Abstract
DNA polymerases are essential for genome replication, DNA repair and translesion DNA synthesis (TLS). Broadly, these enzymes belong to two groups: replicative and non-replicative DNA polymerases. A considerable body of data suggests that both groups of DNA polymerases are associated with cancer. Many mutations in cancer cells are either the result of error-prone DNA synthesis by non-replicative polymerases, or the inability of replicative DNA polymerases to proofread mismatched nucleotides due to mutations in 3′-5′ exonuclease activity. Moreover, non-replicative, TLS-capable DNA polymerases can negatively impact cancer treatment by synthesizing DNA past lesions generated from treatments such as cisplatin, oxaliplatin, etoposide, bleomycin, and radiotherapy. Hence, the inhibition of DNA polymerases in tumor cells has the potential to enhance treatment outcomes. Here, we review the association of DNA polymerases in cancer from the A and B families, which participate in lesion bypass, and conduct gene replication. We also discuss possible therapeutic interventions that could be used to maneuver the role of these enzymes in tumorigenesis.
1. Introduction
DNA polymerases conduct DNA synthesis by incorporating deoxynucleoside monophosphate (dNMP) using deoxynucleoside triphosphate (dNTP) as a substrate. These enzymes are indispensable for genome replication, integrity and repair. Because DNA polymerases are necessarily involved in the introduction and amplification of mutations, an understanding of their structure and function is crucial in elucidating the early events of tumor formation. DNA polymerases have been divided into two groups based on their function: (i) replicative and (ii) non-replicative DNA polymerases. Replicative DNA polymerases are required only during cell division to replicate the genome, while the non-replicative DNA polymerases are needed throughout the life-cycle of the cell. An individual cell withstands a daily barrage of endogenous and exogenous DNA modifying agents that cause nearly 70,000 DNA lesions per day [1,2,3]. The ability of cells to tolerate and repair these lesions is dependent on an elaborate DNA repair machinery, which is accomplished in large part through the activity of DNA polymerases.
Since the discovery of the first DNA polymerase (E. coli DNA polymerase I) [4,5,6,7,8], many additional DNA polymerases with distinct biochemical properties have been identified. Based upon the conserved sequences, DNA polymerases have been grouped into A, B, C, D, X, Y, RT (reverse transcriptase) and AEP (archaeo-eukaryotic primase superfamily) families [9,10,11,12,13,14,15,16] (Table 1). DNA polymerases that share sequence homology with E. coli DNA polymerase I, II, and III have been assigned to the A, B and C families, respectively [9]. DNA polymerases lacking sequence homology with the A, B and C families were grouped into the X-family [9,17]. The D-family polymerases are specific to Archaea [10], whereas Y-family DNA polymerases are found in all kingdoms of life [13]. A separate category of DNA polymerase capable of synthesizing DNA using RNA as a template is the reverse transcriptases (RT) [18,19]. These enzymes are found in retroviruses and humans (human telomerases RT or hTERT).

Table 1.
Polymerase families and representative DNA polymerases.
The Klenow fragment (KF) [20] of E. coli DNA polymerase I (pol I) is the prototypical DNA polymerase that has been used to understand the biochemical mechanism of DNA synthesis [21,22,23,24,25,26,27,28,29,30,31,32,33,34,35]. The availability of over expression systems for both KF and pol I [36] facilitated the determination of the first crystal structure of a DNA polymerase [37], and analyses of the kinetic intermediates of DNA polymerase reactions (reviewed in [38]). Importantly, the presence of different conformational states of enzyme/substrate complexes of DNA polymerases seen in the crystal structures [39,40,41] confirmed earlier models based on kinetic studies [38]. The crystal structures of human immunodeficiency virus type I (HIV-1) reverse transcriptase (RT) showed that these structures resembled a half-open, right hand [42,43]. This right-hand configuration has been observed in all DNA polymerases whose structure has been solved. Due to the resemblance with half-open right-hand, individual structural units have been referred to as the thumb, palm and fingers subdomains [42,44,45]. While early structures of DNA polymerases revealed the location of the active site and conserved motifs among DNA polymerases, it was the ternary complex (enzyme/template-primer/nucleoside triphosphate) structure of DNA polymerase β [46,47], which provided insights into the divalent, cation-mediated, nucleotidyltransferase reaction mechanism. Ternary complex structures of T7 DNA polymerase [41] and HIV-1 RT [40] further enhanced our understanding of divalent-mediated nucleotide incorporation.
To date, at least 17 human DNA polymerases have been discovered. These polymerases have been classified into five groups: A, B, X, Y and AEP (archaeo-eukaryotic primase superfamily) [48,49,50,51]. Of these, AEP is the most recently discovered family of DNA polymerases [16]. The AEP family members of polymerases are multitasking enzymes since they can initiate de novo DNA-dependent RNA synthesis, DNA-dependent DNA synthesis, translesion DNA synthesis (TLS), and origin-independent re-priming (reviewed in [51]). TLS is mainly conducted by the Y-family of DNA polymerases. However, recent studies suggest that polymerases belonging to other families can also conduct TLS. In this review, we focus on the A family of TLS polymerases and the B family of DNA polymerases, with an emphasis of their associations in cancer.
2. Family A DNA Polymerases
E.coli DNA polymerase I typifies A family DNA polymerases. There are three known human DNA polymerases that belong to A Family. These are pol γ, pol θ and pol ν. DNA polymerase γ is the major replicase of mitochondrial DNA, however, another mitochondrion-related DNA polymerase has been recently discovered [51]. Pol γ mutations have been associated with several mitochondrial diseases, and these are reviewed elsewhere [52,53,54,55,56,57].
2.1. DNA Polymerase θ
DNA polymerase θ is encoded by the mammalian POLQ gene [58,59]. Human DNA polymerase θ consists of 2590 amino acids and has three domains: (i) the N-terminal ATPase/helicase-like domain, (ii) a central region, and (iii) the C-terminal polymerase domain [60]. The C-terminal polymerase domain shares ~30% sequence homology with KF [61] and possesses each of the conserved A, B and C polymerase motifs [59]. The structures of the helicase domain (residues 1–891) and the polymerase domain (residues 1792–2590) have been determined [62,63]. The DNA-dependent ATPase activity of polymerase θ has been shown but the helicase activity is yet to be demonstrated [64]. Polymerase θ also has a 5′-dRP lyase activity that is required for short patch base excision repair (BER) [65]. In regards to TLS, DNA polymerase θ efficiently bypasses apurinic/apyrimidinic (AP) sites by preferentially incorporating adenine (A) opposite to the AP site, and thymine glycols [66]. In vitro studies have shown that polymerase θ can efficiently extend mismatched termini resulting from the error-prone dNMP insertion by Y-family DNA polymerase ι [67], and nucleotides inserted against cyclobutane pyrimidine dimers (CPDs) or pyrimidine–6/4-pyrimidone photoproducts ([6,4]PP) [66]. Knockdown of polymerase θ in mouse CH12 B lymphoma cells has been shown to increase the sensitivity to cross-linking agents (mitomycin C and cisplatin), an alkylating agent (methyl methanesulphonate) as well as UV irradiation [68]. Animal model studies have shown that polymerase θ mutant mice cells were vulnerable to radiation-induced micronuclei formation, but were still viable [69]. The POLQ-defective, bone marrow stromal cells were not only sensitive to ionizing radiation and bleomycin, but also showed an increase in micronuclei in red blood cells [70,71]. A comparison of POLQ mRNA in tumor tissues and matched control tissues from the same individuals showed higher relative POLQ expression in stomach, lung and colon cancers [72]. Furthermore, a study of colorectal cancer patients found that cancer patients with higher expression of a group of 47 DNA-replication-related genes which includes POLQ in tumors correlated with poorer patient survival [73]. An analysis on human breast cancers found that of the 14 nuclear DNA polymerase genes, only POLQ expression was significantly higher in the cancer tissues in comparison to normal tissues [74]. Another report found highest POLQ expression in Estrogen Receptor (ER)-negative and high-grade tumors, with higher POLQ levels correlating with shorter relapse-free survival times [75]. POLQ is upregulated in oral squamous cell carcinomas [75], and higher expression of POLQ was associated with poor outcome in patients with early to mid-stage non small-cell lung cancers [76]. Similarly, POLQ gene expression in ovarian carcinoma shows that its expression correlates with tumor grade [77]. Collectively, these studies have raised a possibility that polymerase θ may be a driver of cancer. Further research will shed light on the precise role of different polymerase θ variants in oncogenesis.
