using SPSS Statistics19 using Duncan’s test. We used GraphPad (Version 8.3.0) for graph preparation and presentation. A p < 0.05 was considered to be statistically significant.
3. Results
3.1. In Silico Analysis of the GmMYB183 Gene
Based on data from quantitative phosphoproteomics of TBS, a protein with significantly up-regulated phosphorylation induced by Al stress was screened [44]. The full-length coding sequences (CDSs) of GmMYB183 were amplified using cDNA from TBS. The sequencing results showed that the CDS of GmMYB183 is 885 bp, encoding 294 amino acids, with a MW of 31.87 kDa. The gene sequence was consistent with the sequence of soybean genomic MYB183 (NM_001249070.1). Furthermore, NetNES 1.1 analysis showed that the nuclear export signal was located at the C-terminal of 257–263 amino acids (Figure S3a), and CDD analysis revealed that a SANT conserved the domain of 77–126 amino acids in the GmMYB183 protein, with an E value of 1.30 × 10−14 (Figure S3b). Prediction of the GmMYB183 secondary structure by SOPMA showed that α-helix accounted for 21.77%, extension chain accounted for 20.75%, β-corner accounted for 6.46%, and irregular crimping accounted for 51.02% (Figure S3c). In addition, the three-dimensional (3d) structure was successfully established by PDB software (Figure S3d).
Homology analysis of GmMYB183 amino acid sequences revealed a typical SANT MYB domain, with high similarities in the N-terminal DNA-binding domain, as well as in the C-terminal sequence (Figure 1). In addition, evolutionary relatedness, as evaluated by MEGA7.0, revealed the highest homology (92.83%) between the GmMYB183 of TBS and soybean GmMYB143, and were clustered into one branch with MYB transcription factors of other legumes (Figure S4).

Figure 1.
Homologous alignment analysis of GmMYB183. Red arrow represents phosphorylation sites and the underscore presents the SANT domain. The accession number of each protein is listed in Table S2.
3.2. Expression Profiles of GmMYB183 in Tissues and Pertubations by Aluminum Stress
To evaluate the expression shifts of the TBS GmMYB183 gene under Al stress, we performed qRT-PCR analysis. Expression levels of GmMYB183 were found to be significantly suppressed within 6–24 h treatment of 50 µmol/L Al3+ in root tips, suggesting that GmMYB183 may be involved in the Al stress response (Figure 2a). Furthermore, expression levels of GmMYB183 in stems and leaves were found to be elevated when compared to those in the roots (Figure 2b). However, there were no significant differences in the expression levels in roots, stems and leaves compared with the control after Al stress for 24 h (Figure 2b).
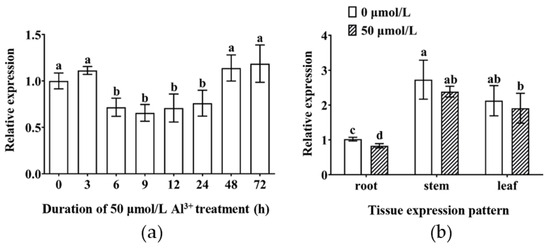
Figure 2.
Expression patterns of GmMYB183 in tissues and under Al3+ treatment. (a) Temporal expression pattern of GmMYB183 in root tips under Al3+ treatment. Seedlings were exposed to 50 µmol/L AlCl3, and the 2 cm long roots were taken from the seedlings at the processing time nodes of 0, 3, 6, 9, 12, 24, 48 and 72 h. (b) Tissue expression patterns of GmMYB183. Samples of roots, stems, and leaves are from seedlings treated with 0 or 50 µmol/L AlCl3 for 24 h. Column bars represent means ± standard errors (n = 3). Different letters represent significant differences (Duncan, p < 0.05).
3.3. Subcellular Localization of the GmMYB183 Protein
To evaluate the localization of the GmMYB183 protein, a full-length GmMYB183 gene was fused to the N-terminal of the eGFP (enhanced green fluorescent protein) reporter gene under a CaMV35S promoter. The eGFP fluorescence signals were analyzed in Arabidopsis root cells via Agrobacterium-mediated transformation. It was found that 35S-eGFP control and GmMYB183-eGFP were localized in both the nucleus and cytoplasmic membrane (Figure 3a). Furthermore, for the mutated Ser36 GmMYB183-S36A-eGFP, the signal was predominantly in the nucleus (Figure 3a).
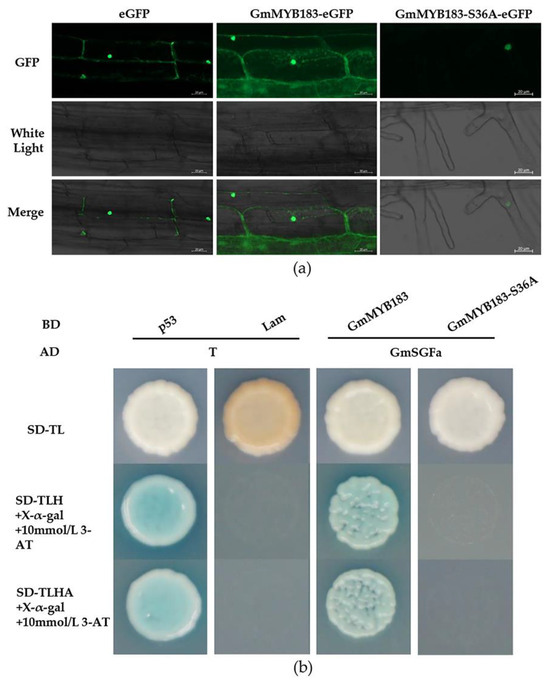
Figure 3.
Subcellular localization analysis of the GmMYB183 protein in Arabidopsis root cells. (a) Subcellular localization of the GmMYB183 and GmMYB183-S36A protein. (b) Evaluation of how GmMYB183 or GmMYB183-S36A interacts with GmSGFa using yeast two hybrid.
Previous studies have shown that 14-3-3 proteins regulate the subcellular localization of phosphorylated proteins [49]. In addition, the Al stimuli enhanced the interaction between 14-3-3 protein and phosphorylated plasma membrane H+-ATPase, thereby promoting citrate secretion [50,51]. Furthermore, our previous data showed that Al stress significantly increased the expression of the 14-3-3a gene (GmSGFa) in TBS roots. We speculated that, through interaction with GmSGFa, GmMYB183 might change its subcellular localization. The interactions of GmMYB183 with GmMYB183-S36A and GmSGFa were studied using the yeast two-hybrid system, and as expected, the GmMYB183 protein was shown to interact with GmSGFa, but not GmMYB183-S36A (Figure 3b).
