1. Introduction
One of the primary functions of the skin, the largest organ in the human body, is to act as a physical barrier to pathogenic microbes and toxic substances, thus making it one of the first lines of defence against infection [
1]. The quality of an individual’s skin is influenced by a multitude of internal and external factors [
1,
2]. External factors include the environment (sun exposure, wind, humidity, air conditioning), physical abrasions, microbes (bacteria, fungi, viruses), and exposure to toxins. Internal factors include genetics, medications (immunosuppressant drugs, birth control), antibiotics, and diet. Other factors that directly exert an effect on the skin are the types of cosmetics, skincare products, washing detergent, soap, and perfume an individual may use [
3].
A combination of these factors alongside switches in microbial diversity can lead to dysbiosis [
4]: a state of microbial imbalance within areas of the body, sometimes impacting the vitality and health status of those areas [
1,
2]. Many human skin disorders and diseases have been linked to changes within the skin microbiome, including acne, atopic dermatitis, and psoriasis [
3,
5,
6,
7,
8,
9]. Unsurprisingly, this has led to a new area of research looking at how to mediate the skin microbiome in both dermatological and cosmetic fields for common conditions such as acne, body odour, and atopic dermatitis [
3,
5,
6,
7,
8,
9].
The recent advancement of technologies has allowed researchers to evaluate the human microbiome in greater depth to better understand the connections between specific microbes and a variety of disease presentations, leading to the development of many novel and emerging therapies to treat commonly occurring skin issues [
10]. The influx of information relating to the skin microbiome and its use in pharmaceuticals has stimulated the conversation and development around probiotics.
It is known that probiotics interfere with the activity of pathogenic bacteria by producing antimicrobial substances including metabolites, peptides, and bacteriocins [
10,
11]. Further to the direct inhibition of the microorganism, they can also create an unfavourable environment for the pathogen as they can alter pH, compete for nutrients, and inhibit pathogen adhesion to epithelial cells by adhering to these cells themselves [
12].
As we have previously reported, the current topical probiotic options are restricted. Many strains used in gut-health products or classified as postbiotics lack evidence to support their role as skin commensals or their ability to survive on the skin [
3]. Recently, a live probiotic hydration serum containing live
M. luteus Q24 was introduced in New Zealand.
M. luteus Q24 is a skin commensal probiotic bacterium that resides on the skin of healthy human adults and has been identified to produce an interesting antimicrobial spectrum that is inhibitory towards pathogenic bacteria associated with many skin diseases and has been found to be safe for topical applications in humans [
12].
The primary aim of this study was to carry out a cosmetic trial to measure changes in (1) skin quality parameters using an advanced handheld skin analyser and (2) align this to any changes in microbial diversity on the skin, using a whole-genome sequencing platform, following twice-daily application of live M. luteus Q24 probiotic serum, directly applied onto the face for 25 days in healthy adult participants.
2. Materials and Methods
Live probiotic hydration serum was supplied by Unconditional Skincare Co., Blis Technologies Limited, Dunedin, New Zealand. The product packaging is composed of two chambers: chamber A containing serum: medium-chain triglyceride, silica dimethyl silylate, polysorbate 80 (T), and BLIS Q24TM (M. luteus Q24—DSMZ 17,172); and chamber B containing the hydrator: aqua, coco-caprylate, glycerin, cetearyl alcohol, glyceryl stearate SE, cetearyl wheat straw glycosides, sodium stearoyl glutamate, xanthan gum, phenoxyethanol, and ethylhexylglycerin. The skin analyser device (dpViso) and software (Dermo Bella Expert Version 3.11.0 PMX) was purchased from Chowis Co. Ltd., Yongin-si, Republic of Korea. A Samsung Tablet 10.1 was purchased from PB Technologies, Auckland, New Zealand. Skin swabs were collected using a DNA/RNA Shield Collection Tube kit (Zymo Research, Irvine, CA, USA).
2.1. Cosmetic and Colonisation Efficacy Trial
A cosmetic trial involving the topical application of a live probiotic hydration serum product was conducted in healthy adult participants.
