Abstract
Automatic skin lesion segmentation is essential for early melanoma diagnosis, yet the scarcity and limited diversity of annotated training data hinder progress. We introduce a two-stage framework that first employs a denoising diffusion probabilistic model (DDPM) enhanced with dilated convolutions and self-attention to synthesize unseen, high-fidelity dermoscopic images. In the second stage, segmentation models—including a dilated U-Net variant that leverages dilated convolutions to enlarge the receptive field—are trained on the augmented dataset. Experimental results demonstrate that this approach not only enhances segmentation accuracy across various architectures with an increase in DICE of more than 0.4, but also enables compact and computationally efficient segmentation models to achieve performance comparable to or even better than that of models with 10 times the parameters. Moreover, our diffusion-based data augmentation strategy consistently improves segmentation performance across multiple architectures, validating its effectiveness for developing accurate and deployable clinical tools.
1. Introduction
Skin cancer, particularly malignant melanoma, represents one of the most aggressive and life-threatening malignancies [1]. The prognosis for patients is starkly dependent on the timing of diagnosis; melanoma detected in its early stages is often curable with simple excision, while late-stage, metastatic melanoma causes a lower survival rate [2]. This clinical reality underscores the importance of accurate and early-stage diagnosis. Dermoscopy has become the frontline, non-invasive diagnostic tool for dermatologists. Providing a magnified, illuminated view of the skin reveals subsurface structures and patterns invisible to the naked eye [3]. However, interpreting dermoscopic images remains highly subjective, susceptible to inter-observer variability, and demands extensive specialized training. The growing volume of cases puts additional strain on clinical resources with objective, reliable, and scalable support.
Deep learning has transformed medical image analysis, with skin lesion segmentation becoming a core component of modern computer-aided diagnosis (CAD) systems. An accurate delineation of the lesion boundary is the foundational step for all subsequent analyses, including the extraction of clinically relevant biomarkers (such as asymmetry, border irregularity, and color variegation) and ultimate classification [4]. Early deep learning approaches successfully used convolutional neural networks (CNNs) to extract powerful high-level features for classification tasks [5,6]. However, for segmentation, the U-Net architecture [7], with its symmetric encoder–decoder structure and skip connections, rapidly became the dominant paradigm in biomedical imaging. Its ability to integrate multi-scale contextual information with high-resolution spatial details proved highly effective for delineating complex structures like skin lesions. Subsequent research produced some variants, such as UNet++ [8], which uses nested and dense skip connections to bridge the semantic gap, and models incorporating attention mechanisms to focus on salient features [9] or integrating Transformer modules for enhanced global context modeling (e.g., TransUNet [10]). Despite this remarkable progress, the development of truly robust segmentation models is severely hampered by two intertwined challenges: chronic data scarcity and the profound complexity of lesion morphology.
First, the performance of deep learning model scales depends heavily on the diversity of the training data. While in the medical domain, assembling large, high-quality datasets is a formidable obstacle. Strict patient privacy regulations (e.g., HIPAA, GDPR), ethical considerations, and the immense cost and labor involved in having clinical experts meticulously annotate create a data bottleneck [11]. Not only the quantity, but we also often underrepresent certain skin types (e.g., on the Fitzpatrick scale), and rare but clinically significant lesion subtypes also exhibit poor generalization, failing when faced with the diversity of real-world clinical cases.
Second, skin lesion segmentation is a challenging computer vision task due to wide variations in size and shape, ambiguous boundaries, and real-world imaging artifacts like hairs and reflections [12]. Class imbalance further biases standard models toward the background. While specialized loss functions (e.g., Dice, Focal) help, the inherent complexity remains a significant challenge. Despite recent progress, traditional DDPMs still face three key limitations in medical image generation. First, they often fail to reconstruct fine-grained anatomical textures—such as microvascular patterns—due to a fixed receptive field, leading to local artifacts and mode collapse [13,14]. Secondly, medical images have inherently highly imbalanced classes, with rare lesions occupying a negligible proportion in the high-dimensional diffusion space [15]. As a result, the Markov chain tends to converge toward common patterns and aggravate class imbalance [16]. Lastly, the stacking of multi-layer convolution was used for expanding the receptive field. To address this, we employ dilated convolutions [17,18], which enlarge the receptive field efficiently while maintaining computational feasibility.
In this work, we confront the dual challenges of data scarcity and segmentation complexity head-on. We propose an innovative two-stage framework that first synthesizes a large corpus of high-fidelity dermoscopic images using a specially enhanced DDPM and then leverages this augmented data to train highly accurate and efficient segmentation models.
The primary contributions of this paper are threefold:
- An enhanced DDPM architecture for high-fidelity dermoscopic image synthesis. We design a DDPM whose U-Net backbone is specifically modified with dilated convolutions and self-attention layers to capture the unique characteristics of diffuse borders and complex internal textures.
- A powerful data augmentation framework that boosts model efficiency. We systematically demonstrate that our synthetic data augmentation strategy provides consistent and substantial performance gains across a broad spectrum of segmentation architectures, from lightweight to complex models. Our framework enables compact, computationally efficient models to achieve accuracy on par with, or even exceeding, those models with more parameters, which is beneficial for deploying accurate models in resource-constrained environments.
- Rigorous and comprehensive experimental validation. We conduct a thorough evaluation of our framework on standard benchmark datasets. Statistical validation of segmentation improvements across multiple architectures, and a deep dive into the trade-offs between model complexity and performance, proving the efficacy and generalizability of our approach.
2. Related Works
This section presents relevant prior research pertinent to our work for a better understanding of the methods in this paper. This section introduces the advancements and challenges in deep learning-based skin lesion segmentation, the generative models for data augmentation, and relevant concepts for feature representation in the medical domain.
