Simple Summary
The Red Sea is a challenging marine environment with very harsh environmental conditions. However, its ecosystems support a rich diversity of organisms, including marine sponges that maintain close associations with diverse microbial communities. In this study, we investigated two Brevibacterium strains isolated from Red Sea sponges to explore their genetic adaptations to these challenging conditions and to assess their capacity for producing bioactive compounds. The metabolites from the two strains exhibited moderate antimicrobial activity. Whole-genome sequencing revealed genes associated with tolerance to salinity and nutrient limitation, as well as genetic pathways for resistance to toxic compounds. Furthermore, some biosynthetic gene clusters were identified, indicating a capacity to produce structurally diverse secondary metabolites with potential pharmaceutical and industrial applications. These findings provide new insights into the adaptive mechanisms of sponge-associated bacteria in extreme marine habitats and highlight their potential as a source of novel bioactive molecules. The study advances our understanding of microbial survival strategies in harsh marine environments and underscores the importance of such microorganisms for both ecological functions and biotechnological innovation.
Abstract
Marine-derived Actinomycetota have emerged as promising sources of bioactive natural products, particularly filamentous actinomycetes (e.g., Streptomyces). However, members from non-filamentous genera have showed potential biotechnological importance. In this study, we performed a comprehensive genomic characterization of two bioactive Brevibacterium strains, Brevibacterium luteolum (B. luteolum) 26C and Brevibacterium casei (B. casei) 13A, isolated from two Red Sea sponges. Whole-genome sequencing and taxonomic analysis confirmed species-level identification, marking the first documented report of these species within the Red Sea ecosystem. The two strains displayed antimicrobial activity against Staphylococcus aureus, Escherichia coli, and Candida albicans. Additionally, functional annotation revealed multiple genomic islands (GIs) enriched with genes conferring heavy metal resistance, DNA repair enzymes, nutrient acquisition, and mobile genetic elements, highlighting potential evolutionary adaptations to the harsh physicochemical conditions of the Red Sea. Genome mining identified biosynthetic gene clusters, including those encoding ε-poly-L-lysine, tropodithietic acid, ectoine, and carotenoids. The comparative analysis of orthologous gene clusters from both strains and their counterparts from terrestrial ecosystems highlighted potential marine adaptive genetic mechanisms. This study highlights the biosynthetic potential of B. luteolum 26C and B. casei 13A and their ecological role as active competitors and potential defensive associates within the sponge microbiome.
1. Introduction
The Red Sea is a unique marine ecosystem characterized by high salinity, elevated temperatures, and oligotrophic conditions, creating a challenging yet biologically rich environment [1,2]. Its coral reefs and sponge communities harbor diverse microbial symbionts that have adapted to these challenging physicochemical stressors, making them valuable models for studying marine microbial ecology and evolution [3]. Marine sponges, in particular, are recognized as prolific sources of bioactive natural products, largely due to their complex symbiotic microbiomes [4,5]. These microbial consortia contribute to host defense, chemical communication, and environmental adaptation through the production of bioactive metabolites with antibacterial, antifungal, antiviral, and anticancer properties [6,7]. Focusing on the Red Sea sponge-associated microbiome, our lab has previously reported several microbial strains with interesting bioactivities and potential genetic adaptation mechanisms [8,9,10].
Marine Actinomycetota associated with sponges, particularly filamentous actinomycetes (e.g., Streptomyces), have attracted attention for their biosynthetic versatility and capacity to yield novel natural products [11,12]. However, other non-filamentous genera such as Brevibacterium, although less explored compared to Streptomyces, are increasingly reported from various marine habitats, including sediments, seawater, and sponge microbiomes. Representative species include B. marinum (from seawater) [13]; B. oceani (from deep-sea sediments, Chagos Trench in the Indian Ocean) [14]; B. profundi (from deep-sea sediments, Western Pacific Ocean); and B. spongiae (from marine sponges) [15]. These different species have emerged as a promising reservoir of unique bioactive compounds, including pigments and biosurfactants, with potential applications in biotechnology and bioremediation [13,14,15]. These capabilities, combined with frequent reports of occurrence in diverse marine niches, underscore the ecological adaptability and applied potential of this genus.