M. luteus was previously assessed for safety as a probiotic [
12]. This product is safe, was marketed in New Zealand, and contains the skin commensal probiotic
M. luteus Q24. All participants provided written informed consent before participating in this cosmetic efficacy study. The study protocol adhered to the ethical principles outlined in the Declaration of Helsinki. The study protocol was submitted to the Health and Disability Ethics Committee (HDEC) (New Zealand) for their review of this cosmetic product study. However, the HDEC advised that their review process focuses on research with potential health-related outcomes. Since this study investigates the cosmetic effects of the product, it fell outside the HDEC’s review scope. Therefore, formal ethics committee approval was not required.
2.2. Trial Design
Ten females participated in a single-site, open-label, non-randomised, baseline-controlled trial. The study design is shown in
Figure 1. The study included healthy adult females between 18 and 60 years old with generally healthy skin but experiencing concerns like mild-to-moderate breakouts, fine lines, wrinkles, uneven skin tone, dullness, or redness. Participants were specifically interested in improving their facial appearance. To ensure participant safety, those with open cuts, active infections, autoimmune disease history, or current use of antibiotics, topical steroids, or anti-inflammatory medications were excluded. Participants were asked to apply two pumps equating to >1 × 10
8 cfu/dose (at the date of manufacturing) of the serum product on the face twice daily (morning and night) for 25 days.
Measurements of skin parameters from the face were taken at baseline and days 11, 18, and 25, using a novel skin analyser device. Skin swab samples were also collected from the same site after skin quality analysis at each sample point. Additional swab samples were also collected at 8 h after the first application and day 7 after the last application of the serum. Participants were instructed not to change their skincare routine or try out new products during the trial except to replace their current moisturiser with the probiotic serum product. Participants were also asked to report adverse events, if any.
2.3. Measurement of Skin Quality
Quantitative measurement of changes in various skin parameters such as hydration/moisture, pores, spots (pigmentation), wrinkles, and impurities (porphyrins) were carried out using a handheld skin analyser (dP/Viso, Chowis, Yongin-si, Republic of Korea). The device utilises advanced optic technology with interchangeable lenses and AI-powered analysis to allow for an in-depth measurement of different skin quality parameters. At each time point, the device camera/sensor was placed on the left cheek in line with the corner of the eye and the tip of the nose. This method was kept consistent so that the triplicate readings were taken from approximately the same area on the cheek. The data were captured as scores or images and analysed in the related app Dermo Bella Expert Version 3.11.0 PMX.
2.4. Statistical Analysis of Skin Analyser Data
Statistical analyses were performed using one-way repeated measures ANOVA, with data visualised in graphs generated with Prism 9.4.0 (GraphPad Software).
2.5. Microbial Analysis
2.5.1. Sample Collection
At each time point, after skin quality parameter measurement, skin swab samples were collected. Briefly, a sterile swab from the DNA/RNA Shield Collection Tube kit (Zymo Research, Irvine, CA, USA) was used to collect the sample by swabbing the cheek in a zig–zag manner, covering about a 4 cm × 4 cm area. The tip of the swab was then cut and placed into the collection tube prefilled with the DNA/RNA Shield reagent. The tube was then capped and inverted several times to ensure homogeneity and stored at −20 °C until analysed. The following samples were sent to an external independent laboratory COSMOS ID, Germantown, MD, USA for whole-genome sequencing (WGS) and bioinformatic analysis: A1, pre-sample day 0 (n = 10); A2, 8 h (n = 6); A3, day 11 (n = 10); A4, day 25 (n = 10); and A5, day 32 (7 days after last serum application) (n = 6).
2.5.2. DNA Extraction
Microbial DNA was isolated from skin swab samples using the ZymoBIOMICS Micro Prep kit (Zymo Research, Irvine, CA, USA), following the manufacturer’s instructions. Extracted DNA was quantified using a Qubit 4 fluorometer with the Qubit™ dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). An Illumina Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, USA) was used to prepare libraries with 1 ng of total DNA input and IDT Unique Dual Indexes (Integrated DNA Technologies, Coralville, IA, USA). Briefly, this involved fragmenting genomic DNA using the Nextera XT enzyme, followed by the addition of unique dual indexes and 12 PCR cycles for library amplification. Libraries were purified using AMPure XP magnetic beads (Beckman Coulter, Indianapolis, IN, USA) and eluted in QIAGEN EB buffer (Qiagen, Venlo, The Netherlands). Final library quantification was performed with the Qubit 4 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and Qubit™ dsDNA HS Assay Kit.