2.1. Skin Lesion Segmentation with Deep Learning
Automated skin lesion segmentation has been studied for decades as a foundational step in computer-aided diagnosis (CAD). Some existing challenges include data scarcity, labeling complex, ambiguous boundary features, and diverse visual variability between and within lesion types. Early stage methods using traditional image processing and handcrafted features with traditional machine learning algorithms. While providing initial solutions, these methods typically rely on the features designed and lack robustness. The advent of deep learning, particularly convolutional neural networks (CNNs), enabled the extraction of powerful deep visual features for more accurate classification and segmentation [5,6].
The U-Net architecture [7], with a symmetric encoder–decoder structure, was designed explicitly for and rapidly became the dominant paradigm of image segmentation in skin lesions and other biomedical image segmentation tasks. Its ability to fuse multi-scale features proved highly effective for delineating lesion boundaries. Consequently, numerous variants have been developed upon it for better performance: PsLSNet, using deeper networks [19], and others proposed, using full-resolution features or high-resolution features in leveraging full-resolution feature maps to refine boundaries [20,21], and integrating attention mechanisms to focus on salient features [9]. Further advancements involve redesigning skip connections with nested or dense pathways (e.g., UNet++ [8]) to bridge the semantic gap between the encoder and decoder [22]. More recent work has explored integrating novel modules like transformers (e.g., TransUNet [10]) and MLP-based blocks (e.g., U-NeXt [23], Rolling-UNet [24]) into U-Net-like frameworks.
A longstanding challenge in medical segmentation, especially for skin lesions smaller than the image size, is class imbalance. Standard loss functions tend to be biased towards the dominant background class. To address this issue, specialized region-based losses like the dice or IoU loss directly optimize the overlap between the predicted and ground-truth one. Other functions, such as Tversky loss and Focal Loss, provide mechanisms to differentially weight false positives/negatives and help the model focus on hard-to-classify pixels. In line with established practices, our work employs a combined Binary Cross-Entropy (BCE) and Dice loss to balance pixel-level classification accuracy and the quality of segmentation overlap, promoting accurate boundary definition [25]. While our framework adopts the commonly used BCE+Dice loss without introducing a novel imbalance-specific objective, the primary contribution towards mitigating class imbalance lies in the data-space augmentation stage. By generating diverse synthetic samples, especially for underrepresented lesion types, the proposed method indirectly alleviates imbalance and enhances segmentation robustness.
2.2. Generative Models for Medical Data Augmentation
The performance of deep learning models is tied to the scale and diversity of training data, a significant bottleneck in the medical field. Generative models offer a powerful solution by synthesizing novel and realistic samples from existing data, surpassing the limitations of traditional augmentations like geometric transforms.
Early generative methods like Hidden Markov Models (HMMs) [26] and Gaussian Mixture Models (GMMs) [27] struggled with high-dimensional data. After 2010, with the development of deep learning, variational autoencoders (VAEs) [28] and generative adversarial networks (GANs) [29] were proposed one after another, gaining widespread attention for synthesizing images, including human face and skin lesion data or other medical images [30,31]. Although hard to train, GANs can suffer from training instability and mode collapse through some techniques, and both GANs and VAEs may fail to capture the fine-grained details necessary for high-quality medical applications.
Denoising Diffusion Probabilistic Models (DDPMs) have recently emerged as a powerful class of generative models, showing their powerful performance in generating high-fidelity, diverse images. Operating through a forward noising process and learning the reverse denoising process, the DDPMs transform Gaussian noise into data. The paradigm implies that DDPMs offer more stable training, diverse, and superior quality than GANs and VAEs. As a result, more researchers [32] have begun to use DDPMs in medical imaging. Our work leverages and enhances this advanced generative framework for the specific task of skin lesion synthesis. Recognizing the drawbacks of standard DDPM, its configurations fail to model the intricate textures and boundary characteristics of dermoscopic images. To address this, we introduce targeted architectural improvements within the DDPMs, mitigating these drawbacks and enhancing performance.
In addition to generative model-based augmentation strategies, recent advances in deep feature fusion have demonstrated strong potential across various domains. For instance, the Multi-level Fusion Swin Transformer (MFST) [33] integrates a multi-level feature merging module and an adaptive feature compression module, effectively narrowing semantic gaps and improving classification performance in remote sensing scene understanding. In the medical imaging domain, the multi-feature fusion CNN and Bi-level Routing Attention Transformer Network (MCBTNet) [34] combine convolutional layers with Transformer-based global modeling in a U-shaped architecture, further enhanced by a frequency channel–spatial attention mechanism on skip connections. These methods concentrate on combining local and global contextual cues for better representation. Unlike the above approaches, our framework focuses on data-space augmentation via an enhanced DDPM to obtain a larger receptive field and generate high-fidelity, diverse synthetic images, thereby enriching training distributions before feature extraction.
3. Proposed Method
Our methodology is a synergistic two-stage framework engineered to address the concurrent challenges of data scarcity and morphological complexity in skin lesion segmentation. As illustrated in Figure 1, the framework consists of (1) a data synthesis stage, where we introduce an architecturally enhanced denoising diffusion probabilistic model (DDPM) to generate a diverse set of high-fidelity synthetic images, and (2) a segmentation stage, where a specialized U-Net variant leverages this augmented dataset for precise lesion delineation. The unifying principle of our framework is the strategic deployment of dilated convolutions in both stages, which expands the model’s receptive field to effectively capture the multi-scale features and ambiguous boundaries characteristic of skin lesions.