Whole-genome sequencing (WGS) has significantly advanced our understanding of the diverse Brevibacterium genus, providing valuable insights into their genetics, metabolism, and ecological roles. Excluding the genomes analyzed in this study, as of August 2025, the NCBI genome database contained 38 genomes of B. casei and 19 genomes of B. luteolum. The predominant isolation sources of these species were cheese, dairy products, fermented foods, or human/animal-associated sources. In contrast, isolates derived from marine-associated environments represent only a minor proportion, indicating their relative scarcity within current genomic datasets. Beyond basic genome statistics, WGS projects identify genes involved in essential functions like carbon utilization, nitrogen and phosphate metabolism, metal transport, and resistance to antibiotics and toxic compounds [16]. Furthermore, WGS studies have illuminated the potential of Brevibacterium species to produce a variety of secondary metabolites, including biosurfactants and other bioactive compounds, which hold promise for various biotechnological applications [17]. Thus, genomic analyses enhance our understanding of microbial functional capacity, ecological adaptation, and the potential for natural product discovery. This study aims to investigate the genomic features of two Red Sea sponge-associated Brevibacterium isolates to elucidate their biosynthetic potential, ecological adaptations, and capacity to produce biotechnologically beneficial compounds.
2. Materials and Methods
2.1. Strains Isolation and Antimicrobial Activity Screening
The strains 26C and 13A were previously isolated in our laboratory from two Red Sea sponges [9]. For metabolic extract preparation, the glycerol stocks (−80 °C) of both isolates were plated on Reasoner’s 2A agar (R2A) (DifcoTM, Detroit, MI, USA) supplemented with 2% NaCl. The plates were incubated at 30 °C for 1–2 days. The isolates were inoculated in flasks containing 100 mL R2A broth medium made with deionized water type 1. Following 7 days of incubation at 25 °C on an incubator shaker at 220 rpm, the fermented broths were extracted twice with double volume ethyl acetate (200 mL × 2). The solvent extracts were concentrated to dryness under reduced pressure using a rotary evaporator at 40 °C and 170 mbar.
For antimicrobial activity screening, the extracted residues were dissolved in 15% dimethylsulfoxide (DMSO) at a concentration of 1 mg/mL. The standardized well-diffusion method was used to investigate the antimicrobial activity using 100 μL of each metabolic extract against Gram-negative strains (Escherichia coli ATCC 10536 and Pseudomonas aeruginosa ATCC25619), a Gram-positive strain (Staphylococcus aureus ATCC 9144), and a yeast (Candida albicans ATCC 90028). Additionally, 15% DMSO was used as a negative control. Ceftazidime and imipenem were used as positive controls for the Gram-negative stains, ampicillin was used as a positive control for S. aureus growth, and nystatin was used as a positive control for C. albicans.
2.2. DNA Extraction and Whole-Genome Sequencing
2.2.1. Reads Preprocessing and Assembly
Trimmomatic version 0.39 was used for quality-based filtration of the raw reads [18]. An Illumina adaptor clipping option, sliding window trimming of a minimum of 4 bases, and an average required quality of 20 were adjusted as parameters for the filtration process. After that, the filtered reads were assembled using the Unicycler assembler [19] on the BV-BRC server (https://www.bv-brc.org) accessed on 24 May 2025.
2.2.2. Strain Typing and Phylogeny
The strain typing was carried out using the GTDB-Tk version 2.1.0 toolkit against release 220 (28 October 2024) of the Genome Taxonomy Database (GTDB) [20]. A phylogenomic-based genome analysis between the genomes of isolates 26C, 13A, and top related type strains was conducted on the Type Strain Genome Server (TYGS) (Leibniz Institute DSMZ, Braunschweig, Germany) [21]. To ensure the taxonomic affiliations of both isolates, the average nucleotide identity (ANI) and DNA–DNA hybridization (DDH) were calculated using the JSpecies server (Leibniz Institute DSMZ, Braunschweig, Germany) [22] and Genome-to-Genome Distance Calculator (Leibniz Institute DSMZ, Braunschweig, Germany) [23]. The average amino acid identity (AAI) was calculated using the AAI calculator tool (Kostas Lab, Georgia Institute of Technology, Atlanta, GA, USA) [24].