2.2. DNA Polymerase ν
The third A family human DNA polymerase is polymerase ν, which is encoded by the POLN gene. The C-terminal domain of polymerase ν protein has ~29% sequence homology with human DNA polymerase θ [61], and contains conserved A, B and C motifs [9,11]. A domain with structural homology to the 3′-5′ exonuclease domain of E.coli DNA polymerase is also present in polymerase ν; however, the metal-coordinating residues required for proofreading activity are not present in this enzyme. In vitro experiments have shown that polymerase ν is a highly error-prone enzyme [78,79], and bypasses thymine glycol efficiently [79]. Nearly 50% of breast carcinomas have mutations within the POLN gene, suggesting that polymerase ν may be associated with breast cancer [80]. A recent report suggests that polymerase ν has some ability to bypass the major groove peptide adducts and residues of the DNA crosslink repair [81]. However, inactivation of POLN in mouse embryonic fibroblasts had no effect on cellular sensitivity to mitomycin C, cisplatin, or aldehydes [81]. In human cells, shRNA or siRNA-mediated depletion of POLN did not change cellular susceptibility to mitomycin C or alter the frequency of mitomycin C-induced, radial chromosomes [81]. On the surface, these results may suggest a limited involvement of polymerase ν in DNA damage; however, more research is needed to establish the extent to which this polymerase is involved in cancer.
Mutations introduced during DNA replication or DNA repair are hallmarks of cancer. Hence, the fidelity of DNA polymerases plays a central role in tumorigenesis. DNA polymerases that have associated 3′-5′ exonuclease activity have the ability to remove errors during DNA synthesis and therefore, have greater fidelity. However, both polymerases (θ and ν) lack 3′-5′ exonuclease activity, which may be a reason for the reduced fidelity of these enzymes. In simplest terms, the fidelity of DNA polymerases is defined as the ratio of polymerase efficiencies [defined as the ratio of catalytic rate (kpol) and dNTP substrate binding affinity (Kd.dNTP)] of correct and mismatched nucleotide incorporation. The recently reported crystal structures of the ternary complex (enzyme/DNA/ddNTP) of polymerase θ [63] and the binary complex (enzyme/DNA) of polymerase ν [82] offer insights into the active site conformation, and an explanation for the error-prone DNA synthesis by these two enzymes. In addition, the ternary complex of polymerase θ in complex with a template-primer containing the AP-site analog tetrahydrofuran provides the first glimpse of the TLS by an A Family DNA polymerase [63] (Figure 1). These structures show that the enzymes retain the core architecture of bacterial A family polymerases, but contains additional loops and inserts for specific functions. Comparison of the crystal structures of polymerase θ in complex with template-primer containing tetrahydrofuran (THF) analog and ddNTP with the template-primer and ddNTP showed that the O-helix adopts different conformations depending upon the templating sequence [63]. It is possible that the conformational flexibility of O-helix of DNA polymerase θ permits TLS and error-prone DNA synthesis by this enzyme.
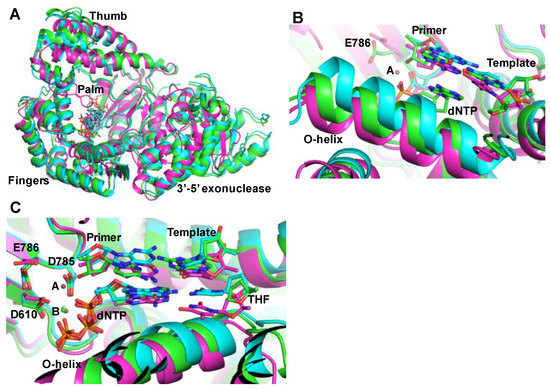
Figure 1.
Structures of family A DNA polymerases. (A) superposition of the ternary complex crystal structures of polymerase θ [63] (green, tetrahydrofuran-ddATP; cyan, dTMP-ddATP) and the ternary complex of KlenTaq (magenta, Protein Data Bank file 1QSY, Li et al., [83]); (B) This figure shows three different conformations of O-helix. Depending upon the template, polymerase θ assumes different O-helix conformation to conduct translesion synthesis; (C) close-up of the active site in three crystal structures. Only metal B, which is Ca2+ (shown as green ball) was seen in the crystal structures of polymerase θ. Metal A (pink ball) as seen in the crystal structure of KlenTaq is also shown here. The three active site residues of KlenTaq (D610, D785 and E786) are also shown in this figure. For simplicity, the residues positions of only KlenTaq are marked.
3. Family B DNA Polymerases
Human B Family polymerases include α, δ, ε and ζ. The majority of nuclear DNA replication is conducted by α, δ and ε polymerases. These three (α, δ and ε) polymerases function as individual, multi-subunit complexes.
3.1 DNA Polymerase α
DNA polymerase α functions as a heterotetrameric complex. The primase active site resides in a p49/p58 complex while the polymerase active site resides in the p180 subunit. Using its primase activity, polymerase α incorporates 7–12 ribonucleoside monophosphates (NMPs) that are extended up to 20–30 nucleotides by the p180 subunit [84]. The polymerase α complex lacks 3′-5′ exonuclease activity, and has moderate fidelity. Mismatched nucleotides incorporated by polymerase α are corrected by the 3′-5′ exonuclease activity of polymerase δ [85,86,87] or the mismatch repair machinery (MMR) [88].
3.2 DNA Polymerase δ and ε
Polymerases δ and ε jointly conduct the replication of the entire nuclear genome. Both polymerases function as holoenzymes composed of multiple subunits. Mammalian polymerase δ is a heterotetramer. The large subunit (p125), which is encoded by the POLD1 gene, harbors both the polymerase and the 3′-5′ exonuclease domain. The other three subunits, namely p50, p68 (also called p66), and p12 are regulatory proteins, and they are encoded by POLD2, POLD3 and POLD4, respectively [89]. DNA polymerase δ is responsible for synthesizing the lagging strand [90]. Recent reports suggest that polymerase δ may also be involved in replicating the leading strand [91]. Human polymerase ε also exists as a heterotetramer. The large catalytic subunit (p261), which is encoded by the POLE1 gene, contains both the polymerase and the 3′-5′ exonuclease activity. The other three subunits are encoded by POLE2, POLE3 and POLE4 genes. These subunits act as regulatory proteins or bind dsDNA [89]. Polymerase ε is known to replicate the leading strand of the replication fork [88,92].
Both polymerases δ and ε carry out high fidelity DNA synthesis, which, in part, is facilitated, by their 3′-5′ exonuclease activity. It has been shown that the 3′-5′ exonuclease activities of the two enzymes can function in trans [93], and any mutation affecting proof-reading activity can lead to genome instability. In line with this notion, the polymerase mutator alleles have been shown to increase the risk of human cancer [94].