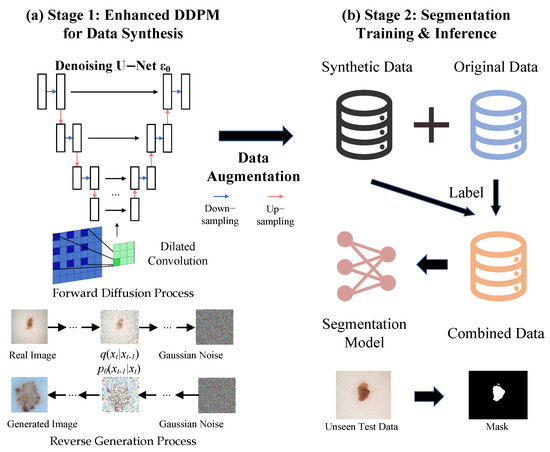
Figure 1.
Overview of our proposed two-stage framework. Stage 1: An architecturally enhanced DDPM generates high-quality synthetic skin lesion images. Stage 2: Segmentation models, including our proposed Dilated-UNet, are trained on the augmented dataset.
3.1. Stage 1: Enhanced DDPM for Data Synthesis
To overcome data scarcity limitations while preserving data diversity, we develop a generative model based on the DDPM paradigm. This paradigm is defined by a fixed forward (noising) process and a learned reverse (denoising) process.
The forward process, q, introduces Gaussian noise to a real skin lesion image over T timesteps, forming a Markov chain:
where denotes a predefined variance schedule. A key property of this process is that it admits a closed-form expression, which allows for direct sampling of a noisy image at any timestep t:
The reverse process, , aims to invert the noising operation by transforming pure noise back into a clean image . The process is accomplished by training a neural network to predict the original noise from the corrupted image . A simplified objective function optimizes the network:
The inference speed of the generative process heavily depends on the design of the noise prediction network . We construct this network as a U-Net with integrated architectural enhancements, which begins with an initial convolution to project the input image into a high-dimensional feature space (). The U-Net encoder then processes these features through four resolution levels with channel multipliers .
Each network level is constructed using residual blocks (ResBlocks), incorporating our key framework enhancements. Specifically, the main architectural modification we performed on the DDPM is the integration of dilated convolutions into the residual blocks of the noise prediction U-Net. This modification enlarges the receptive field without increasing parameters, allowing the model to better capture multi-scale lesion context and ambiguous boundaries. Although conventional DDPM models can be adapted for segmentation and often outperform GAN-based frameworks, they have a limited receptive field due to fixed-size convolutions. This restricts their ability to capture multi-scale lesion context and diffuse boundaries. We address this by integrating dilated convolutions into the DDPM backbone, enlarging the receptive field without adding parameters, thereby generating higher-quality, more diverse dermoscopic images for downstream segmentation. To capture long-range spatial dependencies, we use dilated convolutions [17] in the two deepest levels of ResBlocks. A 2D dilated convolution, , with a filter F and dilation rate r is defined as
where is a pixel location and is the kernel domain. Using rates expands the receptive field without adding parameters, which is critical for capturing the diffuse nature of skin lesions.
Further, to make the network aware of the noise level, we use timestep conditioning. The discrete timestep t is mapped to a vector by sinusoidal positional embeddings [35] of dimension :
where i indexes the embedding dimension. This vector is processed by an MLP and added to the feature maps within each ResBlock, enabling the network to adapt its denoising strategy dynamically.
At the U-Net bottleneck ( resolution), we embed a multi-head self-attention mechanism [35] into the ResBlocks, helping model global correlations and ensure the synthesis of coherent structures.
After the training process, image generation proceeds by reversing the diffusion process. Starting with , we iteratively sample for using the learned network :
where and is the noise variance at step t.
3.2. Stage 2: Segmentation Training and Inference
Following the introduction of our enhanced DDPM, we proceed to demonstrate the efficacy and viability of the overall framework. To accomplish this, we will validate its performance using several baseline segmentation models trained on the augmented dataset. Furthermore, this section describes the strategies employed to address the challenges inherent in the training process, with a particular focus on resolving data imbalance.
3.2.1. Model Architecture: The Dilated-UNet
Our proposed Dilated-UNet is a variant of the U-Net architecture, which retains the characteristic symmetric encoder–decoder structure and skip connections. The fundamental innovation of our model is located in its bottleneck. Here, we have replaced the standard convolutional layers with two sequential dilated convolutions, each with a dilation rate of . This targeted modification significantly expands the model’s receptive field at its most abstract stage, enabling it to aggregate wide-ranging contextual information better to interpret ambiguous lesion borders. To validate the superiority of this approach, our subsequent experiments will focus on the Dilated-UNet architecture.
3.2.2. Loss Function and Training
The model’s final layer uses a convolution and a sigmoid function to output a pixel-wise probability map . To handle class imbalance, we use a hybrid loss function combining Binary Cross-Entropy (BCE) and Dice loss.
Given a predicted probability map and the ground truth mask over N pixels, the BCE loss is defined as
The Dice loss is defined as
where is a small constant for numerical stability. The final segmentation loss is their sum,
Models are trained on a composite dataset of real and synthetic images, enhancing generalization and robustness.
4. Experiments and Results
4.1. Dataset
We utilized the popular ISIC 2017 dataset [36] for training DDPM for image generation in this study, and the ISIC 2018 dataset [37] for evaluating the performance of skin lesion segmentation to avoid the risk of data leakage.
The ISIC 2017 dataset includes three classes of dermatological images: melanoma (MEL), seborrheic keratosis (SK), and Nevus (NV). The ISIC 2018 dataset comprises 2594 images and 13,970 corresponding ground truth response masks. Moreover, all dermoscopic images of ISIC 2017 were used for training the DDPM, and 90% of the ISIC 2018 images were used for segmentation model training, while 10% were used for validation.
4.2. Experimental Setup
All the experiments were implemented based on PyTorch (version 1.11.0 with CUDA 11.3 support) and conducted on NVIDIA GeForce RTX 3090 GPUs with an Intel® Xeon® Silver 4210R CPU @ 2.40 GHz (dual-socket configuration, 40 logical cores in total). The software runs on Linux, with NumPy version 1.24.4, and the CUDA driver version is 11.7.