2.2.3. Reference-Guided Scaffolding and Genome Annotation
RagTag version 2.1.0 was used for reference-guided scaffolding using the default parameters [25]. The Rapid Annotations using Subsystems Technology (RAST) [26] and Prokka [27] were used to annotate the genomes of strains 26C and 13A. The mobile OG-db was used to annotate the bacterial mobile genetic elements (MGEs) [28]. IslandViewer 4 (Simon Fraser University, Burnaby, BC, Canada) was used to analyze the genomic islands (GIs) within the genomes of strains 26C and 13A [29].
2.2.4. Metabolic Pathway Reconstruction and Investigation of Biosynthetic Gene Clusters (BGCs)
To investigate the main metabolic processes in the isolated strains, KEGG pathway annotation and reconstruction were performed using GhostKOALA version 3.1, which automatically assigned KEGG Orthology (KO) identifiers and mapped the predicted protein products from the coding sequences (CDSs) to metabolic pathways [30]. The BGCs responsible for secondary metabolite production and their similarities to known clusters were identified using antiSMASH bacterial version 8.0.1 [31].
2.2.5. Comparative Orthologous Cluster Analysis
To gain an overview of potential marine adaptive genetic elements in B. luteolum 26C and B. casei 13A, a comparative orthologous cluster analysis was performed against related genomes from non-marine niches (Table 1). The analysis was conducted using the OrthoVenn3 web server (https://orthovenn3.bioinfotoolkits.net/home accessed on 30 August 2025) [32].

Table 1.
Genomes used in the orthologous cluster analysis with corresponding NCBI accession numbers, isolation sources, genome sizes, and assembly levels.
3. Results
3.1. The Isolated Strains’ Phenotypic Characteristics
The colony morphologies of B. casei 13A and B. luteolum 26C are shown in Figure 1a,c and described in Table 2.
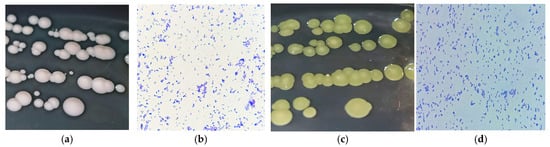
Figure 1.
(a,c) Colony morphologies of B. casei 13A and B. luteolum 26C, respectively, on incubation on marine agar at 30 °C for 1–2 days; (b,d) microscopical examination of B. casei 13A and B. luteolum 26C, respectively.

Table 2.
B. luteolum 26C and B. casei 13A colony morphology.
3.2. Antimicrobial Activities of B. luteolum 26C and B. casei 13A
The metabolic extracts of the strains 26C and 13A showed broad-spectrum antimicrobial activity against S. aureus, E. coli, and C. albicans (Table 3).

Table 3.
The results of the antimicrobial activity screening (measured as inhibition zone diameter in mm) of the metabolic extracts of strains 26C and 13A.
3.3. Genome Characteristics and Strain Typing
For isolate 26C, genome sequencing yielded 29 contigs with an N50 value of 206,763 bp, a GC content of 67.05%, and a total genome length of 2,920,920 bp. Strain typing analysis based on the relative evolutionary divergence (RED) and average nucleotide identity (ANI) criteria was conducted using GTDB-Tk against the GTDB database, which identified the isolate 26C as B. luteolum, a high GC-content, Gram-positive bacterium within the phylum Actinomycetota, order Micrococcales, and family Brevibacteriaceae.
On the other hand, the isolate 13A genome assembly resulted in 61 contigs with an N50 of 111,271 bp, a GC content of 68.27%, and a total size of 3,689,383 bp. GTBD-Tk classified this isolate as B. casei. A phylogenomic tree depicting the evolutionary relationships of both strains and their closely related type strains, made by the TYGS server, is presented in Figure 2.
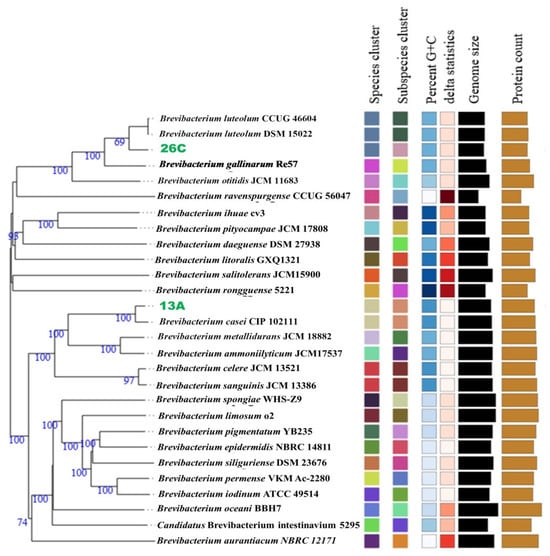
Figure 2.