2.5.3. Library Preparation and Sequencing
Microbial DNA libraries were constructed from the extracted DNA using the Illumina Nextera XT DNA Library Prep Kit with IDT Unique Dual Indexes. The protocol involved fragmenting the genomic DNA with a specific amount of Nextera XT enzyme, followed by the addition of unique dual-index sequences to each sample. Twelve cycles of PCR amplification were then performed to enrich the libraries. Finally, the libraries were purified using AMPure XP magnetic beads (Beckman Coulter) and eluted in QIAGEN EB buffer. The library quality and quantity were assessed using the Qubit 4 fluorometer and Qubit™ dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing was performed on an Illumina HiSeq X platform (Illumina, San Diego, CA, USA), generating 150 bp paired-end reads.
2.5.4. Bioinformatics Analysis
Unassembled sequencing reads were directly analysed by the CosmosID-HUB Microbiome Platform (CosmosID Inc., Germantown, MD, USA) [
13,
14,
15,
16] for multi-kingdom microbiome analysis. Briefly, the system utilises curated genome databases and a high-performance data-mining algorithm that rapidly disambiguates hundreds of millions of metagenomic sequence reads into the discrete microorganisms engendering the particular sequences.
2.5.5. Statistical Analysis of Bioinformatic Analysis Data
Statistical analysis was performed using Prism 9.4.0 (GraphPad Software). Data were analysed by one-way analysis of variance (ANOVA).
4. Discussion
The community of microorganisms that inhabit the human skin is commonly referred to as the skin microbiome and includes pathogenic and commensal bacteria, fungi, viruses, and skin mites [
3,
17,
18,
19]. Human microbiome colonization begins at birth, with the delivery method significantly impacting the initial microbial composition [
1,
4]. Vaginal delivery exposes newborns to microbes from the mother’s vagina and faeces, while caesarean sections introduce them primarily to skin microbiota from the mother and hospital environment [
20,
21]. From this initial state, the microbiome is shaped by a complex interplay of internal and external factors, leading to diverse microbial communities between individuals (interpersonal variation) and even within a single person over time (intrapersonal variation).
Several commensal and pathogenic bacteria co-exist among different niches on the surface of the skin [
5]. Commensal microbes residing on different skin regions contribute to a symbiotic relationship with the host [
5]. However, this homeostasis can be disrupted, leading to a diseased state. The factors triggering this shift remain under investigation, and they likely involve a complex interplay between microbes, their metabolites, and environmental factors [
21,
22,
23]. Potential contributors include physical damage to the skin barrier, disruption of the microbiota by antimicrobials, expression of specific virulence factors by microbes, and increased access to the site by opportunistic pathogens [
21,
22,
23]. However recent evidence supports the claim that a synergistic relationship between the species of bacteria that inhabit the skin influences the pathological progression of skin diseases [
19,
22].
A specific strain of probiotic bacteria,
M. luteus Q24, that resides on the skin of healthy human adults has been identified to produce a unique antimicrobial spectrum that is inhibitory towards pathogenic bacteria associated with many skin diseases [
23]. Resident skin commensals likely possess a competitive advantage, making it harder for microbes not naturally found on the skin’s surface to survive or colonise efficiently. Reasons for this include that the skin presents a continually shedding environment, is more suitable for salt-tolerant microbes, and is exposed to many environmental factors [
3]. This means that topical application of gut-associated probiotics may offer some transient benefits similar to those of prebiotics or postbiotics, but these effects are likely short lived and would not result in long-term colonization like those achieved with probiotics specifically chosen for the skin microbiome [
3,
12]. This highlights the importance of selecting and using live microbes naturally found on the skin for optimal efficacy.