Several mutations within the DNA polymerase δ catalytic subunit have been reported in colorectal cancer [95], colon cancer [96] and in a rat hepatoma cell line [97]. Mutation D502A was found in the normal colon of a patient who did not develop colon cancer until the age of 70 [95]. Another mutation R506H has been found in the colorectal cancer cell lines DLD-1/HCT15 [95]. The polymerase domain mutation R648Q was detected in rat hepatoma cells [97]. Of the six conserved exonuclease motifs (Exo I to Exo VI) that have been identified [98,99,100], amino acid residues 502 and 506 belong to the Exo III motif of polymerase δ, whereas R648 is located in the ‘fingers’ subdomain. The activity of partially purified human polymerase δ containing R648Q mutation reduced the fidelity of DNA synthesis [101,102], suggesting a role of R648 in the recognition of correct nucleotide. In addition, a frameshift mutation in the HCT116 colorectal cancer cell line has been identified [96]. This polymerase δ variant that lacks two conserved carboxy terminus DNA binding domains. This polymerase δ variant is also expressed at decreased levels in mutant cells [96]. As depletion of polymerase δ is known to affect chromosomal instability in yeast [103,104], it is likely that mutations in polymerase δ that reduce fidelity or its abundance may contribute to DNA changes that accompany tumorigenesis.
As with polymerase δ, mutations in the 3′-5′ exonuclease domain of polymerase ε has also been identified in different tumors [105,106,107]. Sequencing of genomic DNA encoding the 3′-5′ exonuclease domain of POLE from a set of 76 colorectal carcinomas and six colorectal cell lines identified F376S mutation in one patient [108]. F367 is a conserved residue within the Exo II domain in family B DNA polymerases. Topologically, the F367 equivalent residue in bacteriophage RB69 replicase is located adjacent to the metal B coordinating carboxylate (D222) [109]. A study reporting comprehensive molecular characterization of human colon and rectal cancers by the Cancer Genome Atlas Network showed that about 4% of tumors classified as microsatellite stable had mutations in the exonuclease domain of POLE [110,111]. Several other studies have reported mutations in polymerase ε in different types of tumors including colorectal adenomas and carcinomas [112,113,114,115]. Screening of the 3′-5′ exonuclease domains of POLE (residues 268–471) in 173 endometrial cancers resulted in the identification of 13 non-synonymous variants in POLE [116]. In addition, the percentage of endometrial cancers with polymerase ε somatic mutations was around 8% [116]. Of the 13 mutations, somatic mutations D275V and V411L and germline mutation R311C are specifically interesting. D275 is an active site aspartate of 3′-5′ exonuclease activity and coordinates with metal A, whereas V411 is at the DNA binding interface. Germline mutation R311C is in the vicinity of the Exo I conserved motif.
Numerous studies have demonstrated that DNA replication fidelity is critical in cancer susceptibility and development. Inhibition of the proofreading activity of DNA polymerase δ contributes to a distinctive spectrum of cancers as compared to inhibiting DNA polymerase ε. Polymerase δ and ε proofreading deficient mice showed different survival rates and surprisingly tissue-specific tumor susceptibility [117]. Mice with a POLD1 mutation exhibited thymic lymphomas and skin sarcomas, whereas mice with POLE mutation had histiocytic sarcomas and nodal lymphomas [117]. In one of the earlier studies by Flohr et al. [96] several colorectal cancer cell lines (DLD-1, HCT 116, SW 620, SW 480, SW 48, HT 29) and samples from colorectal cancer patients were screened for a mutation in DNA polymerase δ. Most of these cell lines and patient samples had a mutation in the mRNA of DNA polymerase δ that was expected to modify the structure of the enzyme and causing a defect in the proofreading activity potentially contributing to a high mutation rate commonly observed in colorectal cancers [96].
POLD1 and POLE are the largest domains of polymerase δ and polymerase ε respectively, in humans that have catalytic proofreading exonuclease activity. Recently, many exonuclease domain mutations (EDMs) in human POLD1 and POLE have been identified that directly correlate and predispose to “polymerase proofreading associated polyposis” (PPAP), a disease characterized by multiple carcinomas [106]. Some of the mutations are enlisted in Table 2. The structure of POLD1 and the location of residues that are mutated along with evidence in pathogenicity, molecular characteristics and occurrence in tumor types have been nicely reviewed recently [118]. Over the past 5 years, studies have shown that germline mutations in these domains predispose to colorectal cancers and other malignancies [108]. Cancer genomes of children′s that acquired biallelic mismatch repair deficiency (bMMRD) exhibited massive amounts of mutations than all childhood and most cancer genomes that were analyzed in the study. All the bMMRD cancers had a somatic driver mutation in either polymerase δ or polymerase ε [119]. The somatic and germline mutations in humans and their correlation to cancer, especially colorectal and endometrium cancer, is a recent finding and is very intriguing [113]. However, more work needs to be done to understand the molecular basis on how these mutations enhance the process of tumorigenesis with an endgoal of developing novel treatment strategies. Additionally, sequence analysis for similar mutations in other forms of cancer and classification based on their origin (spontaneous or hereditary) will allow for better genetic testing and clinical surveillance ultimately leading to better clinical outcomes.

Table 2.
Mutations in 3′-5′ exonuclease domain of polymerase δ and ε and their predisposition to the cancer type.
3.3. Polymerase ζ
Human DNA polymerase ζ was initially discovered as a dimeric enzyme consisting of catalytic subunit Rev3 and the structural subunit Rev7 [120,121,122]. Subsequently, a four-subunit polymerase ζ complex containing Rev3, Rev7, p50 and p66 was discovered [123,124,125]. Subunits p50 and p66 are also part of the polymerase δ holoenzyme. An interaction between the polymerase ζ structural subunit Rev7 and Rev1 has also been documented [126], and this interaction appears to be functionally important for TLS across a (6–4) TT photoproduct [127]. Polymerase ζ is a low fidelity polymerase, and does not possess 3′-5′ exonuclease activity. Initially polymerase ζ was considered a TLS polymerase, a concept that was based on the observation that the yeast polymerase ζ was able to perform DNA synthesis past a cis-cyn TT dimer [120]. However, later studies showed that polymerase ζ itself was unable to synthesize DNA past a lesion [49,122]. It is now believed that polymerase ζ is an “extender polymerase”, which extends the lesion bypassed by Y-family polymerases such as η, ι or κ [49].
The relevance of polymerase ζ in cancer is associated with its role in TLS. It has been shown that polymerase ζ is involved in bypassing cisplatin-GG [128], OBPDE-GG [128], (6,4) TT photoproduct [128,129,130], AP sites [128] and thymine glycol [131]. Polymerase ζ has also been demonstrated as a major determinant for resistance to platinum based anti-cancer compounds [132]. Both in vivo and in vitro studies have demonstrated that decreased REV3 expression increased the sensitivity of lymphoma to cisplatin [133]. Knockout of REV3, the gene encoding the catalytic subunit of polymerase ζ in mouse embryonic fibroblasts, increased chromosomal instability in a p53-dependent manner [134]. Loss of REV3 enhanced spontaneous tumorigenesis in a p53-deficient background [135]. Interestingly, overexpression of REV3 may have the same effect since it was found to increase breast cancer tumor cell migration and invasion. Singh et al hypothesized that REV3 could be protecting the tumors from DNA damage or creating mutations through TLS [136]. A breast cancer epidemiology study found that REV 1 and REV 3 single nucleotide polymorphisms (SNPs) not only affected the kinds of tumors that Swedish patients had but also their chances of surviving. Specifically, minor alleles of the REV1 SNPs rs3792142 and rs6761390 were associated with larger tumors and advanced stage cancer. Meanwhile, the CC variants of the REV 3 SNPs rs11153292 and rs462779 reduced patients’ odds of surviving [137].