A phylogenomic tree constructed by the Type Strain Genome Server (TYGS) based on the genomes of Brevibacterium casei 13A, Brevibacterium luteolum 26C, and their top related type strains. Confidence values are displayed near the nodes.
To confirm the taxonomic positions of both isolates, dDDH, AAI, and ANI comparisons were performed with their closest type strains. For isolate 26C, the type strain B. luteolum CCUG 46604 (NCBI accession: GCF_013004595.1) exhibited the closest relationship, with dDDH and ΔCG values of 71.1% and 0.02%, respectively. The ANIm and AAI values between the genomes were 96.77% and 96.31%, respectively.
In the case of isolate 13A, the type strain B. casei CIP 102111 (NCBI accession: GCF_900169275.1) showed the closest relationship, with dDDH and ΔCG values of 82.4% and 0.23%, respectively. The ANIm and AAI values between these genomes were 98.13% and 97.79%, respectively. These results confirm the taxonomic affiliations of both isolates.
3.4. B. luteolum 26C and B. casei 13A Genome Annotation and Genome Mapping
The Similar Genome Finder tool (www.bv-brc.org/app/GenomeDistance accessed on 24 May 2025) identified B. luteolum strain NEB1784 (GenBank: CP035810.1) as the closest complete genome to 26C, with a distance of 0.029898, making it suitable for reference-guided scaffolding. Using RagTag, the 26C genome was scaffolded into a single chromosome of 3,025,421 bp, covering 96.1% of the reference genome. On the other hand, the genome of B. casei FDAARGOS_1100 (GenBank: CP068173) was the closest complete genome to the genome of B. casei 13A, with a distance of 0.012729, so it was selected as the reference for reference-guided scaffolding. The scaffolding of B. casei 13A resulted in one main scaffold of 3,917,481 bp (93% coverage) and five smaller contigs ranging from 5495 to 13,337 bp. The RAST and Prokka pipelines were used to annotate the genomes of B. luteolum 26C and B. casei 13A. For both, while RAST returned more annotations with functional assignments, Prokka was able to call a greater number of annotations with EC assignments (Table 4). The genome maps of B. luteolum 26C and B. casei 13A are illustrated in Figure 3.

Table 4.
A summary of the genome annotation results of Brevibacterium luteolum 26C and Brevibacterium casei 13A.
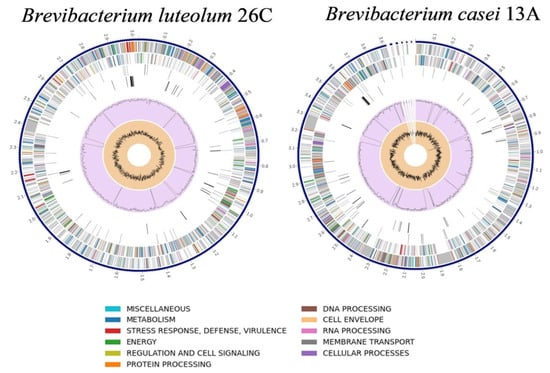
Figure 3.
Circular diagrams representing the genome maps of Brevibacterium luteolum 26C and Brevibacterium casei 13A.
A total of 81.49% and 81.05% of genome-derived proteins were assigned to a COG functional category for B. luteolum 26C and B. casei 13A, respectively. The comparative analysis of COG functional annotations between B. luteolum 26C and B. casei 13A reveals distinct differences in their metabolic and functional capacities. Notably, B. casei 13A generally exhibits higher counts in all COG functional categories except the category X (Mobilome: prophages, transposons), with 29 annotations for strain 26C compared to 9 for strain 13A (Figure 4).