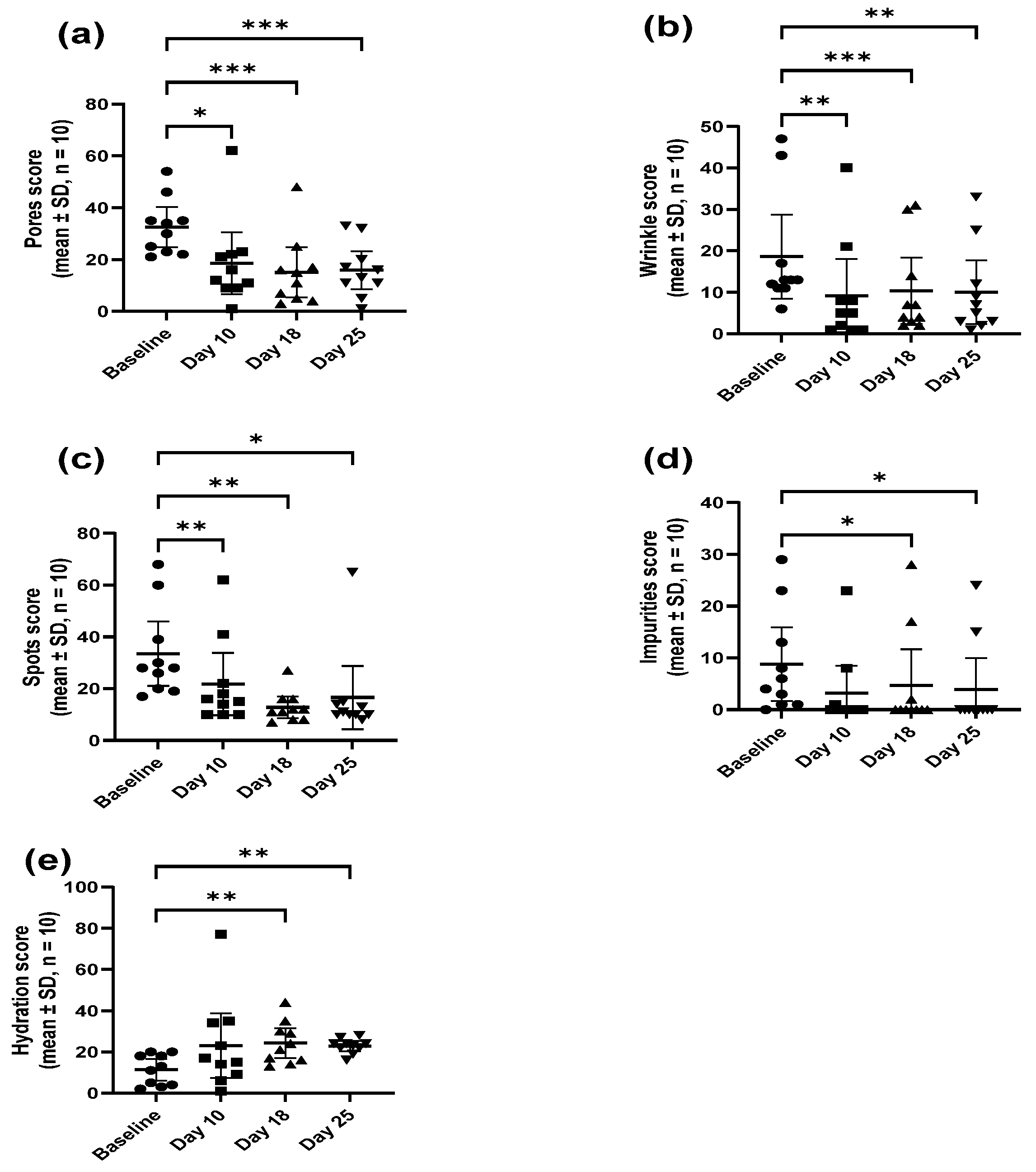
M. luteus Q24 was found to interact with the skin to enhance skin moisture and reduce pores, spots, wrinkles, and impurities [
12]. Unhealthy undernourished skin with poor skin quality is a result of a lack of hydration (moisture) that leads to dryness, and a loss of skin elasticity and smoothness. One of the main reasons for dry skin is the damaged skin barrier. The skin barrier functions not only by maintaining the water content but also by protecting against the penetration of microorganisms, allergens, irritants, etc. [
24]. In this study, a significant improvement in skin hydration was observed following the application of the probiotic serum. This can be attributed to the interaction of the probiotic with the skin layers by hydrating them for repair and improvement of the skin barrier function [
25]. Further, the probiotic serum, due to its nature, forms a protective occlusive layer on the skin to create a hydrophobic barrier over the skin and block the epidermal water loss, thereby maintaining the integrity of the skin [
24,
25]. It is possible that the active metabolites produced by
M. luteus Q24 penetrate deeper into the skin tissues to positively influence the structure and function of the skin barrier, avoiding desiccation of the underlying skin layers, and thus also helping to reduce the pore size and coarse and fine wrinkles, the key signs of skin ageing [
24,
26]. Hyperpigmentation or spots is also a common condition of ageing skin that makes some areas of skin darker than others. The common exogenous factor responsible for skin spots is the photodamage caused by the sun, other factors could include injury to the skin, acne, and cuts. A significant improvement in the skin spots following the application of probiotic serum observed in this study is not surprising as
Micrococcus luteus has been reported to be UV resistant, with antioxidant, and ultraviolet protective properties [
27].