4. Potential Therapeutic Interventions
Accumulation of a higher number of mutations in cancer can usually stimulate the production of “non-self” antigens [138]. This mechanism of producing non-self antigens makes the tumor vulnerable to attack by immune cells. However, cancer cells acquire escape machinery by expressing protein molecules that allow them to remain undetected by the immune surveillance system [138]. The expression of immune checkpoint protein molecules, Programmed Death 1 (PD1) on T regulatory immune cells and PD-L1 and PD-L2 on normal and cancer cells allows the cells to pass the checkpoint and survive. Thus, immune checkpoint blockade has offered remarkable success in treating many forms of cancer [139,140,141,142,143].
A recent study assessed the expression of PD-L1 expression in mismatch repair deficient endometrial tumors and showed a significant increase in its expression than other mismatch repair proficient counterparts [144]. Importantly, blocking the immune checkpoint has recently been shown to be a promising treatment in colorectal cancer patients with mismatch repair deficiency (dMMR). In this study, pembrolizumab an anti-PD1 inhibitor significantly benefitted the cancer patients with mismatch repair deficiency than the patients with tumors that have mismatch repair proficiency [145]. Another recent phase II trials- Checkmate 142 assessed the use of a well-tolerated Nivolumab anti-PD1 inhibitor in patients with metastatic colorectal cancers and mismatch repair deficiency [146]. Considering the stark similarities between the cancers with (dMMR) and the ones with a defect in proofreading exonuclease activity [147,148,149], the immune checkpoint blockade offers a unique opportunity in the treatment of cancers that have mutations in polymerases lacking proofreading activity. However, more work needs to be done to identify predictive biomarkers that would select patients for such kind of immunotherapy and spare nonresponders from potential side-effects. Also, the novel antigens that are produced by large accumulations of mutations in tumors caused by mismatch repair deficiency is an obvious weakness that can be exploited using a combination of exome based identification of novel antigens and selectively enhancing the activity of cytotoxic T cells against these antigens in tumors [150].
Recently, it was demonstrated that DNA polymerases δ and ε could be a potential candidate as targets for gene therapy in hepatocellular carcinoma (HCC). In this study, microRNAs targeting DNA polymerases δ and ε were used to block the proliferation of HCC cells using a controlled tumor-specific promoter system. This approach might be used to block the function of mutated/dysregulated polymerases specifically in cancer cells potentially avoiding the off-target specificity [151]. Another strategy that could be employed to treat cancers with polymerase proofreading deficiency is to increase the number of mutations in tumors to a level that exceeds a threshold of the cancer cells not allowing them to grow. In support of this concept, cadmium has been shown to increase the lethality of yeast that expresses a proofreading deficient polymerase δ with no effect on WT strains [152]. It has also been demonstrated that altering the dNTP pools significantly affects the sensitivity of yeast strains that express the Pol δ and Pol ε exonuclease domain mutations [153,154]. Hence, in tumor types with mild mutations, inhibiting the dNTP pools might result in a reduction in tumor adaptation. Conversely, dNTP pools can be increased to levels that exceed the mutation threshold in tumors with a higher number of mutations. However, DNA damage response could be different in yeast and humans and hence more work is warranted to test certain mutagenic compounds that might exacerbate the lethality of cancer cells leading to better clinical outcomes.
Many mutations that cause a defect in the exonuclease activity of polymerases δ and ε have been identified in in vitro and in vivo models. With the advent of CRISPR Cas-9 gene editing system [155] gain of function mutation could be achieved. It is too early to predict the clinical outcomes using this technology, however, this approach could also be used in near future to regain the proofreading activity of polymerases that have a crucial role in suppressing the activation of tumorigenesis. Targeting the polymerase using small molecule inhibitors to suppress its function can be an additional therapeutic intervention in cancer.
5. Concluding Remarks
There is growing appreciation for the various ways in which DNA polymerases play a role in genome instability. In the case of non-replicative DNA polymerases such as polymerase θ, small molecule inhibitors can be developed to specifically target these enzymes in tumor cells. However, different strategies like gene therapy or gene editing would be required to correct subunit specific SNPs in order to restrict tumor growth. For tumors associated with mutations in replicative polymerases immunotherapy approaches could be utilized for better recognition of tumor cells by the host immune systems. Additionally, CRISPR Cas-9 gene editing system can be applied to restore normal 3′-5′ exonuclease function. Further in vitro and in vivo characterization of known and newly discovered DNA polymerases will provide additional insights into their function in normal and tumor cells, which can lead to new therapeutic interventions against variety of cancers.
Acknowledgments
Authors acknowledge financial support from the Bond Life Sciences Center, University of Missouri. The authors thank Dr. Michael Petris for critical reading of this manuscript. We apologize to researchers whose work could not be cited due to the need for brevity and space constraint.
Author Contributions
Vinit Shanbhag and Kamalendra Singh conceived and wrote the article. Jacqueline A Flores and Shrikesh Sachdev and Mukund J. Modak contributed in writing at various stages of the review article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tubbs, A.; Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Barnes, D.E. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 2000, 65, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Wood, R.D. Quality control by DNA repair. Science 1999, 286, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Bessman, M.J.; Kornberg, A.; Lehman, I.R.; Simms, E.S. Enzymic synthesis of deoxyribonucleic acid. Biochim. Biophys. Acta 1956, 21, 197–198. [Google Scholar] [PubMed]
- Kornberg, A. Biologic synthesis of deoxyribonucleic acid. Science 1960, 131, 1503–1508. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, A.; Lieberman, I.; Simms, E.S. Enzymatic synthesis and properties of 5-phosphoribosylpyrophosphate. J. Biol. Chem. 1955, 215, 389–402. [Google Scholar] [PubMed]
- Lehman, I.R. Discovery of DNA polymerase. J. Biol. Chem. 2003, 278, 34733–34738. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C. The eureka enzyme: The discovery of DNA polymerase. Nat. Rev. Mol. Cell Biol. 2006, 7, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Delarue, M.; Poch, O.; Tordo, N.; Moras, D.; Argos, P. An attempt to unify the structure of polymerases. Protein Eng. 1990, 3, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Komori, K.; Cann, I.K.; Koga, Y. A novel DNA polymerase family found in archaea. J. Bacteriol. 1998, 180, 2232–2236. [Google Scholar] [PubMed]