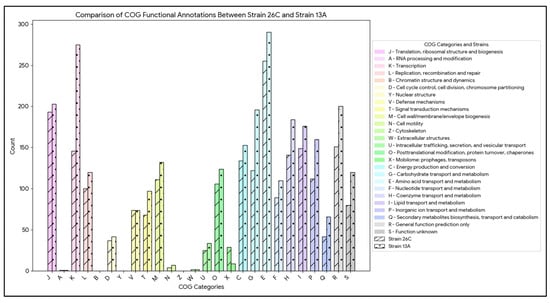
Figure 4.
Bar chart comparing the number of genome-derived protein counts per COG functional category in Brevibacterium luteolum 26C and Brevibacterium casei 13A.
IslandViewer 4 revealed the presence of multiple genomic islands (GIs) within the genomes of both B. luteolum 26C (12 GIs) and B. casei 13A (14 GIs) (Figure 5). A consistent theme across both strains is the prevalence of genes associated with metal detoxification and resistance, including specific transporters for nickel, cadmium, cobalt, zinc, copper, arsenic, and chromate. Furthermore, both strains exhibit robust DNA repair and maintenance systems, with various glycosylases, helicases, and recombinases (Table 5 and Table 6).
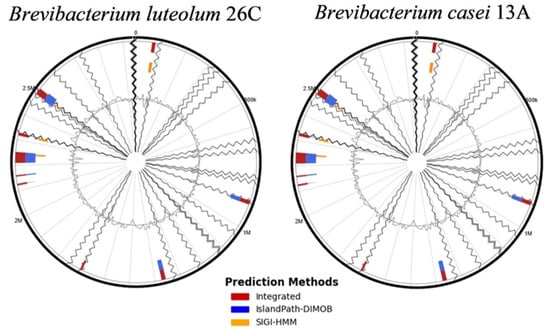
Figure 5.
The genomic islands (GIs) identified within the genomes of Brevibacterium luteolum 26C and Brevibacterium casei 13A using the IslandViewer 4 server.

Table 5.
A summary of the genomic islands (GIs) identified within the genome of Brevibacterium luteolum 26C and the genes identified within each GI, excluding hypothetical proteins.

Table 6.
A summary of the genomic islands (GIs) identified within the genome of Brevibacterium casei 13A and the genes identified within each GI, excluding hypothetical proteins.
Moreover, numerous mobile genetic elements (MGEs), including a variety of transposases and insertion sequences, were identified in both genomes. B. luteolum 26C harbors a greater number of MGE-associated genes compared to B. casei 13A, with a total of 67 genes versus 49, respectively (Table 7).

Table 7.
A summary of the mobile-genetic-element-related genes identified within the genomes of Brevibacterium luteolum 26C and Brevibacterium casei 13A.
3.5. Metabolic Pathways and Biosynthetic Gene Clusters (BGCs)
From 3449 and 2672 CDs, there were 1552 (~45%) and 1353 (~51%) protein products annotated on the KEGG database for B. casei 13A and B. luteolum 26C, respectively. The annotated proteins were incorporated into 232 KEGG pathways for B. casei 13A and 221 for B. luteolum 26C (Figure 6).
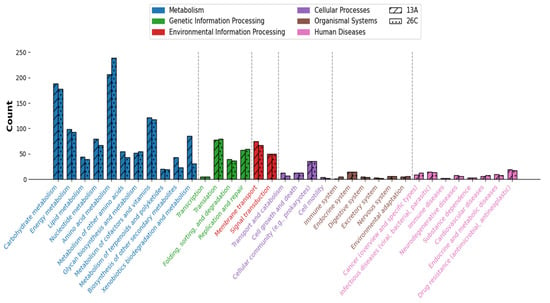
Figure 6.
Comparison between the numbers of genes annotated under each KEGG subcategory in Brevibacterium luteolum 26C and Brevibacterium casei 13A. Subcategories under the same top-level category are colored the same.
There were a total of 44 and 51 complete KEGG modules identified within the genomes of B. luteolum 26C and B. casei 13A, respectively. While the majority of these modules were shared between both strains, there were some unique modules. For instance, the biosynthesis of ectoine (M00033) module was identified within the genome of B. casei 13A, while not found within the genome of B. luteolum 26C.