For the DDPM for medical image generation, the U-Net backbone comprised four resolution levels with channel multipliers [1, 2, 2, 2] and self-attention at the second level. Timestep embedding was used for better representation using a sinusoidal positional encoding after fully connected layers. The U-Net was optimized using the AdamW optimizer with weight decay with a cosine-annealing learning rate. Data augmentation included normalization and random flipping, and gradient clipping was employed for stable training with a maximum norm of 1.0. Moreover, the batch size was set to 8. The segmentation model FCN-ResNet50 was initialized with pre-trained parameters. Models were trained using RMSprop optimizers, with momentum = 0.9, and initial learning rate with weight decay = . For computational efficiency, the dermoscopic images of skin lesions were resized to pixels. The original dataset was split into training and validation sets, with 10% for validation and 90% for the additional DDPM dataset used for training. The training was configured for up to 50 epochs with a batch size of 8. The details are in Table 1.

Table 1.
Key hyperparameter settings for experiments.
For the real images from the ISIC 2017 and ISIC 2018 datasets, the corresponding ground truth masks were provided with the datasets. However, the synthetic images generated by the enhanced DDPM did not have corresponding labels. To ensure they could be used for supervised segmentation training, we manually annotated all synthetic images using the Labelme software, following the same annotation protocol as the original datasets. Experienced annotators performed the manual annotations and subsequently reviewed them to ensure consistency and accuracy.
We generated 5000 synthetic dermoscopic images at resolution using the enhanced DDPM and used a subset of 1000 for training. Because DDPM does not provide labels, we first conducted manual quality control (QC) to discard visually implausible samples (e.g., unrealistic borders or textures) and then created masks using Labelme. To reduce subjectivity, we additionally applied lightweight automated checks (FID percentile screening and LPIPS outlier filtering) before annotation; only images passing these checks were retained.
4.3. Evaluation Metrics
The main evaluation metrics used for this study are the Dice similarity coefficient (DICE) and Fréchet inception distance (FID). FID is mainly used to assess the quality and diversity of generated images and distribution differences, and DICE is used to evaluate the segmentation accuracy.
where
- are the mean and covariance of the real image features.
- are the mean and covariance of the generated image features.
- is the squared Euclidean distance between the two mean vectors.
- denotes the trace of a matrix (sum of diagonal elements).
- is the matrix square root of the product of the two covariance matrices.
4.4. Composite Image Quality Assessment
To validate the utility and diversity of the synthetic skin lesion images generated by our enhanced denoising diffusion probabilistic model (DDPM), we aim to demonstrate that the generated images are realistic and diverse enough to augment the training data for the downstream segmentation task effectively. A lower FID score indicates that the distribution of features of the generated images is closer to that of the real images, which means a higher quality and diversity.
Qualitatively, we provide visual examples of the synthetic skin lesion images in Figure 2. Both examples showcase the realism and variations in appearance produced by the diffusion model.
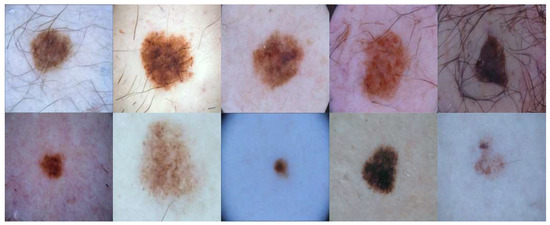
Figure 2.
Examples of synthetic skin lesion images generated by the enhanced DDPM. The five in the above line are generated using a dilated convolution layer, while those in the line below are generated using a normal convolution layer.
Although both DDPM models are good at generating some usable photos, we must admit that not all generated photos are usable. Due to the nature of the diffusion model itself and the limitations of our dataset, we still generated some less usable photos during our experiment, as shown in Figure 3.
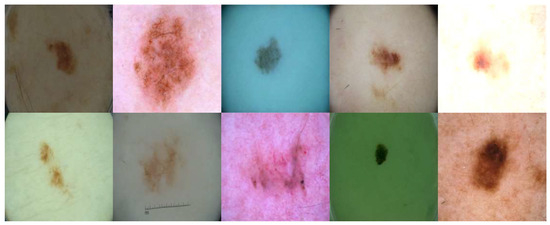
Figure 3.
Examples of poor-quality synthetic skin lesion images generated by the enhanced DDPM. The five in the above line are generated using a dilated convolution layer, while those in the line below are generated using a normal convolution layer.
Both examples showcase the realism and variations in appearance produced by the diffusion model. Furthermore, to quantitatively evaluate the fidelity and diversity of the generated images, we conducted an assessment using the Fréchet inception distance (FID). As summarized in Table 2, the FID score for the traditional DDPM was computed using 5000 generated images with no manual filtering against a reference set of 1000 authentic images from the ISIC 2017 dataset. The same evaluation protocol was also applied to the modified DDPM architecture. Lower FID scores indicate better distributional similarity between the generated and real image sets, with the results showing a slight performance improvement when dilated convolutions are integrated into the model.

Table 2.
Fréchet inception distance (FID) between generated synthetic images and real images on the ISIC 2017 dataset.
This assessment confirms the capability of our diffusion-based model. It is capable of producing high-quality and diverse images, showing its potential as a promising and valuable resource for augmenting the training data. Most of the generated images are usable, while our model still generates some poor quality images, as shown in Figure 3. This implies that the addition of synthetic images needs to be manually reviewed beforehand.
Qualitative examples Figure 2 show that the enhanced DDPM preserves key lesion characteristics such as boundary shape and color distribution, making the images visually close to authentic dermoscopic images. Quantitatively, the low FID scores in Table 2 indicate a small distribution gap. Minor differences remain in fine vascular textures and rare lesion types Figure 3, so manual quality control is applied before training.