3.4. Phenotypic Identification of GmMYB183 Transgenic Arabidopsis
According to the above results, we speculate that GmMYB183 plays an important role in the regulation of the tolerance to Al stress. To evaluate the role of GmMYB183 in Al tolerance, six homozygous T3 transgenic Arabidopsis lines were carefully chosen for subsequent phenotypic and physiological analysis (Figure S5). Relative root growth is one of the most important indices for evaluating Al tolerance in plants [52]. Exposure to Al stress exerted a concentration-dependent inhibition of Arabidopsis root growth (Figure 4a). The root growth of transgenic Arabidopsis plants was diminished. In addition, under Al stress, the root growth of transgenic plants overexpressing GmMYB183-S36A were shorter than those of WT plants. However, unlike WT plants, transgenic plants overexpressing GmMYB183 exhibited relatively longer root growth under Al stress (Figure 4b). Moreover, compared to WT, upon hematoxylin staining, transgenic plant roots overexpressing GmMYB183 or GmMYB183-S36A showed lighter or deeper staining, respectively (Figure 4c). In addition, citrate secretions in transgenic plants overexpressing GmMYB183 were significantly higher compared to those of WT (Figure 4d), consistent with the expression of the citrate transporter gene (Figure S6). These findings imply that overexpression of GmMYB183 enhanced citrate secretion to alleviate Al toxicity in Arabidopsis.
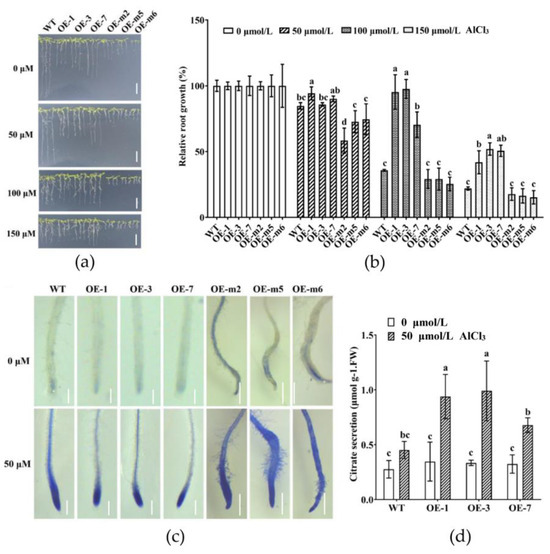
Figure 4.
Overexpression of the GmMYB183 gene improved Al tolerance in Arabidopsis. (a) After germination in 1/2 MS medium, three-day-old seedlings were transferred to solid medium containing 0, 50, 100, and 150 μmol/L AlCl3 (500 μmol/L CaCl2, pH 4.5) for 7 days (scale bar = 1 cm). (b) Relative root growth after treatment with various AlCl3 concentrations for 7 days. (c) Hematoxylin staining for roots treated with 50 μmol/L AlCl3 (scale bar = 1 mm). (d) Citrate secretion of roots treated with various AlCl3 concentrations for 24 h. OE: Overexpression of GmMYB183 in Arabidopsis; OE-m: Overexpression of GmMYB183-S36A in Arabidopsis. Columns and bars represent means ± standard errors (n = 3). Different letters represent significant differences (Duncan, p < 0.05).
3.5. Overexpression of GmMYB183 in Soybean Hairy Roots Confers Al Tolerance
To further evaluate the role of GmMYB183 in Al stress responses, 35S::Red and 35S::GmMYB183::Red constructs were introduced into soybean hairy roots (Figures S7 and S8) [53]. The hematoxylin staining degree of the OX root tip was lighter than that of EV (Figure 5a). Furthermore, citrate secretion from the OX root tip was found to be significantly higher than those of EV (Figure 5b), consistent with findings from Arabidopsis. In addition, under Al stress, expression levels of Al-tolerance genes such as GmMATE75 in OX were significantly higher than that in EV (Figure 5c), implying that GmMYB183 enhanced citrate secretion by promoting GmMATE75 expression.
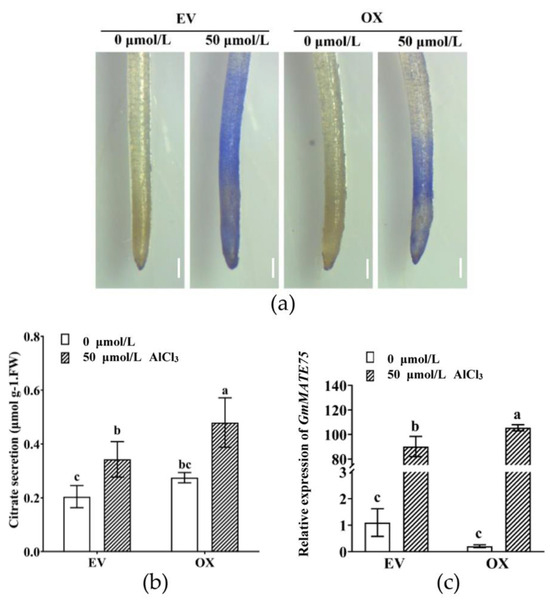
Figure 5.
Overexpression of the GmMYB183 gene improves Al tolerance in soybean hairy roots. (a) Hematoxylin staining of hairy roots after 50 μmol/L AlCl3 treatment (scale bar = 1 mm). (b) Citrate secretion of hairy roots treated with various AlCl3 concentrations for 24 h. (c) Relative expression of GmMATE75 in hairy roots. EV: empty vector; OX: overexpression of GmMYB183 in soybean hairy root. Bars on columns represent means ± standard errors (n = 3). Different letters represent significant differences (Duncan, p < 0.05).
3.6. GmMYB183 Binds to the GmMATE75 Promoter
GmMATE75, a gene encoding the citrate transporter, has been shown to enhance Al tolerance through Al-induced citrate efflux [23]. Using the 5′ rapid amplification of the cDNA ends (RACE) technique, we identified a transcriptional initiation site of the GmMATE75 gene, and analyzed MYB binding sites in the GmMATE75 gene promoter using PlantCare software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 19 April 2019) (Figure S9). Interactions between GmMYB183 and the GmMATE75 gene promoter were tested using the double luciferase detection system (Figure 6a). It was found that GmMYB183 exhibited much higher activity of the luciferase reporter gene compared to the empty vector control (Figure 6b). The yeast one-hybrid (Y1H) assay was also used to determine whether GmMYB183 binds MYB regions of the GmMATE75 promoter (Figure 6c). However, the yeast strain, EGY48, co-transformed with pB42AD-GmMYB183 and P3-reporter was the only one that showed the blue color (Figure 6d). Therefore, GmMYB183 positively regulates GmMATE75 by binding the P3 segment of the GmMATE75 promoter.