- Braithwaite, D.K.; Ito, J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993, 21, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Cann, I.K.; Ishino, Y. Archaeal DNA replication: Identifying the pieces to solve a puzzle. Genetics 1999, 152, 1249–1267. [Google Scholar] [PubMed]
- Ohmori, H.; Friedberg, E.C.; Fuchs, R.P.; Goodman, M.F.; Hanaoka, F.; Hinkle, D.; Kunkel, T.A.; Lawrence, C.W.; Livneh, Z.; Nohmi, T.; et al. The y-family of DNA polymerases. Mol. Cell 2001, 8, 7–8. [Google Scholar] [CrossRef]
- Patel, P.H.; Loeb, L.A. Getting a grip on how DNA polymerases function. Nat. Struct. Biol. 2001, 8, 656–659. [Google Scholar] [CrossRef] [PubMed]
- Sandalli, C.; Singh, K.; Modak, M.J.; Ketkar, A.; Canakci, S.; Demir, I.; Belduz, A.O. A new DNA polymerase i from geobacillus caldoxylosilyticus tk4: Cloning, characterization, and mutational analysis of two aromatic residues. Appl. Microbiol. Biotechnol. 2009, 84, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Koonin, E.V.; Leipe, D.D.; Aravind, L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm–domain proteins: Structural insights and new members. Nucleic Acids Res. 2005, 33, 3875–3896. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Braithwaite, D.K. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991, 19, 4045–4057. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D. RNA-dependent DNA polymerase in virions of rna tumour viruses. Nature 1970, 226, 1209–1211. [Google Scholar] [CrossRef] [PubMed]
- Temin, H.M.; Mizutani, S. RNA-dependent DNA polymerase in virions of rous sarcoma virus. Nature 1970, 226, 1211–1213. [Google Scholar] [CrossRef] [PubMed]
- Klenow, H.; Henningsen, I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from escherichia coli b by limited proteolysis. Proc. Natl. Acad. Sci. USA 1970, 65, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Polesky, A.H.; Dahlberg, M.E.; Benkovic, S.J.; Grindley, N.D.; Joyce, C.M. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J. Biol. Chem. 1992, 267, 8417–8428. [Google Scholar] [PubMed]
- Polesky, A.H.; Steitz, T.A.; Grindley, N.D.; Joyce, C.M. Identification of residues critical for the polymerase activity of the klenow fragment of DNA polymerase I from Escherichia coli. J. Biol. Chem. 1990, 265, 14579–14591. [Google Scholar] [PubMed]
- Astatke, M.; Ng, K.; Grindley, N.D.; Joyce, C.M. A single side chain prevents escherichia coli DNA polymerase I (klenow fragment) from incorporating ribonucleotides. Proc. Natl. Acad. Sci. USA 1998, 95, 3402–3407. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.M., Jr.; Grindley, N.D.; Joyce, C.M. Interaction of DNA polymerase I (klenow fragment) with the single-stranded template beyond the site of synthesis. Biochemistry 2003, 42, 2373–2385. [Google Scholar] [CrossRef] [PubMed]
- Joyce, C.M.; Steitz, T.A. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994, 63, 777–822. [Google Scholar] [CrossRef] [PubMed]
- Bermek, O.; Grindley, N.D.; Joyce, C.M. Prechemistry nucleotide selection checkpoints in the reaction pathway of DNA polymerase I and roles of Glu710 and Tyr766. Biochemistry 2013, 52, 6258–6274. [Google Scholar] [CrossRef] [PubMed]
- Lam, W.C.; Thompson, E.H.; Potapova, O.; Sun, X.C.; Joyce, C.M.; Millar, D.P. 3′-5′ exonuclease of klenow fragment: Role of amino acid residues within the single-stranded DNA binding region in exonucleolysis and duplex DNA melting. Biochemistry 2002, 41, 3943–3951. [Google Scholar] [CrossRef] [PubMed]
- Pandey, V.N.; Williams, K.R.; Stone, K.L.; Modak, M.J. Photoaffinity labeling of the thymidine triphosphate binding domain in Escherichia coli DNA polymerase I: Identification of histidine-881 as the site of cross-linking. Biochemistry 1987, 26, 7744–7748. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.D.; Pandey, V.N.; Modak, M.J. Properties of tyrosine 766→serine mutant of Escherichia coli DNA polymerase I: Template-specific effects. Biochemistry 1994, 33, 11868–11874. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Modak, M.J. A unified DNA- and dNTP-binding mode for DNA polymerases. Trends Biochem. Sci. 1998, 23, 277–281. [Google Scholar] [CrossRef]
- Tuske, S.; Singh, K.; Kaushik, N.; Modak, M.J. The J-helix of Escherichia coli DNA polymerase I (Klenow fragment) regulates polymerase and 3′-5′-exonuclease functions. J. Biol. Chem. 2000, 275, 23759–23768. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Modak, M.J. Presence of 18-a long hydrogen bond track in the active site of escherichia coli DNA polymerase i (klenow fragment). Its requirement in the stabilization of enzyme-template-primer complex. J. Biol. Chem. 2003, 278, 11289–11302. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Modak, M.J. Contribution of polar residues of the J-helix in the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I (Klenow fragment): Q677 regulates the removal of terminal mismatch. Biochemistry 2005, 44, 8101–8110. [Google Scholar] [CrossRef] [PubMed]
- Kukreti, P.; Singh, K.; Ketkar, A.; Modak, M.J. Identification of a new motif required for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I (Klenow fragment): The RRRY motif is necessary for the binding of single-stranded DNA substrate and the template strand of the mismatched duplex. J. Biol. Chem. 2008, 283, 17979–17990. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Srivastava, A.; Patel, S.S.; Modak, M.J. Participation of the fingers subdomain of Escherichia coli DNA polymerase I in the strand displacement synthesis of DNA. J. Biol. Chem. 2007, 282, 10594–10604. [Google Scholar] [CrossRef] [PubMed]
- Joyce, C.M.; Grindley, N.D. Construction of a plasmid that overproduces the large proteolytic fragment (Klenow fragment) of DNA polymerase i of Escherichia coli. Proc. Natl. Acad. Sci. USA 1983, 80, 1830–1834. [Google Scholar] [CrossRef] [PubMed]
- Ollis, D.L.; Kline, C.; Steitz, T.A. Domain of E. coli DNA polymerase I showing sequence homology to T7 DNA polymerase. Nature 1985, 313, 818–819. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.A. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993, 62, 685–713. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Korolev, S.; Waksman, G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 1998, 17, 7514–7525. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chopra, R.; Verdine, G.L.; Harrison, S.C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science 1998, 282, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Doublie, S.; Tabor, S.; Long, A.M.; Richardson, C.C.; Ellenberger, T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 a resolution. Nature 1998, 391, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Kohlstaedt, L.A.; Wang, J.; Friedman, J.M.; Rice, P.A.; Steitz, T.A. Crystal structure at 3.5 a resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 1992, 256, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Das, K.; Hsiou, Y.; Sarafianos, S.G.; Clark, A.D., Jr.; Jacobo-Molina, A.; Tantillo, C.; Hughes, S.H.; Arnold, E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 a resolution. J. Mol. Biol. 1998, 284, 1095–1111. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Smerdon, S.J.; Jager, J.; Kohlstaedt, L.A.; Rice, P.A.; Friedman, J.M.; Steitz, T.A. Structural basis of asymmetry in the human immunodeficiency virus type 1 reverse transcriptase heterodimer. Proc. Natl. Acad. Sci. USA 1994, 91, 7242–7246. [Google Scholar] [CrossRef] [PubMed]