4.5. Generalization of Data Augmentation to Other Segmentation Architectures
To demonstrate the broad applicability and model-agnostic benefits of our synthetic data generation strategy, we extended our augmentation approach to a diverse set of established segmentation architectures: Standard U-Net [7], Dilated-UNet (our U-Net variant with dilated convolutions, as described in Section 3.2.1), FCN-ResNet50, FCN-ResNet101 [38,39], DeepLabV3-ResNet50, DeepLabV3-ResNet101 [40], MEFP-Net [41], and ChimeraNet + LAMA [42]. Each architecture was trained with real data only (R) and real data augmented by synthetic images from our Enhanced DDPM (R+S). All models were trained under consistent hyperparameter settings for their families and evaluated on the test set. Performance metrics are presented in Table 3, with a visual summary in Figure 4.

Table 3.
Segmentation performance (DICE/IoU) with updated results. Best R + S results per metric in bold. Statistical significance between R and R + S is assessed using a paired t-test: * , ** , n.s. denotes no significant difference.
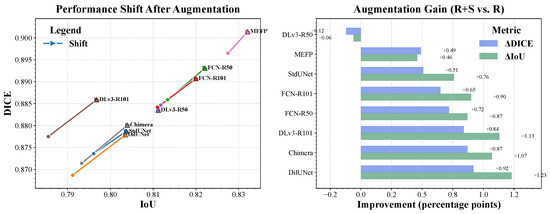
Figure 4.
Visual summary of segmentation performance across different architectures with and without synthetic data augmentation. This combined figure includes both the absolute DICE and IoU scores, as well as the performance gains (in percentage points) due to augmentation.
In addition to assessing the visual fidelity and FID scores of the synthetic images, we further validate their utility by examining the impact on downstream segmentation performance. As shown in Figure 5, models trained with the augmented dataset (real + synthetic) consistently produce sharper boundaries and more complete lesion masks compared to those trained with real data only. In particular, our proposed Dilated-UNet demonstrates more accurate delineation of ambiguous lesion regions, where the segmentation masks are visually closer to the ground truth. This qualitative comparison highlights that the synthetic data, despite including a small portion of low-quality samples, can substantially enhance the effectiveness of lesion segmentation.
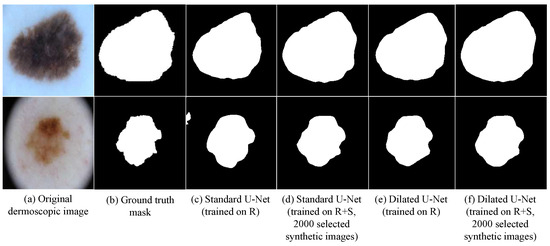
Figure 5.
Qualitative comparison of segmentation results. (a) Original image, (b) ground-truth mask, (c) U-Net (R), (d) U-Net (R+S), (e) Dilated U-Net (R), (f) Dilated U-Net (R+S). Incorporating synthetic data (R+S) improves boundary sharpness and completeness, especially for ambiguous lesion regions.
Both results demonstrate the value of augmenting training data with synthetic images from our Enhanced DDPM for most architectures, as five of six models showed improvements in DICE and IoU scores (Table 3). The exception was DeepLabV3-ResNet50, which experienced marginal degradation. Notably, the Dilated-UNet showed the most substantial gains from augmentation (IoU = +1.23 pp), suggesting its dilated convolutions synergize effectively with synthetic data. Among ResNet-based models, FCN variants benefited most significantly, with FCN-ResNet50 achieving the highest absolute performance (DICE: 0.8931, IoU: 0.8221) when augmented. This represents a key shift from our initial findings, where DeepLabV3 variants previously dominated. The results highlight that architectural differences significantly influence how effectively models leverage synthetic data, with FCN architectures showing particular compatibility with our augmentation approach. It is worth noting that the DeepLabV3-ResNet50 architecture exhibited a slight performance decrease after augmentation (Table 3). We attribute this to stochastic variation inherent in the training process, as the performance differences are within the range of typical run-to-run fluctuations observed in our experiments. Additionally, this architecture may inherently benefit less from the type of diversity introduced by our synthetic data, leading to marginal or no gain in certain runs. Additionally, this comparison can also be interpreted as an ablation study on the effect of dilated convolutions.
4.6. Impact of Synthetic Data on Segmentation Performance
We conducted a series of experiments to validate the effect of incorporating generated synthetic data on the performance of a U-Net-based focal point segmentation model. We first compared the model performance when trained using five distinct data configurations: (1) only real data, (2) only synthetic data generated using dilated convolutions, (3) only synthetic data generated using ordinary (regular) convolutions, (4) a combination of real data and synthetic data from dilated convolutions, and (5) a combination of real and synthetic data from ordinary convolutions. This initial comparison, summarized in Table 4, highlights the overall contribution of synthetic data and demonstrates the differing impact of the two convolution approaches for synthetic data generation, particularly in scenarios where real data might be limited. We evaluated segmentation performance using the DICE coefficient, where a higher value indicates better accuracy. All evaluations were performed on a held-out real test set.

Table 4.
Segmentation performance (DICE) on the real test set using different initial training data configurations on Dilated-UNet. Synthetic data, when used in mixed configurations, consisted of 1000 images.
Furthermore, we conducted additional experiments to assess how the proportion of real and synthetic data affects the final segmentation performance. For these, our model consistently generated synthetic images enhanced with dilated convolutional layers. The training data configurations for this evaluation were
- Full Synthetic Data: 0 real images + 1000 synthetic images.
- Real/Synthetic (1:10): 100 real images + 1000 synthetic images.
- Real/Synthetic (1:2): 500 real images + 1000 synthetic images.
- Real/Synthetic (1:1): 1000 real images + 1000 synthetic images.