The antiSMASH identified a total of four and five BGCs within the genomes of B. luteolum 26C and B. casei 13A, respectively (Table 8). There was only one cluster highly similar to ε-poly-L-lysine, having high similarity between the two strains (Figure 7).

Table 8.
Biosynthetic gene clusters (BGCs) identified within the genomes of Brevibacterium luteolum 26C and Brevibacterium casei 13A using antiSMASH version 8.
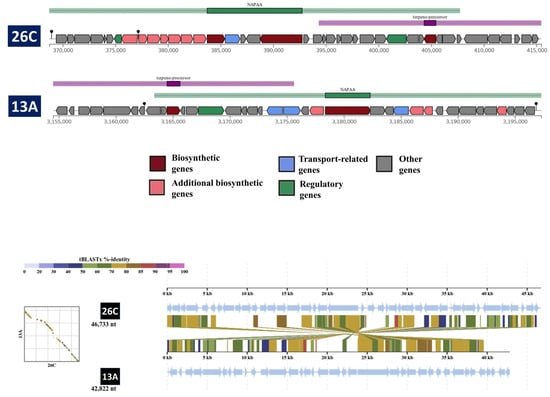
Figure 7.
A comparison between the compositions of the NPAA cluster in Brevibacterium luteolum 26C and Brevibacterium casei 13A. The tblastx alignment between the two clusters was carried out by the DiGAlign server.
3.6. Comparative Orthologous Cluster Analysis
The orthologous clustering analysis conducted with OrthoVenn3 compared B. luteolum strain 26C against the related strains DMY-1 and NEB1784. A total of 2447 clusters were detected in 26C, 2415 in DMY-1, and 2459 in NEB1784. Among these, 2178 clusters were conserved across all three strains, representing the stable backbone of essential cellular functions. Strain 26C also harbored a distinct set of orthologous clusters associated with transposition and DNA restriction–modification systems (Figure 8a), which may contribute to genomic plasticity and defense against foreign genetic elements. In addition, the analysis highlighted 202 singleton genes specific to 26C, underscoring its distinct genomic potential. Of these, 61 were annotated as functional genes by Prokka, while the remaining genes were predicted as hypothetical proteins (Supplementary Table S1). The annotated genes spanned diverse categories, including metabolic enzymes (e.g., ilvG, aceB, cadA), regulators (e.g., yurK, nanR), transport systems for amino acids, peptides, and ions, as well as resistance determinants such as abaF, hipA, and merA, and multidrug efflux components (mdtA). A substantial proportion of these singletons corresponded to insertion sequences and transposases, reinforcing the observation of transposition-related clusters and highlighting the role of genome mobility in shaping the evolutionary trajectory of strain 26C. Together, these features suggest that strain 26C has evolved specialized capabilities for adaptability, resistance, and stress tolerance in its marine habitat.
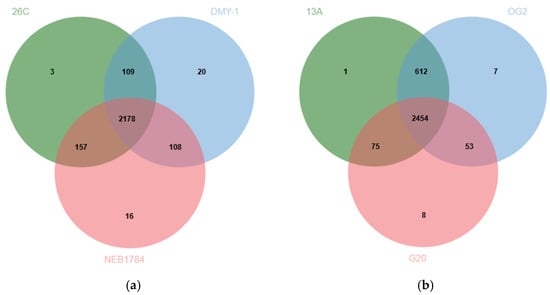
Figure 8.
(a) A Venn diagram indicating the number of shared orthologous protein clusters between the genomes of B. luteolum strain 26C and the related B. luteolum strains DMY-1 and NEB1784. (b) A Venn diagram indicating the number of shared orthologous protein clusters between the genomes of B. casei strain 13A and the related B. casei strains OG2 and G20.
Similarly, orthologous clustering analysis compared B. casei strain 13A with the related strains OG2 and G20. A total of 3142 clusters were detected in 13A, 3126 in OG2, and 2590 in G20. Among these, 2454 clusters were shared across all three strains, reflecting the conserved backbone of essential cellular functions. Strain 13A also contained a unique orthologous cluster associated with a membrane transporter protein YrkJ (Figure 8b). Moreover, OrthoVenn identified 277 singleton genes specific to 13A. Of these, 38 were annotated as functional proteins by Prokka, while the remaining genes were predicted as hypothetical proteins (Supplementary Table S2). The annotated proteins included metabolic enzymes (e.g., poxB, sir), regulators (e.g., betI, slyA, cdhR), and transport systems for amino acids, dicarboxylates, and ions (yjeH, genK, sdcS, srpC). Singletons corresponding to insertion sequences and transposases (IS3 family), alongside resistance determinants such as arsC2 (arsenate resistance) and bspRIM (modification methylase), were identified.