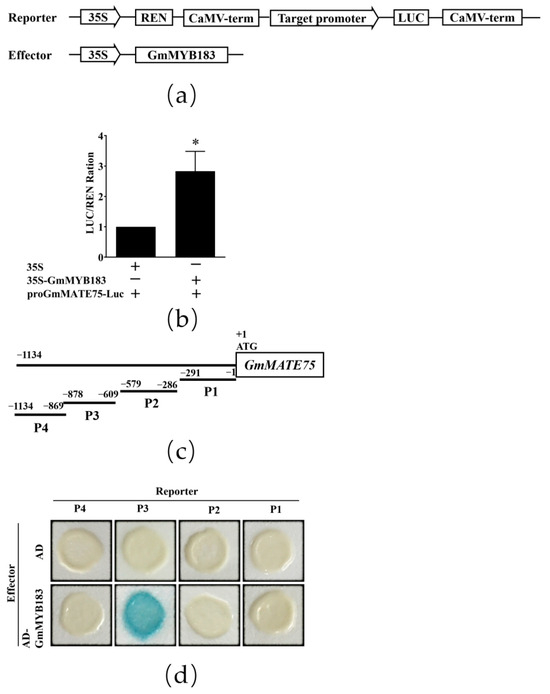
Figure 6.
Dual-luciferase and yeast one-hybrid analysis of GmMYB183 and promoters of the GmMATE75 gene. (a) Vector construction of transient expression assay. (b) Dual-luciferase assays between GmMYB183 and promoters of the GmMATE75 gene. (c) The GmMATE75 promoter region was divided into four fragments (P1, P2, P3 and P4). (d) Yeast one-hybrid assays. LUC, Firefly luciferase activity; REN, Renilla luciferase activity (control). Bars on columns represent means ± standard errors (n = 3). * above columns represent a significant difference (Duncan, p < 0.05).
To further analyze the transcriptional regulation of GmMATE75 by GmMYB183, we designed mutation primers Mut-proGmMATE75-1 or Mut-proGmMATE75-2 to mutate two MYB sites in the P3 segment of the GmMATE75 promoter, and generated promoter-Luc reporter constructs (pGreenII 0800-proGmMATE75-m1-Luc, pGreenII 0800-proGmMATE75-m2-Luc and pGreenII 0800-proGmMATE75-m1m2-Luc) (Figure 7a). Double luciferase activity detection revealed that GmMYB183 activates the expression of GmMATE75, and the expression is a function of the two MYB sites (Figure 7b).
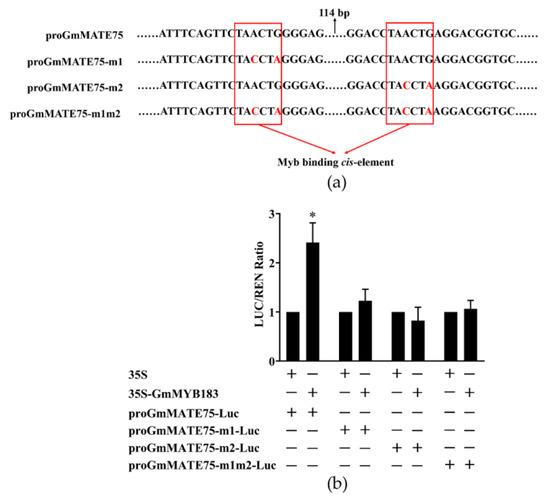
Figure 7.
Dual-luciferase analysis of GmMYB183 and promoters of the GmMATE75 gene, without or with mutation sites. (a) Sequences of GmMATE75 gene promoters without or with mutation site. (b) Dual-luciferase assays between GmMYB183 and P3 segments of the GmMATE75 promoter. LUC, Firefly luciferase activity; REN, Renilla luciferase activity. Columns and bars represent means ± standard errors (n = 3). * above columns represent a significant difference (Duncan, p < 0.05).
3.7. GmMYB183 Regulates GmMATE75-Dependent S36 Phosphorylation
To investigate the importance of Ser36 phosphorylation in the GmMYB183-specific regulation of GmMATE75, Ser36 was mutated to alanine after which the activity of the luciferase gene was analyzed using the double luciferase detection system (Figure 8a). Whereas GmMYB183 promoted the activity of the GmMATE75 promoter, the mutated GmMYB183-S36A did not (Figure 8b). These data show that GmMYB183 activity may depend on the phosphorylation of its Ser36 in regulating the expression of GmMATE75.
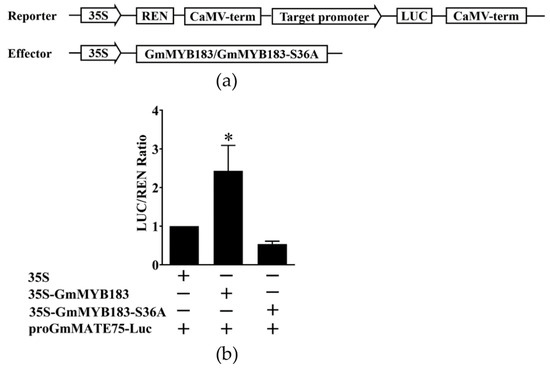
Figure 8.
Dual-luciferase analysis of GmMYB183, GmMYB183-S36A and promoters of the GmMATE75 gene. (a) Dual-luciferase assays between GmMYB183 and promoters of the GmMATE75 gene. (b) Analysis of double luciferase activity. LUC, Firefly luciferase activity; REN, Renilla luciferase activity. Bars on columns represent means ± standard errors (n = 3). * above columns represent a significant difference (Duncan, p < 0.05).