- Real/Synthetic (2:1): 2000 real images + 1000 synthetic images.
We conducted experiments with different random seeds eight times. These experiments, detailed in Table 5, aim to quantify the contribution of synthetic data at different mixing ratios and to determine if an optimal balance between real and synthetic samples can be identified for improved segmentation accuracy.

Table 5.
Impact of real-to-synthetic data ratios on segmentation performance (DICE) on Dilated-UNet. Synthetic data was generated using dilated convolutions. Evaluation is on the real test set (10% of the real set).
We present qualitative examples in Figure 6 for a visual understanding. We compare the predicted segmentation masks obtained from models trained under different configurations, specifically contrasting a model trained on real data only with one trained on a combination of real and synthetic data.
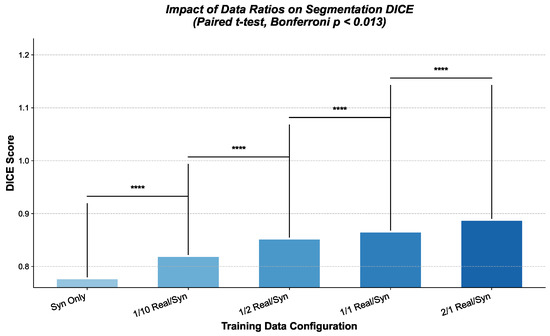
Figure 6.
Qualitative comparison of segmentation results on example skin lesion images on Dilated-UNet. Average accuracy and statistical significance ( ****: p < 0.001, t-test).
These comprehensive experiments demonstrate that incorporating synthetic data, particularly incorporated by the plot enhanced by DDPM with dilated convolutions, significantly improves the performance of the U-Net-based segmentation model. Statistical analysis using a t-test also highlights the contribution of the synthetic data to the final results in Figure 6. Our proposed framework mitigates the challenges of data scarcity in medical image analysis tasks. The solidity experiment across different real-to-synthetic data ratios further underscores how it optimizes this balance for specific applications.
4.7. Analysis of Augmentation Efficacy and Model Efficiency
Building upon the performance results in Section 4.5, this section provides a deeper analysis of the interplay between model architecture, complexity, and the efficacy of our proposed data augmentation framework. By contextualizing the performance gains with the parameter counts of each model, we demonstrate that a primary advantage of our method is its ability to enhance model efficiency significantly. The performance improvements shown in Table 3 are not uniform across all models, suggesting that the model’s architecture dictates its capacity to leverage our synthetic data. For instance, the Dilated-UNet, a moderately sized architecture, registered the most significant gains (IoU + 1.23 pp). This indicates that our framework is particularly effective at providing valuable, generalizable features for models that might otherwise struggle to teach them from a limited real dataset. Conversely, the minor performance degradation in DeepLabV3-ResNet50 suggests a potential “feature conflict,” where the synthetic data distribution may interfere with the specific learning trajectory of this architecture–backbone combination. The fact that its deeper ResNet101 counterpart benefited substantially highlights the nuanced sensitivity of model performance to architecture and data augmentation. To frame this discussion more concretely, Table 6 details the complexity of each model, as measured by trainable parameters. The models span a vast complexity range, from the ultra-lightweight DFF-UNet (0.19M) to the heavyweight DeepLabV3-ResNet101, providing an ideal landscape to evaluate our framework’s impact.

Table 6.
Comparison of model complexity and performance impact of our framework. The table lists trainable parameters, baseline and augmented IoU scores, and the change in percentage points (pp). Our framework generally provides a significant performance boost, especially for models like Dilated-UNet and FCN-ResNet50.
A cross-comparison between performance (Table 3) and complexity (Table 6), visually summarized in Figure 7, reveals two key insights into our framework’s value. First, it empowers lightweight models to achieve heavyweight performance. The most striking example is the DFF-UNet. With only 0.19M parameters, its baseline IoU (0.8175) is respectable but not top-tier. However, when trained with our augmented data (R+S), its IoU surges to 0.8219. This result is remarkable, as it effectively matches the performance of the augmented FCN-ResNet50 (0.8221), a model with 185 times more parameters (35.31 M vs. 0.19 M). This result validates that our framework can compensate for the limited intrinsic capacity of smaller models by providing diverse, information-rich training examples, helping them to punch far above their weight. Second, it allows moderately sized models to outperform their larger counterparts, enhancing their cost-effectiveness. The augmented FCN-ResNet50 not only achieves the highest IoU (0.8221) among all tested models but does so with significantly fewer parameters than the DeepLabV3-ResNet101 (35.31 M vs. 60.99 M), which proves that our framework can obviate the need for larger, more computationally expensive models by using moderately sized architectures more efficiently. In conclusion, our data augmentation framework is a powerful tool for improving model efficiency. It breaks the convention that top-tier performance requires massive parameter counts. For resource-constrained environments, it enables lightweight models to deliver state-of-the-art results. Our framework provides a more compact and efficient architecture without sacrificing performance for the application, paving the way for developing highly accurate segmentation tools in real-world deployment.
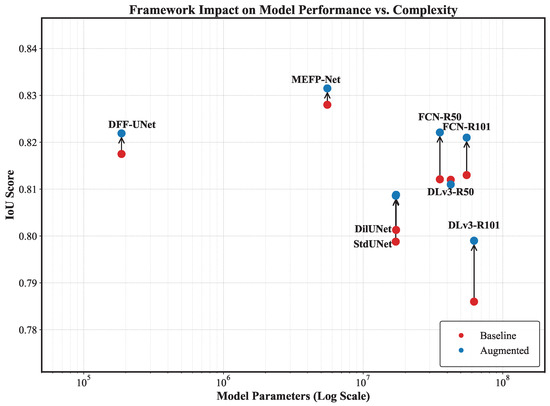
Figure 7.