4. Discussion
The Red Sea represents one of the most unique marine ecosystems globally, characterized by high salinity and pronounced thermal and nutrient gradients, which exert strong selective pressures on resident organisms [33]. These challenging environmental conditions support a remarkable diversity of marine microorganisms with specialized stress-tolerance and metabolic capabilities uniquely adapted to such stressors [34]. Within these microbial communities, the genus Brevibacterium, a member of the phylum Actinomycetota, has garnered increasing attention due to its biotechnological potential [35]. In this study, we present a comprehensive genomic analysis of two Brevibacterium strains, B. luteolum 26C and B. casei 13A, isolated from Red Sea sponges. Through the integration of phenotypic characterization, whole-genome sequencing, and genome mining, the study provides valuable insights into the ecological adaptation and biosynthetic potential of these marine-derived isolates.
Previous studies have reported the successful isolation of Brevibacterium sp. from various Red Sea environments, including sediments, coral reefs, and sponges [36,37,38]. However, to our knowledge, no prior study has confirmed the presence of B. casei or B. luteolum at the species level in the Red Sea. This study is the first to report and validate the occurrence of these two species in the Red Sea, based on whole-genome sequencing analysis. The ANI, AAI (96.77% and 96.31%, respectively, for B. luteolum 26C; 98.13% and 97.79%, respectively, for B. casei 13A), and dDDH (>70%) values meet accepted thresholds for species-level classification.
As an adaptation to the challenging marine environment, marine bacteria have been reported to possess biosynthetic and antistress genetic mechanisms that may not be present in their terrestrial counterparts [8,39]. These adaptations are generally acquired through horizontal gene transfer mediated by MGEs and GIs. Genomic islands are substantial, distinct segments of DNA within a genome, frequently acquired via HGT and commonly harboring genes that confer adaptive advantages to the host bacterium. In contrast, mobile genetic elements are smaller DNA sequences capable of relocating within or between genomes, such as plasmids and transposons [40,41]. Within this context, multiple GIs were identified in both strains (12 in 26C and 14 in 13A), many harboring genes associated with heavy metal resistance (e.g., cobalt, nickel, zinc, cadmium, and arsenic transporters). These genes likely contribute to the strains’ ability to withstand the fluctuating chemical and metal stressor characteristics of marine environments [42,43,44,45,46,47]. In addition, both strains also encoded multiple DNA repair mechanisms (e.g., helicases, glycosylases), suggesting mechanisms to counter genomic damage caused by marine environmental stressors such as UV radiation and desiccation, enhancing genome stability and ensuring survival under harsh marine conditions [48,49]. Notably, B. casei 13A harbored additional oxidative stress response genes (e.g., ahpD, thioredoxins), further supporting resilience under oxidative stress [50].
Beyond stress adaptation, the GIs in both strains encode genes associated with broad metabolic versatility such as those involved in allantoin catabolism (allE, allB), potentially facilitating nutrient acquisition and processing from the surrounding environment. Allantoin, derived from host excretion or from the surrounding seawater, represents a valuable nitrogen source in an oligotrophic environment. The ability to catabolize this compound suggests potential mutualistic benefits to the sponge host through nutrient recycling and broadens the nitrogen acquisition capability of these strains, especially in nitrogen-limited oligotrophic waters [51,52].
Genomic islands also encoded traits that may promote competitive adaptability and symbiotic roles within the sponge microbiome. Notably, quorum-quenching genes such as ahlD were identified, encoding an N-acyl-homoserine lactonase implicated in the disruption of microbial communication and biofilm formation [43,53]. Of particular ecological significance, GIs harboring multidrug transporter-related genes (e.g., mdtL) were identified in B. casei 13A, which may confer resilience against competitors’ antimicrobials, environmental toxins, and self-produced metabolites [54]. Furthermore, the genome of B. casei 13A harbored the complete hcnABC operon, responsible for hydrogen cyanide (HCN) biosynthesis, whereas B. luteolum 26C contained only the hcnC gene. Given the well-documented antimicrobial and antifungal activities of HCN, this difference may suggest that B. casei 13A plays a more pronounced role in microbial competition and host defense within the sponge microbiome [44,55,56,57]. These features collectively highlight the potential role of these strains, particularly B. casei 13A, as defensive symbionts in the highly competitive sponge microenvironment.