- Jacobo-Molina, A.; Clark, A.D., Jr.; Williams, R.L.; Nanni, R.G.; Clark, P.; Ferris, A.L.; Hughes, S.H.; Arnold, E. Crystals of a ternary complex of human immunodeficiency virus type 1 reverse transcriptase with a monoclonal antibody Fab fragment and double-stranded DNA diffract x-rays to 3.5-a resolution. Proc. Natl. Acad. Sci. USA 1991, 88, 10895–10899. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, H.; Sawaya, M.R.; Kumar, A.; Wilson, S.H.; Kraut, J. Structures of ternary complexes of rat DNA polymerase β, α DNA template-primer, and ddCTP. Science 1994, 264, 1891–1903. [Google Scholar] [CrossRef] [PubMed]
- Sawaya, M.R.; Pelletier, H.; Kumar, A.; Wilson, S.H.; Kraut, J. Crystal structure of rat DNA polymerase β: Evidence for a common polymerase mechanism. Science 1994, 264, 1930–1935. [Google Scholar] [CrossRef] [PubMed]
- Hubscher, U.; Maga, G.; Spadari, S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002, 71, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Washington, M.T. Translesion synthesis: Insights into the selection and switching of DNA polymerases. Genes 2017, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.S.; Takata, K.; Wood, R.D. DNA polymerases and cancer. Nat. Rev. Cancer 2011, 11, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Rudd, S.G.; Bianchi, J.; Doherty, A.J. Primpol—A new polymerase on the block. Mol. Cell. Oncol. 2014, 1, e960754. [Google Scholar] [CrossRef] [PubMed]
- Copeland, W.C.; Longley, M.J. Mitochondrial genome maintenance in health and disease. DNA Repair 2014, 19, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Copeland, W.C. Inherited mitochondrial diseases of DNA replication. Annu. Rev. Med. 2008, 59, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.J.; Naviaux, R.K.; Brunetti-Pierri, N.; Zhang, Q.; Schmitt, E.S.; Truong, C.; Milone, M.; Cohen, B.H.; Wical, B.; Ganesh, J.; et al. Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum. Mutat. 2008, 29, E150–E172. [Google Scholar] [CrossRef] [PubMed]
- Saneto, R.P.; Naviaux, R.K. Polymerase gamma disease through the ages. Dev. Disabil. Res. Rev. 2010, 16, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C. Genetics: Mitochondrial DNA in evolution and disease. Nature 2016, 535, 498–500. [Google Scholar] [CrossRef] [PubMed]
- Krasich, R.; Copeland, W.C. DNA polymerases in the mitochondria: A critical review of the evidence. Front. Biosci. 2017, 22, 692–709. [Google Scholar]
- Wood, R.D.; Doublie, S. DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair 2016, 44, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Knobel, P.A.; Marti, T.M. Translesion DNA synthesis in the context of cancer research. Cancer Cell Int. 2011, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Malaby, A.W.; Martin, S.K.; Wood, R.D.; Doublie, S. Expression and structural analyses of human DNA polymerase θ (POLQ). Methods Enzymol. 2017, 592, 103–121. [Google Scholar] [PubMed]
- Black, S.J.; Kashkina, E.; Kent, T.; Pomerantz, R.T. DNA polymerase θ: A unique multifunctional end-joining machine. Genes 2016, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.A.; Cooper, C.D.; Aitkenhead, H.; Gileadi, O. Structure of the helicase domain of DNA polymerase theta reveals a possible role in the microhomology-mediated end-joining pathway. Structure 2015, 23, 2319–2330. [Google Scholar] [CrossRef] [PubMed]
- Zahn, K.E.; Averill, A.M.; Aller, P.; Wood, R.D.; Doublie, S. Human DNA polymerase θ grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 2015, 22, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Marini, F.; Wood, R.D. POLQ (Pol θ), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003, 31, 6117–6126. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Longley, M.J.; Sharief, F.S.; Hou, E.W.; Copeland, W.C.; Wilson, S.H. Human DNA polymerase theta possesses 5′-drp lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009, 37, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Masutani, C.; Yang, L.W.; Schuffert, A.; Iwai, S.; Bahar, I.; Wood, R.D. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004, 23, 4484–4494. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.E.; Washington, M.T.; Haracska, L.; Prakash, S.; Prakash, L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 2000, 406, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Ukai, A.; Maruyama, T.; Mochizuki, S.; Ouchida, R.; Masuda, K.; Kawamura, K.; Tagawa, M.; Kinoshita, K.; Sakamoto, A.; Tokuhisa, T.; et al. Role of DNA polymerase θ in tolerance of endogenous and exogenous DNA damage in mouse B cells. Genes Cells 2006, 11, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Shima, N.; Hartford, S.A.; Duffy, T.; Wilson, L.A.; Schimenti, K.J.; Schimenti, J.C. Phenotype-based identification of mouse chromosome instability mutants. Genetics 2003, 163, 1031–1040. [Google Scholar] [PubMed]
- Goff, J.P.; Shields, D.S.; Seki, M.; Choi, S.; Epperly, M.W.; Dixon, T.; Wang, H.; Bakkenist, C.J.; Dertinger, S.D.; Torous, D.K.; et al. Lack of DNA polymerase θ (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat. Res. 2009, 172, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Higgins, G.S.; Prevo, R.; Lee, Y.F.; Helleday, T.; Muschel, R.J.; Taylor, S.; Yoshimura, M.; Hickson, I.D.; Bernhard, E.J.; McKenna, W.G. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010, 70, 2984–2993. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Bahar, R.; Seimiya, M.; Chiyo, M.; Wada, A.; Okada, S.; Hatano, M.; Tokuhisa, T.; Kimura, H.; Watanabe, S.; et al. DNA polymerase θ is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer 2004, 109, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Casademunt, J.; Jimenez-Aquino, J.I.; Sancho, J.M. Decay of unstable states in the presence of colored noise and random initial conditions. I. Theory of nonlinear relaxation times. Phys. Rev. A 1989, 40, 5905–5914. [Google Scholar] [CrossRef]
- Lemee, F.; Bergoglio, V.; Fernandez-Vidal, A.; Machado-Silva, A.; Pillaire, M.J.; Bieth, A.; Gentil, C.; Baker, L.; Martin, A.L.; Leduc, C.; et al. DNA polymerase θ up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl. Acad. Sci. USA 2010, 107, 13390–13395. [Google Scholar] [CrossRef] [PubMed]
- Higgins, G.S.; Harris, A.L.; Prevo, R.; Helleday, T.; McKenna, W.G.; Buffa, F.M. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget 2010, 1, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Allera-Moreau, C.; Rouquette, I.; Lepage, B.; Oumouhou, N.; Walschaerts, M.; Leconte, E.; Schilling, V.; Gordien, K.; Brouchet, L.; Delisle, M.B.; et al. DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis 2012, 1, e30. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Liu, J.C.; Amunugama, R.; Hajdu, I.; Primack, B.; Petalcorin, M.I.; O′Connor, K.W.; Konstantinopoulos, P.A.; Elledge, S.J.; Boulton, S.J.; et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 2015, 518, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Beard, W.A.; Wilson, S.H. Structures of human DNA polymerases ν and θ expose their end game. Nat. Struct. Mol. Biol. 2015, 22, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Takata, K.; Shimizu, T.; Iwai, S.; Wood, R.D. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5s-thymine glycol. J. Biol. Chem. 2006, 281, 23445–23455. [Google Scholar] [CrossRef] [PubMed]