Framework impact on model performance vs. complexity. This scatter plot visualizes the trade-off between model performance (IoU score) and complexity (model parameters, on a log scale). Red points indicate the baseline performance of various segmentation models, while blue points show the performance after applying our data augmentation framework. The vertical arrows highlight the improvement in the IoU score. The chart demonstrates that our framework consistently boosts performance across different architectures, enabling smaller, more efficient models to match or exceed the performance of their larger counterparts. For the same amount of Dilated-UNet and standard U-Net, and a similar performance trained by R+S, the labels were set on the left side of the baseline point.
5. Conclusions
In this work, we addressed a persistent challenge in automated skin lesion segmentation: the scarcity of annotated medical data and the trade-off between inference speed and accuracy. We introduced an innovative two-stage framework to tackle these issues head-on. Our comprehensive experimental validation demonstrated that our two-stage data augmentation framework yields consistent and substantial performance improvements across different amounts and a broad spectrum of segmentation architectures. Our framework paves the way for highly accurate and lightweight segmentation tools for real-world clinical settings, including resource-constrained environments. By obviating the need for massive computational overhead without sacrificing accuracy, it enables the model to approach its performance ceiling more closely rather than being limited by the amount of data. Future work may focus on further improving the model’s performance, extending this framework to other medical imaging modalities, exploring conditional generation to target specific rare lesion types, and developing automated quality filtering mechanisms for synthetic data to streamline the pipeline even further.
Author Contributions
Conceptualization, P.Y.; methodology, P.Y.; software, P.Y.; validation, Z.C.; resources, X.D.; data curation, X.S.; writing—original draft preparation, P.Y.; writing—review and editing, X.D. All authors have read and agreed to the published version of the manuscript.
Funding
The research work described in this paper was fully supported by the Natural Science Foundation Project of Shandong Province (ZR2024QF184).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Flavio, J.J.; Fernandez, S. The Rising Incidence of Skin Cancers in Young Adults: A Population-Based Study in Brazil. Sci. J. Dermatol. Venereol. 2025, 3, 39–53. [Google Scholar]
- Waseh, S.; Lee, J.B. Advances in melanoma: Epidemiology, diagnosis, and prognosis. Front. Med. 2023, 10, 1268479. [Google Scholar] [CrossRef]
- Abbas, Q.; Fondón, I.; Rashid, M. Unsupervised skin lesions border detection via two-dimensional image analysis. Comput. Methods Programs Biomed. 2011, 104, e1–e15. [Google Scholar] [CrossRef]
- Singh, G.; Kamalja, A.; Patil, R.; Karwa, A.; Tripathi, A.; Chavan, P. A comprehensive assessment of artificial intelligence applications for cancer diagnosis. Artif. Intell. Rev. 2024, 57, 179. [Google Scholar] [CrossRef]
- Khouloud, S.; Ahlem, M.; Fadel, T.; Amel, S. W-net and inception residual network for skin lesion segmentation and classification. Appl. Intell. 2022, 52, 3976–3994. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Xia, Y.; Shen, C. Attention residual learning for skin lesion classification. IEEE Trans. Med. Imaging 2019, 38, 2092–2103. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef]
- Kaur, R.; Kaur, S. Automatic skin lesion segmentation using attention residual U-Net with improved encoder-decoder architecture. Multimed. Tools Appl. 2025, 84, 4315–4341. [Google Scholar] [CrossRef]
- Chen, J.; Mei, J.; Li, X.; Lu, Y.; Yu, Q.; Wei, Q.; Luo, X.; Xie, Y.; Adeli, E.; Wang, Y.; et al. TransUNet: Rethinking the U-Net architecture design for medical image segmentation through the lens of transformers. Med. Image Anal. 2024, 97, 103280. [Google Scholar] [CrossRef]
- Bie, Y.; Luo, L.; Chen, H. Mica: Towards explainable skin lesion diagnosis via multi-level image-concept alignment. In Proceedings of the AAAI Conference on Artificial Intelligence, Vancouver, BC, Canada, 20–27 February 2024; Volume 38, pp. 837–845. [Google Scholar]
- El-Shafai, W.; El-Fattah, I.A.; Taha, T.E. Advancements in non-invasive optical imaging techniques for precise diagnosis of skin disorders. Opt. Quantum Electron. 2024, 56, 1112. [Google Scholar] [CrossRef]
- Jiang, H.; Imran, M.; Zhang, T.; Zhou, Y.; Liang, M.; Gong, K.; Shao, W. Fast-DDPM: Fast denoising diffusion probabilistic models for medical image-to-image generation. IEEE J. Biomed. Health Inform. 2025; Early Access. [Google Scholar] [CrossRef]
- Bourou, A.; Boyer, T.; Gheisari, M.; Daupin, K.; Dubreuil, V.; De Thonel, A.; Mezger, V.; Genovesio, A. PhenDiff: Revealing subtle phenotypes with diffusion models in real images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; Springer: Berlin/Heidelberg, Germany, 2024; pp. 358–367. [Google Scholar]
- Huijben, E.M.; Pluim, J.P.; van Eijnatten, M.A. Denoising diffusion probabilistic models for addressing data limitations in chest X-ray classification. Inform. Med. Unlocked 2024, 50, 101575. [Google Scholar] [CrossRef]