4. Discussion
Al toxicity is a crucial factor that significantly restrains plant growth and crop productivity in acidic soils [1]. Recent studies have shed light on the regulation of Al-tolerance-related genes by transcription factors [54,55,56,57,58]. Nonetheless, research exploring the post-translational modification of transcription factors in response to Al stress is scarce. Remarkable shifts in phosphorylation have been observed in GmMYB183 in response to Al stress, without any alterations in the transcript levels of GmMYB183 [44]. Interestingly, the phosphorylation of ALR1, a recently discovered Al receptor, is also regulated by Al stress [59]. With increasing Al concentration, the phosphorylation of ALR1-BAK1 increased, similar to GmMYB183, which may partially explain the concentration-dependent release of organic acids from roots under Al stress. In addition, Al stress induces the kinase activity of MPK4, which interacts with and phosphorylates STOP1 [60]. Hence, it is more probable that GmMYB183 contributes to Al tolerance through their phosphorylation activities. However, the mechanism by which ALR1 regulates GmMYB183 remains unclear. Further studies are necessary to provide a scientific basis for elucidating the mechanism of plant Al tolerance.
Hematoxylin staining has been employed to assess the levels of stress tolerance in soybean. For instance, it was observed that roots overexpressing the GmME1 or GsGIS3 genes displayed minimal hematoxylin staining compared to WT, following exposure to Al stress, indicating a robust resistance to Al [61,62]. In this study, faint staining of the GmMYB183-OE root tips in both Arabidopsis and soybean hairy roots was consistent with the relative growth of roots in Arabidopsis plants (Figure 4). Regarding further conditions, transgenic GmMYB183 plants exhibited significantly higher levels of citrate secretion compared to the WT under Al stress (Figure 5b). These findings demonstrate that overexpression of GmMYB183 enhances citrate secretion, thereby conferring Al tolerance in Arabidopsis and soybean hairy roots. In addition, citrate secretion could potentially alter the apoplastic pH of the root environment, impacting the availability of nutrients and the activity of enzymes involved in root growth. It is worth mentioning that citrate secretion can improve the utilization capacity of soil phosphorus and reduce the use of phosphorus fertilizer in acidic soil. Therefore, the role of GmMYB183 under abiotic stress is also worthy of our investigation. MATE transporters play multiple roles in plants including detoxification, secondary metabolite transport, Al tolerance, and disease resistance. To investigate the potential molecular regulatory mechanisms of GmMYB183 under Al stress, we profiled several Al-tolerance-related genes in WT or transgenic lines. RT-qPCR analysis revealed that, under Al stress, GmMATE75 was upregulated in GmMYB183-OX root tips, which might be responsible for improved Al tolerance in transgenic plants (Figure 5c). GmMATE75, a plasma-membrane-localized citrate transporter, mediates Al-induced citrate efflux in soybean root apices [23]. Our previous plasma membrane proteomics analysis showed that, under Al stress, GmMATE75 is upregulated in Al-tolerant TBS [63]. In this study, we found that GmMYB183 binds the promoter of soybean GmMATE75 genes. Similarly, Li et al. reported that OsWRKY22 activates OsFRDL4 expression and enhances citrate secretion, thereby promoting Al tolerance in rice [36]. These findings suggest that GmMYB183 functions as a regulator of GmMATE75 activity in Al tolerance. In addition, Arabidopsis MATE transporter 30 (AtDTX30) regulates auxin homeostasis in roots, influencing root development and Al tolerance [64]. The dtx30 mutants show reduced elongation in primary roots, root hairs, and lateral roots, which is similar to GmMYB183-OE roots. The altered root architecture could be an adaptive response to metal stress, including Al stress, as root hairs can secrete citrate and create a rhizosphere environment that reduces Al toxicity. Moreover, citrate has been implicated in hormonal signaling pathways, particularly those related to auxin and ethylene, which play crucial roles in root development [65]. Therefore, we hypothesized that GmMYB183 modulates hormone levels in roots to regulate root development and promote Al tolerance.
The 14-3-3 proteins, discrete phosphateserine/threonine-binding modules, play crucial roles in regulating plant growth, development, and stress responses [66]. For instance, TaGRF6-A, which encodes a 14-3-3 protein, enhances salt tolerance in wheat by interacting with TaMYB64 [67]. The interactions between the 14-3-3 protein and MYBS2 at specific serine residues play significant roles in regulating sugar-dependent nucleocytoplasmic shuttling and maintenance in the cytoplasm [68]. Moreover, inhibiting the interactions between 14-3-3 proteins and transcription factor PIF7 accelerates PIF7 dephosphorylation and promotes shade-induced nuclear localization, thereby enhancing the shade phenotype [69]. In this study, GmMYB183 in Arabidopsis roots was found to be located in the nucleus, cytoplasm, and cell membrane, while GmMYB183-S36A was only observed in the nucleus (Figure 3a). Additionally, unlike GmMYB183-S36A, GmMYB183 was found to bind to 14-3-3 protein (Figure 3b). These findings suggest that binding to 14-3-3 proteins alters the localization of GmMYB183. Similarly, the binding of soybean 14-3-3 proteins influences the intracellular localization of GmMYB176 and affects isoflavonoid synthesis [70]. However, the precise function of GmMYB183 after binding with 14-3-3 proteins remains undefined.
The phosphorylation of transcription factors plays a crucial role in the regulation of gene expression and functions [71]. In plants, the ABA-dependent multisite phosphorylation of transcription activator AREB1 is essential for its own activity and the expression of ABA-inducible genes [72]. Freezing tolerance in Arabidopsis requires the MAPK6-mediated phosphorylation of the transcriptional repressor MYB15 [73]. Additionally, the phosphorylation of WRKY33 and ERF6 by MPK3/MPK6 activates defense-related genes and enhances the resistance against Botrytis cinerea [74,75]. Similarly, rhizobia inoculation in soybean inhibits the phosphorylation of GmMYB183 at Ser61 and enhances tolerance to salinity [76]. This study reveals that GmMYB183 regulates the expression of GmMATE75, potentially associated with the phosphorylation of GmMYB183 at Ser36, leading to the promotion of citrate secretion. Furthermore, the regulation of GmMATE75 expression by GmMYB183 depends on two binding sites on the GmMATE75 promoter (Figure 9). These findings highlight the involvement of GmMYB183 in response to different abiotic stresses through the phosphorylation of different sites. Therefore, evaluating the potential functions of GmMYB183-S36A under Al stress and identifying its upstream kinase of GmMYB183 and association with Al receptors are crucial, which provide valuable insights for the development of Al-tolerant crop varieties, contributing to sustainable agriculture and food security.