The impact of probiotic serum on the skin quality parameters was captured using a skin analyser that measures these parameters using advanced optics, sensors, and artificial intelligence in conjunction with advanced software and a complex algorithm to generate quantitative scores that can compare the skin quality before and after the application of the cosmetic product containing an active ingredient. The skin analyser measures the moisture level using a sensor that detects the changes in capacitance, which differ depending on the moisture distribution. Moisture measurement indicates the hydration of the skin. When the level of hydration experienced by the skin is low, this alters confounding issues associated with aging (dark, sunken, and dry skin) [
28]. Pores are small openings on the skin’s surface where sebaceous glands open to the external environment [
28]. The relative size of a pore may appear larger in areas where excess sebum is produced, where the pores are blocked with impurities, or where there is a loss of elasticity due to aging. The skin analyser measures the pore size by using the differences in brightness between the skin’s surface and the pore itself. Using brightness, morphological changes such as size, shape, depth, and elasticity of the pore are measured and used by the device software to calculate the pore index. Spots are markers of hyperpigmentation. This is due to ultraviolet rays, skin infections, or areas of scarring [
29]. Melanin, the pigment responsible for skin colour, is excessively produced with hyperpigmentation; therefore, the difference in brightness allows detection. The numerical value for pigmentation is computed by the analyser using the brightness value. Wrinkles are the folds and creases or ridges in the skin due to the loss of skin elasticity, collagen, and moisture; UV radiation exposure; or a result of aging. Similarly, to the pores, the skin analyser uses light to detect wrinkles on the skin’s surface. The difference in brightness allows the length and depth of the wrinkle to be measured and quantified into a fixed value using AI. Finally, impurities (porphyrins) are an indicator of the amount of porphyrin on the skin, a substance produced by
Cutibacterium acnes [
30]. As a result, the number of impurities identified relates to the skin disease acne. This is identified as an orange colour that can be detected by light response using specific wavelength range detection.
There was a significant enhancement in moisture score observed as early as the 10-day time point with the enhanced score obtained throughout the application of the serum. The enhanced moisture level was complemented by a significant reduction in wrinkles, pores, and impurities, again, from the 10-day time point onwards, showing the potential of the probiotic in providing cosmetic benefits in reducing signs of skin fatigue and ageing.