- Shivapurkar, N.; Sood, S.; Wistuba, I.I.; Virmani, A.K.; Maitra, A.; Milchgrub, S.; Minna, J.D.; Gazdar, A.F. Multiple regions of chromosome 4 demonstrating allelic losses in breast carcinomas. Cancer Res. 1999, 59, 3576–3580. [Google Scholar] [PubMed]
- Takata, K.I.; Reh, S.; Yousefzadeh, M.J.; Zelazowski, M.J.; Bhetawal, S.; Trono, D.; Lowery, M.G.; Sandoval, M.; Takata, Y.; Lu, Y.; et al. Analysis of DNA polymerase ν function in meiotic recombination, immunoglobulin class-switching, and DNA damage tolerance. PLoS Genet. 2017, 13, e1006818. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Gao, Y.; Yang, W. How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis. Nat. Struct. Mol. Biol. 2015, 22, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mitaxov, V.; Waksman, G. Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation. Proc. Natl. Acad. Sci. USA 1999, 96, 9491–9496. [Google Scholar] [CrossRef] [PubMed]
- Thompson, H.C.; Sheaff, R.J.; Kuchta, R.D. Interactions of calf thymus DNA polymerase α with primer/templates. Nucleic Acids Res. 1995, 23, 4109–4115. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, Y.I.; Frahm, C.; Nick McElhinny, S.A.; Niimi, A.; Suzuki, M.; Kunkel, T.A. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr. Biol. 2006, 16, 202–207. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, S.D.; Kunkel, T.A. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008, 18, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Perrino, F.W.; Loeb, L.A. Hydrolysis of 3′-terminal mispairs in vitro by the 3′-5′ exonuclease of DNA polymerase δ permits subsequent extension by DNA polymerase α. Biochemistry 1990, 29, 5226–5231. [Google Scholar] [CrossRef] [PubMed]
- Preston, B.D.; Albertson, T.M.; Herr, A.J. DNA replication fidelity and cancer. Semin. Cancer Biol. 2010, 20, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.; Eckert, K.A. Eukaryotic replicative DNA polymerases. Nucleic Acid Polym. 2014, 17–41. [Google Scholar]
- Larrea, A.A.; Lujan, S.A.; Nick McElhinny, S.A.; Mieczkowski, P.A.; Resnick, M.A.; Gordenin, D.A.; Kunkel, T.A. Genome-wide model for the normal eukaryotic DNA replication fork. Proc. Natl. Acad. Sci. USA 2010, 107, 17674–17679. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.E.; Klassen, R.; Prakash, L.; Prakash, S. A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol. Cell 2015, 59, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Burgers, P.M.J.; Gordenin, D.; Kunkel, T.A. Who is leading the replication fork, Pol ε or Pol δ? Mol. Cell 2016, 61, 492–493. [Google Scholar] [CrossRef] [PubMed]
- Lujan, S.A.; Williams, J.S.; Pursell, Z.F.; Abdulovic-Cui, A.A.; Clark, A.B.; Nick McElhinny, S.A.; Kunkel, T.A. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012, 8, e1003016. [Google Scholar] [CrossRef] [PubMed]
- Loeb, L.A.; Monnat, R.J., Jr. DNA polymerases and human disease. Nat. Rev. Genet. 2008, 9, 594–604. [Google Scholar] [CrossRef] [PubMed]
- da Costa, L.T.; Liu, B.; el-Deiry, W.; Hamilton, S.R.; Kinzler, K.W.; Vogelstein, B.; Markowitz, S.; Willson, J.K.; de la Chapelle, A.; Downey, K.M.; et al. Polymerase δ variants in RER colorectal tumours. Nat. Genet. 1995, 9, 10–11. [Google Scholar] [CrossRef] [PubMed]
- Flohr, T.; Dai, J.C.; Buttner, J.; Popanda, O.; Hagmuller, E.; Thielmann, H.W. Detection of mutations in the DNA polymerase δ gene of human sporadic colorectal cancers and colon cancer cell lines. Int. J. Cancer 1999, 80, 919–929. [Google Scholar] [CrossRef]
- Popanda, O.; Flohr, T.; Fox, G.; Thielmann, H.W. A mutation detected in DNA polymerase δ cDNA from Novikoff hepatoma cells correlates with abnormal catalytic properties of the enzyme. J. Cancer Res. Clin. Oncol. 1999, 125, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Bernad, A.; Blanco, L.; Lazaro, J.M.; Martin, G.; Salas, M. A conserved 3′→5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 1989, 59, 219–228. [Google Scholar] [CrossRef]
- Derbyshire, V.; Pinsonneault, J.K.; Joyce, C.M. Structure-function analysis of 3′→5′-exonuclease of DNA polymerases. Methods Enzymol. 1995, 262, 363–385. [Google Scholar] [PubMed]
- Shevelev, I.V.; Hubscher, U. The 3′→5′ exonucleases. Nat. Rev. Mol. Cell Biol. 2002, 3, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Fox, G.; Popanda, O.; Edler, L.; Thielmann, H.W. Preferential inhibition of DNA polymerases α, δ, and ε from Novikoff hepatoma cells by inhibitors of cell proliferation. J. Cancer Res. Clin. Oncol. 1996, 122, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Fox, G.; Popanda, O.; Thielmann, H.W. Evidence for reduced copying fidelity of DNA polymerases α, δ, and ε from Novikoff hepatoma cells. J. Cancer Res. Clin. Oncol. 1997, 123, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, F.J.; Degtyareva, N.P.; Kokoska, R.J.; Petes, T.D. Reduced levels of DNA polymerase δ induce chromosome fragile site instability in yeast. Mol. Cell. Biol. 2008, 28, 5359–5368. [Google Scholar] [CrossRef] [PubMed]
- Kokoska, R.J.; Stefanovic, L.; DeMai, J.; Petes, T.D. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase δ or by decreases in the cellular levels of DNA polymerase δ. Mol. Cell. Biol. 2000, 20, 7490–7504. [Google Scholar] [CrossRef] [PubMed]
- Hoang, L.N.; McConechy, M.K.; Kobel, M.; Anglesio, M.; Senz, J.; Maassen, M.; Kommoss, S.; Meng, B.; Postovit, L.; Kelemen, L.E.; et al. Polymerase ε exonuclease domain mutations in ovarian endometrioid carcinoma. Int. J. Gynecol. Cancer 2015, 25, 1187–1193. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Tomlinson, I. Replicative DNA polymerase mutations in cancer. Curr. Opin. Genet. Dev. 2014, 24, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Mertz, T.M.; Harcy, V.; Roberts, S.A. Risks at the DNA replication fork: Effects upon carcinogenesis and tumor heterogeneity. Genes 2017, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R.; Miyashita, K.; Inoue, M.; Shimamoto, A.; Yan, Z.; Egashira, A.; Oki, E.; Kakeji, Y.; Oda, S.; Maehara, Y. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur. J. Hum. Genet. 2011, 19, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Shamoo, Y.; Steitz, T.A. Building a replisome from interacting pieces: Sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 1999, 99, 155–166. [Google Scholar] [CrossRef]
- Henninger, E.E.; Pursell, Z.F. DNA polymerase ε and its roles in genome stability. IUBMB Life 2014, 66, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar]
- Cancer Genome Atlas Research, N.; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Palles, C.; Cazier, J.B.; Howarth, K.M.; Domingo, E.; Jones, A.M.; Broderick, P.; Kemp, Z.; Spain, S.L.; Guarino, E.; Salguero, I.; et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013, 45, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Choi, M.; Overton, J.D.; Bellone, S.; Roque, D.M.; Cocco, E.; Guzzo, F.; English, D.P.; Varughese, J.; Gasparrini, S.; et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc. Natl. Acad. Sci. USA 2013, 110, 2916–2921. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, D.A.; Wang, L. From human genome to cancer genome: The first decade. Genome Res. 2013, 23, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Church, D.N.; Briggs, S.E.; Palles, C.; Domingo, E.; Kearsey, S.J.; Grimes, J.M.; Gorman, M.; Martin, L.; Howarth, K.M.; Hodgson, S.V.; et al. DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet. 2013, 22, 2820–2828. [Google Scholar] [CrossRef] [PubMed]