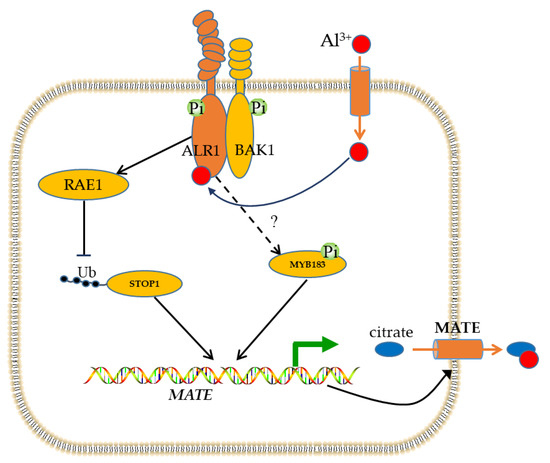
Figure 9.
Schematic presentation of transcriptional regulation of MATE by GmMYB183 transcription factor. In response to Al stress, Al3+ interacts with a receptor (ALR1) on the plasma membrane to initiate a signaling pathway. Al signal phosphorylates GmMYB183 through an unknown pathway. Furthermore, the ALR1-BAK1 complex inhibits F-box protein (RAE1) activity, thereby preventing the degradation of STOP1. Both STOP1 and GmMYB183 activate the expression of the MATE gene, promoting the secretion of citrate and chelating Al ions.
5. Conclusions
In this study, we identified GmMYB183, a R2R3 MYB transcription factor in TBS, is involved in Al tolerance. Overexpression of GmMYB183 in Arabidopsis and soybean hairy roots enhanced plant tolerance to Al stress compared to the wild type, with higher citrate secretion and less Al accumulation. Furthermore, using a dual-luciferase reporter system and yeast one hybrid, the GmMYB183 protein was shown to directly activate the transcription of GmMATE75. These results indicated that GmMYB183 is responsible for Al detoxification by promoting the secretion of citrate, providing valuable insight into the genetic basis for further elucidating the mechanism for improving plant tolerance to Al stress in acid soils.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom14060724/s1, Figure S1: Citrate secretion from roots of TBS induced by Al stress. Figure S2: Transgenic Arabidopsis lines were screened. Figure S3: Bioinformatics for GmMYB183 transcription factor. Figure S4: Phylogenetic trees of GmMYB183 protein. Figure S5: Relative expression level of GmMYB183 in Arabidopsis. Figure S6: Relative expression level of AtMATE in transgenic Arabidopsis. Figure S7: Different stages of the soybean hairy root transformation. Figure S8: Relative expression level of GmMYB183 in hairy roots. Figure S9: Promoter sequence analysis of GmMATE75. Table S1: The primers used in this study. Table S2: The accession number of each protein in homologous alignment analysis.
Author Contributions
Conceptualization, Y.W. and R.H.; writing—original draft preparation, Y.W. and R.H.; Writing—review and editing, Y.W. and R.H.; formal analysis, Y.W. and R.H.; funding acquisition, Y.Y. and R.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National 973 Project of China (2014CB138701) and Peacock Program of Shenzhen (KQTD2017-032715165926).
Institutional Review Board Statement
Ethical review and approval were waived for this study because it did not involve humans or animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Material.
Acknowledgments
We are very grateful to Wei Qian (Southwest University) for providing us the pBin35S-Red3 vector.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kochian, L.V.; Piñeros, M.A.; Liu, J.; Magalhaes, J.V. Plant Adaptation to Acid Soils: The Molecular Basis for Crop Aluminum Resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef]
- Ofoe, R.; Thomas, R.H.; Asiedu, S.K.; Wang-Pruski, G.; Fofana, B. Abbey, Lord Aluminum in plant: Benefits, toxicity and tolerance mechanisms. Front. Plant Sci. 2023, 13, 1085998. [Google Scholar] [CrossRef]
- Wei, Y.M.; Han, R.R.; Xie, Y.H.; Jiang, C.D.; Yu, Y.X. Recent Advances in Understanding Mechanisms of Plant Tolerance and Response to Aluminum Toxicity. Sustainability 2021, 13, 1782. [Google Scholar] [CrossRef]
- Eekhout, T.; Larsen, P.; De Veylder, L. Modification of DNA Checkpoints to Confer Aluminum Tolerance. Trends Plant Sci. 2017, 22, 102–105. [Google Scholar] [CrossRef]
- Yang, L.T.; Qi, Y.P.; Jiang, H.X.; Chen, L.S. Roles of organic acid anion secretion in aluminum tolerance of higher plants. Biomed Res. Int. 2013, 2013, 9–15. [Google Scholar] [CrossRef]
- Yang, J.-L.; Fan, W.; Zheng, S.-J. Mechanisms and regulation of aluminum-induced secretion of organic acid anions from plant roots. J. Zhejiang Univ. Sci. B 2019, 20, 513–527. [Google Scholar] [CrossRef]
- Riaz, M.; Yan, L.; Wu, X.; Hussain, S.; Aziz, O.; Jiang, C. Mechanisms of organic acids and boron induced tolerance of aluminum toxicity: A review. Ecotoxicol. Environ. Saf. 2018, 165, 25–35. [Google Scholar] [CrossRef]
- Gruber, B.D.; Ryan, P.R.; Richardson, A.E.; Tyerman, S.D.; Ramesh, S.; Hebb, D.M.; Howitt, S.M.; Delhaize, E. HvALMT1 from barley is involved in the transport of organic anions. J. Exp. Bot. 2010, 61, 1455–1467. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, X.X.; Wang, J.; Qi, C.; Wang, X.; Wang, G.; Li, M.; Li, X.; Guo, Y.D. BoALMT1, an Al-induced malate transporter in cabbage, enhances aluminum tolerance in Arabidopsis thaliana. Front. Plant Sci. 2018, 8, 2156. [Google Scholar] [CrossRef]
- Park, W.; Kim, H.S.; Park, T.W.; Lee, Y.H.; Ahn, S.J. Functional characterization of plasma membrane-localized organic acid transporter (CsALMT1) involved in aluminum tolerance in Camelina sativa L. Plant Biotechnol. Rep. 2017, 11, 181–192. [Google Scholar] [CrossRef]
- Ligaba, A.; Katsuhara, M.; Ryan, P.R.; Shibasaka, M.; Matsumoto, H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006, 142, 1294–1303. [Google Scholar] [CrossRef]