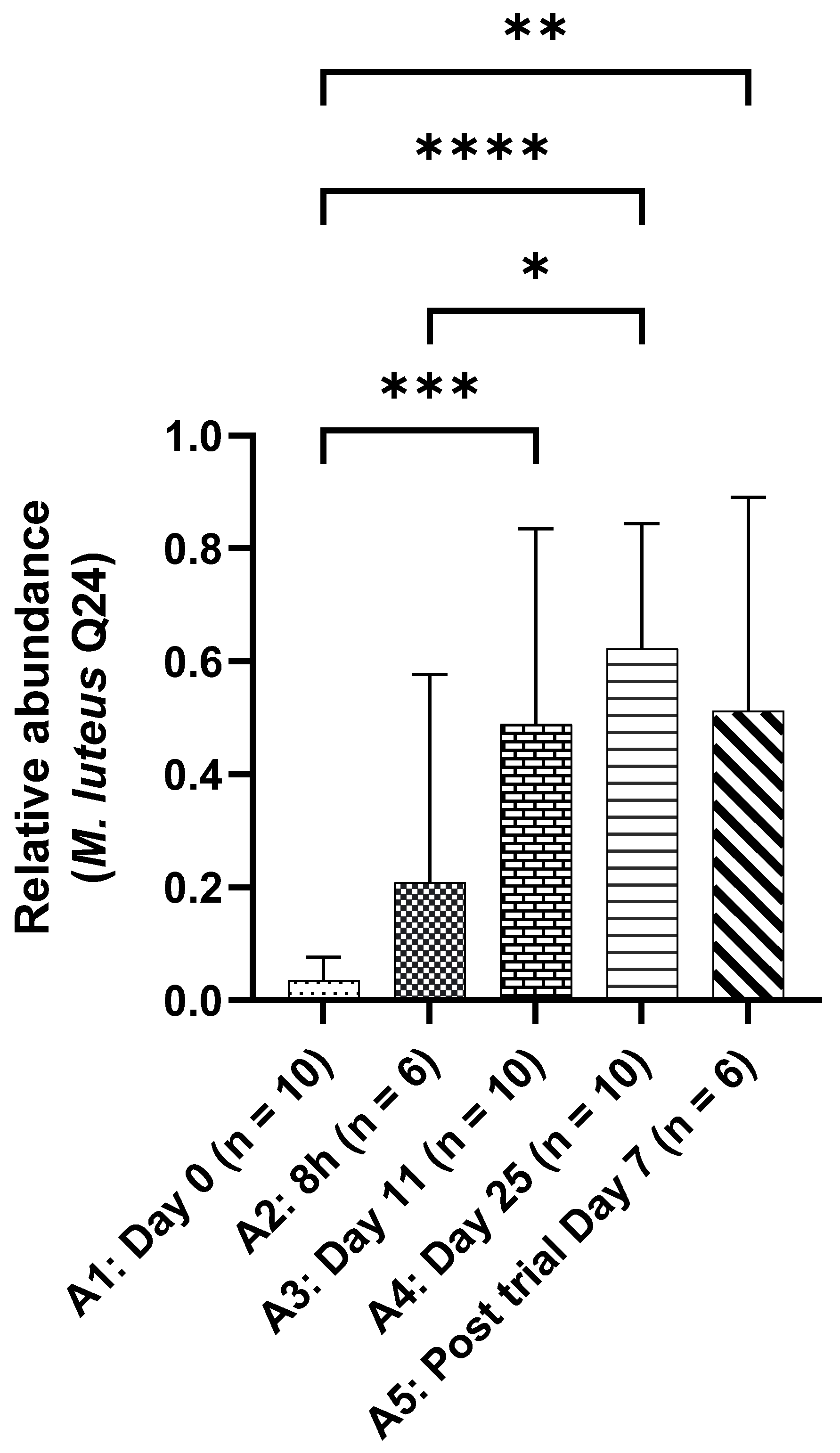
The skin samples were analysed by an independent laboratory using whole-genome sequencing on the swab samples, highlighting the microbiome modulation mediated by the application of
M. luteus Q24 serum. It was interesting but unsurprising to see the prominent relative abundance of
M. luteus Q24 in the skin swab samples with a significant increase throughout the application. The change in the relative abundance of
M. luteus Q24 coincided with the improvement in hydration level and reduction in the skin parameters associated with skin quality, suggesting that the presence of
M. luteus Q24 may have been responsible for underlying mechanisms. Lastly, the preservation of microbial diversity on healthy skin suggests that topical application of the