- Pan, Z.; Xia, J.; Yan, Z.; Xu, G.; Wu, Y.; Jia, Z.; Chen, J.; Shi, Y. Rethinking Medical Anomaly Detection in Brain MRI: An Image Quality Assessment Perspective. arXiv 2024, arXiv:2408.08228. [Google Scholar] [CrossRef]
- Yu, F.; Koltun, V. Multi-scale context aggregation by dilated convolutions. arXiv 2015, arXiv:1511.07122. [Google Scholar]
- Mehta, S.; Rastegari, M.; Caspi, A.; Shapiro, L.; Hajishirzi, H. Espnet: Efficient spatial pyramid of dilated convolutions for semantic segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 552–568. [Google Scholar]
- Dash, M.; Londhe, N.D.; Ghosh, S.; Semwal, A.; Sonawane, R.S. PsLSNet: Automated psoriasis skin lesion segmentation using modified U-Net-based fully convolutional network. Biomed. Signal Process. Control 2019, 52, 226–237. [Google Scholar] [CrossRef]
- Xie, F.; Yang, J.; Liu, J.; Jiang, Z.; Zheng, Y.; Wang, Y. Skin lesion segmentation using high-resolution convolutional neural network. Comput. Methods Programs Biomed. 2020, 186, 105241. [Google Scholar] [CrossRef]
- Al-Masni, M.A.; Al-Antari, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 2018, 162, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Shuai, R.; Ma, L.; Liu, W.; Wu, M. Segmentation of skin lesions image based on U-Net++. Multimed. Tools Appl. 2022, 81, 8691–8717. [Google Scholar] [CrossRef]
- Zeng, Z.; Hu, Q.; Xie, Z.; Li, B.; Zhou, J.; Xu, Y. Small but mighty: Enhancing 3d point clouds semantic segmentation with u-next framework. Int. J. Appl. Earth Obs. Geoinf. 2025, 136, 104309. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, H.; Liu, M.; Yu, H.; Chen, Z.; Gao, J. Rolling-unet: Revitalizing mlp’s ability to efficiently extract long-distance dependencies for medical image segmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, Vancouver, BC, Canada, 20–27 February 2024; Volume 38, pp. 3819–3827. [Google Scholar]
- Tang, H.; Li, Z.; Zhang, D.; He, S.; Tang, J. Divide-and-conquer: Confluent triple-flow network for RGB-T salient object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 47, 1958–1974. [Google Scholar] [CrossRef]
- Eddy, S.R. What is a hidden Markov model? Nat. Biotechnol. 2004, 22, 1315–1316. [Google Scholar] [CrossRef]
- Rasmussen, C.E. The infinite Gaussian mixture model. Adv. Neural Inf. Process. Syst. 1999, 12, 554–560. Available online: https://proceedings.neurips.cc/paper_files/paper/1999/file/97d98119037c5b8a9663cb21fb8ebf47-Paper.pdf (accessed on 20 August 2025).
- Kingma, D.P.; Welling, M. An Introduction to Variational Autoencoders (Foundations and Trends® in Machine Learning); Now Publishers: Norwell, MA, USA, 2019; Volume 12, pp. 307–392. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Qin, Z.; Liu, Z.; Zhu, P.; Xue, Y. A GAN-based image synthesis method for skin lesion classification. Comput. Methods Programs Biomed. 2020, 195, 105568. [Google Scholar] [CrossRef] [PubMed]
- Casti, P.; Cardarelli, S.; Comes, M.C.; D’Orazio, M.; Filippi, J.; Antonelli, G.; Mencattini, A.; Di Natale, C.; Martinelli, E. S3-VAE: A novel Supervised-Source-Separation Variational AutoEncoder algorithm to discriminate tumor cell lines in time-lapse microscopy images. Expert Syst. Appl. 2023, 232, 120861. [Google Scholar] [CrossRef]
- Kazerouni, A.; Aghdam, E.K.; Heidari, M.; Azad, R.; Fayyaz, M.; Hacihaliloglu, I.; Merhof, D. Diffusion models in medical imaging: A comprehensive survey. Med. Image Anal. 2023, 88, 102846. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, N.; Liu, W.; Chen, H.; Xie, Y. MFST: A multi-level fusion network for remote sensing scene classification. IEEE Geosci. Remote Sens. Lett. 2022, 19, 1–5. [Google Scholar] [CrossRef]
- Zhang, B.; Zheng, Z.; Zhao, Y.; Shen, Y.; Sun, M. MCBTNet: Multi-Feature Fusion CNN and Bi-Level Routing Attention Transformer-based Medical Image Segmentation Network. IEEE J. Biomed. Health Inform. 2025, 29, 5069–5082. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar] [CrossRef]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; IEEE: New York, NY, USA, 2018; pp. 168–172. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 1–9. [Google Scholar] [CrossRef]
- Nie, X.; Zhang, C.; Cao, Q. Image segmentation method on quartz particle-size detection by deep learning networks. Minerals 2022, 12, 1479. [Google Scholar] [CrossRef]
- Sang, D.V.; Minh, N.D. Fully residual convolutional neural networks for aerial image segmentation. In Proceedings of the 9th International Symposium on Information and Communication Technology, Danang City, Vietnam, 6–7 December 2018; pp. 289–296. [Google Scholar]
- Heryadi, Y.; Irwansyah, E.; Miranda, E.; Soeparno, H.; Hashimoto, K. The effect of resnet model as feature extractor network to performance of DeepLabV3 model for semantic satellite image segmentation. In Proceedings of the 2020 IEEE Asia-Pacific Conference on Geoscience, Electronics and Remote Sensing Technology (AGERS), Jakarta, Indonesia, 7–8 December 2020; IEEE: New York, NY, USA, 2020; pp. 74–77. [Google Scholar]
- Hao, S.; Yu, Z.; Zhang, B.; Dai, C.; Fan, Z.; Ji, Z.; Ganchev, I. MEFP-Net: A dual-encoding multi-scale edge feature perception network for skin lesion segmentation. IEEE Access 2024, 12, 140039–140052. [Google Scholar] [CrossRef]
- Khan, S.; Khan, A.; Teng, Y. DFF-UNet: A Lightweight Deep Feature Fusion UNet Model for Skin Lesion Segmentation. IEEE Trans. Instrum. Meas. 2025, 74, 5030214. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).