- Ligaba, A.; Katsuhara, M.; Sakamoto, W.; Matsumoto, H. The BnALMT1 Protein that is an Aluminum-Activated Malate Transporter is Localized in the Plasma Membrane. Plant Signal. Behav. 2007, 2, 255–257. [Google Scholar] [CrossRef]
- Ma, X.; An, F.; Wang, L.; Guo, D.; Xie, G.; Liu, Z. Genome-wide identification of aluminum-activated malate transporter (ALMT) gene family in rubber trees (Hevea brasiliensis) highlights their involvement in aluminum detoxification. Forests 2020, 11, 142. [Google Scholar] [CrossRef]
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Ma, J.F. Two MATE transporters with different subcellular localization are involved in Al tolerance in buckwheat. Plant Cell Physiol. 2017, 58, 2179–2189. [Google Scholar] [CrossRef]
- Sawaki, Y.; Kihara-Doi, T.; Kobayashi, Y.; Nishikubo, N.; Kawazu, T.; Kobayashi, Y.; Koyama, H.; Sato, S. Characterization of Al-responsive citrate excretion and citrate-transporting MATEs in Eucalyptus camaldulensis. Planta 2013, 237, 979–989. [Google Scholar] [CrossRef]
- Maron, L.G.; Piñeros, M.A.; Guimarães, C.T.; Magalhaes, J.V.; Pleiman, J.K.; Mao, C.; Shaff, J.; Belicuas, S.N.J.; Kochian, L.V. Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J. 2010, 61, 728–740. [Google Scholar] [CrossRef]
- Li, N.; Meng, H.; Xing, H.; Liang, L.; Zhao, X.; Luo, K. Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus. J. Exp. Bot. 2017, 68, 5669–5683. [Google Scholar] [CrossRef]
- Liu, M.Y.; Chen, W.W.; Xu, J.M.; Fan, W.; Yang, J.L.; Zheng, S.J. The role of VuMATE1 expression in aluminum-inducible citrate secretion in rice bean (Vigna umbellata) roots. J. Exp. Bot. 2013, 64, 1795–1804. [Google Scholar] [CrossRef]
- Fan, W.; Lou, H.Q.; Gong, Y.L.; Liu, M.Y.; Cao, M.J.; Liu, Y.; Yang, J.L.; Zheng, S.J. Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol. 2015, 208, 456–468. [Google Scholar] [CrossRef]
- Yang, X.Y.; Yang, J.L.; Zhou, Y.; Piñeros, M.A.; Kochian, L.V.; Li, G.X.; Zheng, S.J. A de novo synthesis citrate transporter, Vigna umbellata multidrug and toxic compound extrusion, implicates in Al-activated citrate efflux in rice bean (Vigna umbellata) root apex. Plant Cell Environ. 2011, 34, 2138–2148. [Google Scholar] [CrossRef]
- Iguchi, A.; Sanmiya, K.; Watanabe, K. Identification of genes encoding ALMT and MATE transporters as candidate aluminum tolerance genes from a typical acid soil plant, Psychotria rubra (Rubiaceae). PeerJ 2019, 7, e7739. [Google Scholar] [CrossRef]
- Melo, J.O.; Lana, U.G.P.; Piñeros, M.A.; Alves, V.M.C.; Guimarães, C.T.; Liu, J.; Zheng, Y.; Zhong, S.; Fei, Z.; Maron, L.G.; et al. Incomplete transfer of accessory loci influencing SbMATE expression underlies genetic background effects for aluminum tolerance in sorghum. Plant J. 2013, 73, 276–288. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Z.; Gong, L.; Chen, A.; Liu, N.; Li, S.; Sun, H.; Yang, Z.; You, J. Functional characterization of three MATE genes in relation to aluminum-induced citrate efflux from soybean root. Plant Soil 2019, 443, 121–138. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Wang, W.; Gai, J.; Li, Y. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genom. 2016, 17, 223. [Google Scholar] [CrossRef]
- Wang, P.; Yu, W.Q.; Zhang, J.R.; Rengel, Z.; Xu, J.; He, Q.Q.; Chen, L.M.; Li, K.Z.; Yu, Y.X.; Chen, Q. Auxin enhances aluminium-induced citrate exudation through upregulation of GmMATE and activation of the plasma membrane H+-ATPase in soybean roots. Ann. Bot. 2016, 118, 933–940. [Google Scholar] [CrossRef]
- Ma, Q.; Yi, R.; Li, L.; Liang, Z.; Zeng, T.; Zhang, Y.; Huang, H.; Zhang, X.; Yin, X.; Cai, Z.; et al. GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminum tolerance in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 212. [Google Scholar] [CrossRef]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef]
- Kundu, A.; Das, S.; Basu, S.; Kobayashi, Y.; Kobayashi, Y.; Koyama, H.; Ganesan, M. GhSTOP1, a C2H2 type zinc finger transcription factor is essential for aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biol. 2019, 21, 35–44. [Google Scholar] [CrossRef]
- Arun, A.; Yuriko, D.; Sanjib, K.; Panda, K. Characterization of CcSTOP1; a C2H2-type transcription factor regulates Al tolerance gene in pigeonpea. Planta 2017, 247, 201–214. [Google Scholar]
- Yamaji, N.; Huang, C.F.; Nagao, S.; Yano, M.; Sato, Y.; Nagamura, Y.; Ma, J.F. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009, 21, 3339–3349. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, Z.M.; Gong, L.; Liu, R.K.; Sun, H.R.; You, J.F. Molecular characterization of GmSTOP1 homologs in soybean under Al and proton stress. Plant Soil 2018, 427, 213–230. [Google Scholar] [CrossRef]
- Ito, H.; Kobayashi, Y.; Yamamoto, Y.Y.; Koyama, H. Characterization of NtSTOP1-regulating genes in tobacco under aluminum stress. Soil Sci. Plant Nutr. 2019, 65, 251–258. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Guo, J.; Zhou, F.; Singh, S.; Xu, X.; Xie, Q.; Yang, Z.; Huang, C.F. F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 319–327. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, Y.; Gao, H.; Li, S.; Wang, Z.Y.; Huang, C.F. Mutation of HPR1 encoding a component of the THO/TREX complex reduces STOP1 accumulation and aluminum resistance in Arabidopsis thaliana. New Phytol. 2020, 228, 179–193. [Google Scholar] [CrossRef]