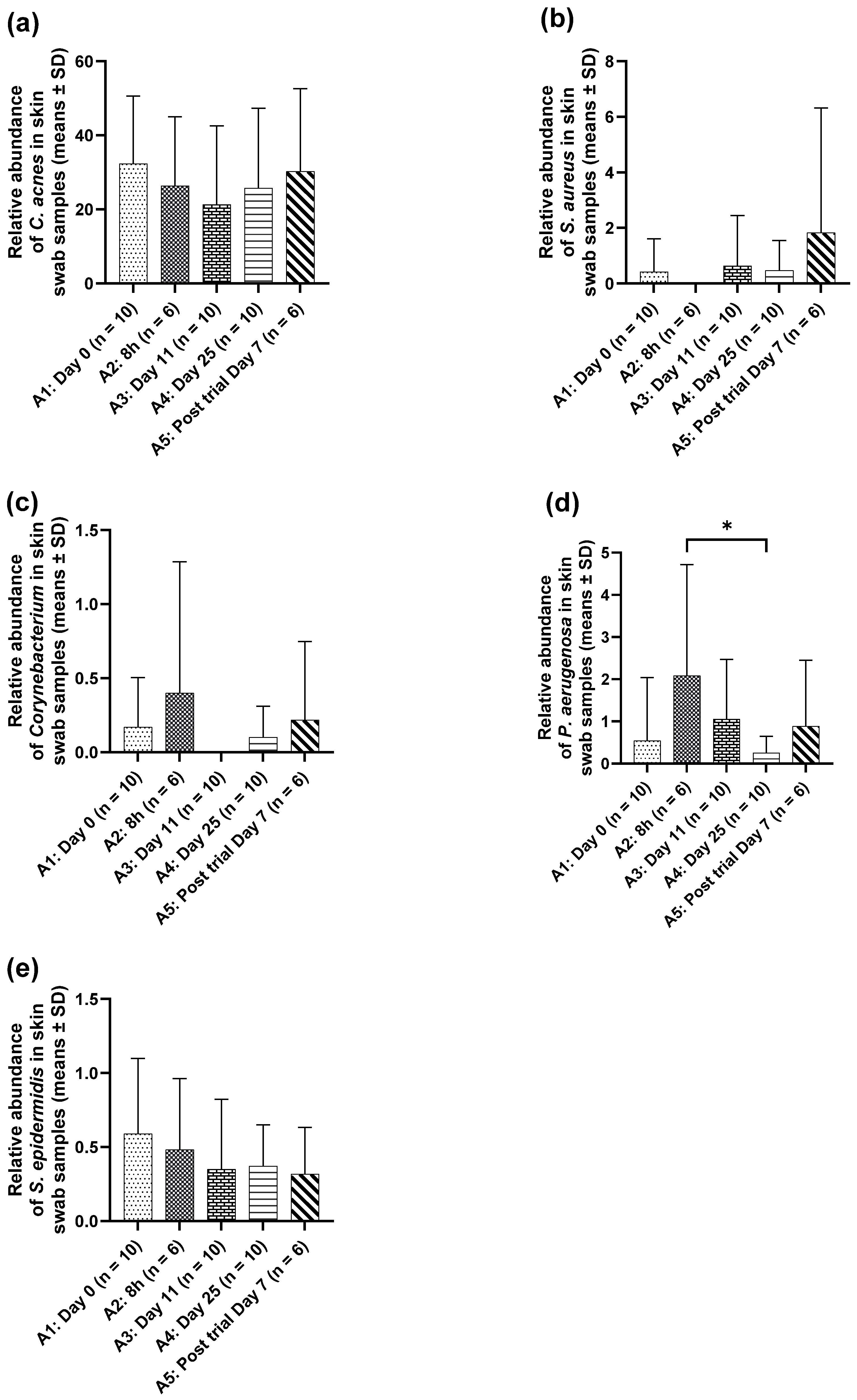
M. luteus Q24 probiotic may not only improve skin quality but also be considered microbiome-friendly. This is supported by the natural presence on human skin (commensal origin) [
3], its established safety profile [
12], its in vitro efficacy evidenced by antimicrobial activity against key skin pathogens [
12], the anti-inflammatory and skin rejuvenation effects in the 3D cell culture models [
31], and the positive outcomes observed in this study. This study has a few limitations such as the lack of a placebo arm and a small number of participants. To address these limitations, future trials should incorporate a placebo arm to account for the placebo effect and recruit a larger participant pool, ensuring the results are more widely applicable.