- Albertson, T.M.; Ogawa, M.; Bugni, J.M.; Hays, L.E.; Chen, Y.; Wang, Y.; Treuting, P.M.; Heddle, J.A.; Goldsby, R.E.; Preston, B.D. DNA polymerase ε and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl. Acad. Sci. USA 2009, 106, 17101–17104. [Google Scholar] [CrossRef] [PubMed]
- Rayner, E.; van Gool, I.C.; Palles, C.; Kearsey, S.E.; Bosse, T.; Tomlinson, I.; Church, D.N. A panoply of errors: Polymerase proofreading domain mutations in cancer. Nat. Rev. Cancer 2016, 16, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Shlien, A.; Campbell, B.B.; de Borja, R.; Alexandrov, L.B.; Merico, D.; Wedge, D.; Van Loo, P.; Tarpey, P.S.; Coupland, P.; Behjati, S.; et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat. Genet. 2015, 47, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.R.; Lawrence, C.W.; Hinkle, D.C. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 1996, 272, 1646–1649. [Google Scholar] [CrossRef] [PubMed]
- Murakumo, Y.; Roth, T.; Ishii, H.; Rasio, D.; Numata, S.; Croce, C.M.; Fishel, R. A human REV7 homolog that interacts with the polymerase ζ catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J. Biol. Chem. 2000, 275, 4391–4397. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Prakash, L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes Dev. 2002, 16, 1872–1883. [Google Scholar] [CrossRef] [PubMed]
- Makarova, A.V.; Stodola, J.L.; Burgers, P.M. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012, 40, 11618–11626. [Google Scholar] [CrossRef] [PubMed]
- Baranovskiy, A.G.; Lada, A.G.; Siebler, H.M.; Zhang, Y.; Pavlov, Y.I.; Tahirov, T.H. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J. Biol. Chem. 2012, 287, 17281–17287. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.E.; Prakash, L.; Prakash, S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc. Natl. Acad. Sci. USA 2012, 109, 12455–12460. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, E.; Hanafusa, T.; Kamei, K.; Song, I.; Tomida, J.; Hashimoto, H.; Vaziri, C.; Ohmori, H. Identification of a novel REV1-interacting motif necessary for DNA polymerase κ function. Genes Cells 2009, 14, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Prakash, L.; Prakash, S. Error-free replicative bypass of (6-4) photoproducts by DNA polymerase ζ in mouse and human cells. Genes Dev. 2010, 24, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Shachar, S.; Ziv, O.; Avkin, S.; Adar, S.; Wittschieben, J.; Reissner, T.; Chaney, S.; Friedberg, E.C.; Wang, Z.; Carell, T.; et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009, 28, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Szuts, D.; Marcus, A.P.; Himoto, M.; Iwai, S.; Sale, J.E. Rev1 restrains DNA polymerase ζ to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo. Nucleic Acids Res. 2008, 36, 6767–6780. [Google Scholar] [CrossRef] [PubMed]
- Jansen, J.G.; Tsaalbi-Shtylik, A.; Hendriks, G.; Verspuy, J.; Gali, H.; Haracska, L.; de Wind, N. Mammalian polymerase ζ is essential for post-replication repair of UV-induced DNA lesions. DNA Repair 2009, 8, 1444–1451. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Bhatia, G.; Prakash, S.; Prakash, L. Error-free replicative bypass of thymine glycol by the combined action of DNA polymerases κ and ζ in human cells. Proc. Natl. Acad. Sci. USA 2010, 107, 14116–14121. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Shah, N.A.; Joiner, A.M.; Roberts, K.H.; Canman, C.E. DNA polymerase ζ is a major determinant of resistance to platinum-based chemotherapeutic agents. Mol. Pharmacol. 2012, 81, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Doles, J.; Hemann, M.T.; Walker, G.C. Error-prone translesion synthesis mediates acquired chemoresistance. Proc. Natl. Acad. Sci. USA 2010, 107, 20792–20797. [Google Scholar] [CrossRef] [PubMed]
- Wittschieben, J.P.; Reshmi, S.C.; Gollin, S.M.; Wood, R.D. Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res. 2006, 66, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Wittschieben, J.P.; Patil, V.; Glushets, V.; Robinson, L.J.; Kusewitt, D.F.; Wood, R.D. Loss of DNA polymerase ζ enhances spontaneous tumorigenesis. Cancer Res. 2010, 70, 2770–2778. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Li, X.; Owens, K.M.; Vanniarajan, A.; Liang, P.; Singh, K.K. Human REV3 DNA polymerase ζ localizes to mitochondria and protects the mitochondrial genome. PLoS ONE 2015, 10, e0140409. [Google Scholar] [CrossRef] [PubMed]
- Varadi, V.; Bevier, M.; Grzybowska, E.; Johansson, R.; Enquist, K.; Henriksson, R.; Butkiewicz, D.; Pamula-Pilat, J.; Tecza, K.; Hemminki, K.; et al. Genetic variation in genes encoding for polymerase ζ subunits associates with breast cancer risk, tumour characteristics and survival. Breast Cancer Res. Treat. 2011, 129, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Nishikawa, H. Roles of regulatory T cells in cancer immunity. Int. Immunol. 2016, 28, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; O'Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Sznol, M.; McDermott, D.F.; Kluger, H.M.; Carvajal, R.D.; Sharfman, W.H.; Brahmer, J.R.; Lawrence, D.P.; Atkins, M.B.; Powderly, J.D.; et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J. Clin. Oncol. 2014, 32, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crino, L.; Eberhardt, W.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Sloan, E.A.; Ring, K.L.; Willis, B.C.; Modesitt, S.C.; Mills, A.M. PD-L1 expression in mismatch repair-deficient endometrial carcinomas, including lynch syndrome-associated and MLH1 promoter hypermethylated tumors. Am. J. Surg. Pathol. 2017, 41, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- van Gool, I.C.; Eggink, F.A.; Freeman-Mills, L.; Stelloo, E.; Marchi, E.; de Bruyn, M.; Palles, C.; Nout, R.A.; de Kroon, C.D.; Osse, E.M.; et al. POLE proofreading mutations elicit an antitumor immune response in endometrial cancer. Clin. Cancer Res. 2015, 21, 3347–3355. [Google Scholar] [CrossRef] [PubMed]
- Howitt, B.E.; Shukla, S.A.; Sholl, L.M.; Ritterhouse, L.L.; Watkins, J.C.; Rodig, S.; Stover, E.; Strickland, K.C.; D'Andrea, A.D.; Wu, C.J.; et al. Association of polymerase e-mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-l1. JAMA Oncol. 2015, 1, 1319–1323. [Google Scholar] [CrossRef] [PubMed]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wei, Q.; Wang, J.; Huang, X.; Li, C.; Zheng, Q.; Cao, J.; Jia, Z. DNA polymerases as targets for gene therapy of hepatocellular carcinoma. BMC Cancer 2015, 15, 325. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.H.; Clark, A.B.; Slebos, R.J.; Al-Refai, H.; Taylor, J.A.; Kunkel, T.A.; Resnick, M.A.; Gordenin, D.A. Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat. Genet. 2003, 34, 326–329. [Google Scholar] [CrossRef] [PubMed]
- Mertz, T.M.; Sharma, S.; Chabes, A.; Shcherbakova, P.V. Colon cancer-associated mutator DNA polymerase δ variant causes expansion of dNTP pools increasing its own infidelity. Proc. Natl. Acad. Sci. USA 2015, 112, E2467–2476. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.N.; Marjavaara, L.; Knowels, G.M.; Schultz, E.M.; Fox, E.J.; Chabes, A.; Herr, A.J. dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase ε variants. Proc. Natl. Acad. Sci. USA 2015, 112, E2457–2466. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).