- Fang, Q.; Zhang, J.; Zhang, Y.; Fan, N.; Burg, H.A.; Huang, C.F. Regulation of Aluminum-Resistance in Arabidopsis Involves the SUMOylation of the Zinc Finger Transcription Factor STOP1. Plant Cell 2020, 32, 3921–3938. [Google Scholar] [CrossRef]
- Li, G.Z.; Wang, Z.Q.; Yokosho, K.; Ding, B.; Fan, W.; Gong, Q.Q.; Li, G.X.; Wu, Y.R.; Yang, J.L.; Ma, J.F.; et al. Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 2018, 219, 149–162. [Google Scholar] [CrossRef]
- Feng, X.; Liu, W.; Dai, H.; Qiu, Y.; Zhang, G.; Chen, Z.-H.; Wu, F. HvHOX9, a Novel Homeobox Leucine Zipper Transcription Factor Revealed by Root miRNA and RNA Sequencing in Tibetan Wild Barley, Positively Regulates Al Tolerance. J. Exp. Bot. 2020, 71, 6057–6073. [Google Scholar] [CrossRef]
- Lou, H.Q.; Fan, W.; Jin, J.F.; Xu, J.M.; Chen, W.W.; Yang, J.L.; Zheng, S.J. A NAC-type transcription factor confers aluminum resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant Cell Environ. 2020, 43, 463–478. [Google Scholar] [CrossRef]
- Ma, R.; Liu, B.; Geng, X.; Ding, X.; Yan, N.; Sun, X.; Wang, W.; Sun, X.; Zheng, C. Biological Function and Stress Response Mechanism of MYB Transcription Factor Family Genes. J. Plant Growth Regul. 2023, 42, 83–95. [Google Scholar] [CrossRef]
- Wei, Q.; Chen, R.; Wei, X.; Liu, Y.; Zhao, S.; Yin, X.; Xie, T. Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genom. 2020, 21, 792. [Google Scholar]
- Shin, D.; Moon, S.J.; Han, S.; Kim, B.G.; Park, S.R.; Lee, S.K.; Yoon, H.J.; Lee, H.E.; Kwon, H.-B.; Baek, D.; et al. Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought tolerance. Plant Physiol. 2011, 155, 421–432. [Google Scholar] [CrossRef]
- Campos, J.F.; Cara, B.; Pérez-Martín, F.; Pineda, B.; Egea, I.; Flores, F.B.; Fernandez-Garcia, N.; Capel, J.; Moreno, V.; Angosto, T.; et al. The tomato mutant ars1 (altered response to salt stress 1) identifies an R1-type MYB transcription factor involved in stomatal closure under salt acclimation. Plant Biotechnol. J. 2016, 14, 1345–1356. [Google Scholar] [CrossRef]
- Wang, H.; Yin, X.; Du, D.; Liang, Z.; Han, Z.; Nian, H.; Ma, Q. GsMYB7 encoding a R2R3-type MYB transcription factor enhances the tolerance to aluminum stress in soybean (Glycine max L.). BMC Genom. 2022, 23, 529. [Google Scholar] [CrossRef]
- Han, R.R.; Wei, Y.M.; Xie, Y.H.; Liu, L.S.; Jiang, C.D.; Yu, Y.X. Quantitative phosphoproteomic analysis provides insights into the aluminum-responsiveness of Tamba black soybean. PLoS ONE 2020, 15, e0237845. [Google Scholar] [CrossRef]
- Min, Y.; Guo, C.L.; Zhao, X.L.; Wang, L.; Yu, Y.X.; Chen, L.M. Adenosine 5′-monophosphate decreases citrate exudation and aluminium resistance in Tamba black soybean by inhibiting the interaction between 14-3-3 proteins and plasma membrane H+-ATPase. Plant Growth Regul. 2018, 84, 285–292. [Google Scholar] [CrossRef]
- Jiang, C.; Liu, L.; Li, X.; Han, R.; Wei, Y.; Yu, Y. Insights into aluminum-tolerance pathways in Stylosanthes as revealed by RNA-Seq analysis. Sci. Rep. 2018, 8, 6072. [Google Scholar] [CrossRef]
- Yu, Y.; Duan, X.; Ding, X.; Chen, C.; Zhu, D.; Yin, K.; Cao, L.; Song, X.; Zhu, P.; Li, Q.; et al. A novel AP2/ERF family transcription factor from Glycine soja, GsERF71, is a DNA binding protein that positively regulates alkaline stress tolerance in Arabidopsis. Plant Mol. Biol. 2017, 94, 509–530. [Google Scholar] [CrossRef]
- Zhang, F.; Fu, X.; Lv, Z.; Lu, X.; Shen, Q.; Zhang, L.; Zhu, M.; Wang, G.; Sun, X.; Liao, Z.; et al. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol. Plant 2015, 8, 163–175. [Google Scholar] [CrossRef]
- Keicher, J.; Jaspert, N.; Weckermann, K.; Möller, C.; Throm, C.; Kintzi, A.; Oecking, C. Arabidopsis 14-3-3 epsilon members contribute to polarity of PIN auxin carrier and auxin transport-related development. eLife 2017, 6, e24336. [Google Scholar] [CrossRef]
- Chen, Q.; Guo, C.L.; Wang, P.; Chen, X.Q.; Wu, K.H.; Li, K.Z.; Yu, Y.X.; Chen, L.M. Up-regulation and interaction of the plasma membrane H+-ATPase and the 14-3-3 protein are involved in the regulation of citrate exudation from the broad bean (Vicia faba L.) under Al stress. Plant Physiol. Biochem. 2013, 70, 504–511. [Google Scholar] [CrossRef]
- Guo, C.L.; Chen, Q.; Zhao, X.L.; Chen, X.Q.; Zhao, Y.; Wang, L.; Li, K.Z.; Yu, Y.X.; Chen, L.M. Al-enhanced Expression and Interaction of 14-3-3 Protein and Plasma Membrane H+-ATPase is Related to Al-induced Citrate Secretion in an Al-resistant Black Soybean. Plant Mol. Biol. Report. 2013, 31, 1012–1024. [Google Scholar] [CrossRef]
- Wagatsuma, T.; Maejima, E.; Watanabe, T.; Toyomasu, T.; Kuroda, M.; Muranaka, T.; Ohyama, K.; Ishikawa, A.; Usui, M.; Hossain Khan, S.; et al. Dark conditions enhance aluminum tolerance in several rice cultivars via multiple modulations of membrane sterols. J. Exp. Bot. 2018, 69, 567–577. [Google Scholar] [CrossRef]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.T.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef]
- Che, J.; Tsutsui, T.; Yokosho, K.; Yamaji, N.; Ma, J.F. Functional characterization of an aluminum (Al)-inducible transcription factor, ART2, revealed a different pathway for Al tolerance in rice. New Phytol. 2018, 220, 209–218. [Google Scholar] [CrossRef]
- Huang, S.; Gao, J.; You, J.F.; Liang, Y.N.; Guan, K.X.; Yan, S.Q.; Zhan, M.Q.; Yang, Z.M. Identification of STOP1-Like Proteins Associated with Aluminum Tolerance in Sweet Sorghum (Sorghum bicolor L.). Front. Plant Sci. 2018, 9, 258. [Google Scholar] [CrossRef]
- Li, C.X.; Yan, J.Y.; Ren, J.Y.; Sun, L.; Xu, C.; Li, G.X.; Ding, Z.J.; Zheng, S.J. A WRKY transcription factor confers aluminum tolerance via regulation of cell wall modifying genes. J. Integr. Plant Biol. 2019, 62, 1176–1192. [Google Scholar] [CrossRef]
- Wu, L.; Guoa, Y.; Caia, S.; Kuanga, L.; Shena, Q.; Wu, D.; Zhanga, G. A zinc finger transcription factor HvATF1 regulates aluminum tolerance in barley. J. Exp. Bot. 2020, 71, 6512–6523. [Google Scholar] [CrossRef]
- Cai, Z.; Xian, P.; Lin, R.; Cheng, Y.; Lian, T.; Ma, Q.; Nian, H. Characterization of the Soybean GmIREG Family Genes and the Function of GmIREG3 in Conferring Tolerance to Aluminum Stress. Int. J. Mol. Sci. 2020, 21, 497. [Google Scholar] [CrossRef]
- Ding, Z.J.; Xu, C.; Yan, J.Y.; Wang, Y.X.; Cui, M.Q.; Yuan, J.J.; Wang, Y.N.; Li, G.X.; Wu, J.X.; Wu, Y.R.; et al. The LRR receptor-like kinase ALR1 is a plant aluminum ion sensor. Cell Res. 2024, 34, 281–294. [Google Scholar] [CrossRef]
- Zhou, F.; Singh, S.; Zhang, J.; Fang, Q.; Li, C.; Wang, J.; Zhao, C.; Wang, P.; Huang, C.F. The MEKK1-MKK1/2-MPK4 cascade phosphorylates and stabilizes STOP1 to confer aluminum resistance in Arabidopsis. Mol. Plant 2023, 16, 337–353. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, Z.; Xu, Y.; Sun, H.; Sun, Z.; Lin, B.; Sun, W.; You, J. Soybean NADP-malic enzyme functions in malate and citrate metabolism and contributes to their efflux under Al stress. Front. Plant Sci. 2018, 8, 2246. [Google Scholar] [CrossRef]
- Liu, Y.T.; Shi, Q.H.; Cao, H.J.; Ma, Q.-B.; Nian, H.; Zhang, X.X. Heterologous expression of a Glycine soja C2H2 zinc finger gene improves aluminum tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 2754. [Google Scholar] [CrossRef]
- Wei, Y.; Jiang, C.; Han, R.; Xie, Y.; Liu, L.; Yu, Y. Plasma membrane proteomic analysis by TMT-PRM provides insight into mechanisms of aluminum resistance in tamba black soybean roots tips. PeerJ 2020, 8, e9312. [Google Scholar] [CrossRef]
- Upadhyay, N.; Kar, D.; Datta, S. A multidrug and toxic compound extrusion (MATE) transporter modulates auxin levels in root to regulate root development and promotes aluminium tolerance. Plant Cell Environ. 2020, 43, 745–759. [Google Scholar] [CrossRef]
- Zahan, M.I.; Karim, M.; Imran, S.; Hunter, C.T.; Islam, S.; Mia, A.; Hannan, A.; Rhaman, M.S. Citric acid-mediated abiotic stress tolerance in plants. Int. J. Mol. Sci. 2021, 22, 7235. [Google Scholar] [CrossRef]
- Yaffe, M.B. How do 14-3-3 proteins work?—Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002, 513, 53–57. [Google Scholar]
- Shao, W.; Chen, W.; Zhu, X.; Zhou, X.; Jin, Y.; Zhan, C.; Liu, G.; Liu, X.; Ma, D.; Qiao, Y. Genome-Wide Identification and Characterization of Wheat 14-3-3 Genes Unravels the Role of TaGRF6-A in Salt Stress Tolerance by Binding MYB Transcription Factor. Int. J. Mol. Sci. 2021, 22, 1904. [Google Scholar] [CrossRef]
- Chen, Y.S.; Ho, T.H.D.; Liu, L.; Lee, D.H.; Lee, C.H.; Chen, Y.R.; Lin, S.Y.; Lu, C.A.; Yu, S.M. Sugar starvation-regulated MYBS2 and 14-3-3 protein interactions enhance plant growth, stress tolerance, and grain weight in rice. Proc. Natl. Acad. Sci. USA 2019, 116, 21925–21935. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, Q.; Jiang, Y.; Yang, C.; Wang, Q.; Li, L. Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. Elife 2018, 7, e31636. [Google Scholar] [CrossRef]
- Li, X.; Chen, L.; Dhaubhadel, S. 14-3-3 proteins regulate the intracellular localization of the transcriptional activator GmMYB176 and affect isoflavonoid synthesis in soybean. Plant J. 2012, 71, 239–250. [Google Scholar] [CrossRef]
- Karin, M. Signal transduction from the cell surface to the nucleus through the phosphorylation of transcription factors. Curr. Opin. Cell Biol. 1994, 6, 415–424. [Google Scholar] [CrossRef]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, H.S.; Bahk, S.; An, J.; Yoo, Y.; Kim, J.Y.; Chung, W.S. Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res. 2017, 45, 6613–6627. [Google Scholar] [CrossRef]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef]
- Meng, X.; Xu, J.; He, Y.; Yang, K.Y.; Mordorski, B.; Liu, Y.; Zhang, S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 2013, 25, 1126–1142. [Google Scholar] [CrossRef]
- Pi, E.; Xu, J.; Li, H.; Fan, W.; Zhu, C.; Zhang, T.; Jiang, J.; He, L.; Lu, H.; Wang, H.; et al. Enhanced Salt Tolerance of Rhizobia-inoculated Soybean Correlates with Decreased Phosphorylation of the Transcription Factor GmMYB183 and Altered Flavonoid Biosynthesis. Mol. Cell. Proteom. 2019, 18, 2225–2243. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).