Moreover, the frequent occurrence of MGEs in both strains, particularly in B. luteolum 26C, which exhibited a higher abundance of integration, excision, and phage-associated genes compared to B. casei 13A, highlights the crucial role of horizontal gene transfer as a mechanism for rapid genomic evolution and the acquisition of new adaptive traits [58]. These features likely contribute to the ability of these strains to successfully colonize and persist within the dynamic and selective environment of marine sponge hosts. The greater representation of MGE-related genes in B. luteolum 26C further suggests a more dynamic or recently active history of genomic rearrangements and horizontal gene acquisition relative to B. casei 13A.
Both B. luteolum 26C and B. casei 13A strains exhibited considerable secondary metabolite biosynthetic potential. The antiSMASH analysis revealed four BGCs in B. luteolum 26C and five in B. casei 13A. Notably, both strains shared a cluster with high similarity to ε-poly-L-lysine biosynthesis, a compound with a broad-spectrum antimicrobial activity and food-preservative properties [59,60]. The presence of an ectoine biosynthesis module in B. casei 13A suggests a specialized role in osmoregulation, supporting its adaptation to marine ecosystems due to its role as a compatible solute and extremolyte [61].
The antimicrobial activity exhibited by B. luteolum 26C and B. casei 13A against S. aureus, E. coli, and C. albicans is strongly supported by their genomic profiles, as revealed by antiSMASH analysis, and the presence of GI-encoded secondary metabolite genes. These observations underscore the ecological role of B. luteolum 26C and B. casei 13A strains as active competitors and potential defensive associates within the sponge microbiome.
The comparative analysis with terrestrial counterparts showed potential genetic adaptation of strains 26C and 13A for the marine environment. For instance, the genome of B. luteolum 26C is characterized by several unique genetic elements, such as merA, abaF, and mdtA. The merA encodes the enzyme mercuric reductase, which is a key component of the mer operon, a gene system that provides resistance to mercury toxicity. The MerA enzyme converts highly toxic ionic mercury into a less toxic form. This process is of interest for bioremediation, as it allows for the cleanup of mercury-contaminated sites [62]. Furthermore, the ability of Brevibacterium to survive in these harsh marine conditions likely depends on its robust detoxification systems, including efflux pumps. The presence of several efflux-system-related genes, such as abaF and mdtA, suggests a strong adaptive potential in this marine strain compared to its terrestrial counterparts. This repertoire of efflux system genes enhances the strain’s ability to export harmful substances, like antibiotics and heavy metals, out of the bacterial cell, a mechanism that is known to confer multidrug resistance [63,64].
On the other hand, the orthologous clustering analysis of B. casei 13A in comparison with the terrestrial counterparts OG2 and G20 highlights both conserved genomic features and distinct adaptive traits, indicating that strain 13A may have developed specialized genomic traits supporting adaptability and stress tolerance in its marine environment. For instance, the presence of singletons related to metabolic enzymes (e.g., poxB, sir) and regulatory proteins (betI, slyA, cdhR) suggests potential modulation of central metabolism and transcriptional networks that could enhance metabolic flexibility under fluctuating marine conditions. In addition, transport-related genes (yjeH, genK, sdcS, srpC) point toward adaptations for nutrient acquisition in nutrient-variable seawater. Moreover, resistance determinants such as arsC2 (arsenate resistance) and bspRIM (modification methylase) highlight the potential for resilience against toxic compounds and phage infection, traits that could provide selective advantages in complex marine microbial communities. Together, these features suggest that B. casei strain 13A has evolved specialized genomic strategies that balance the conservation of essential cellular machinery with the acquisition of novel functions, enabling successful adaptation to the marine environment [65,66,67].
5. Conclusions
This study provides a comprehensive genomic analysis of two Brevibacterium strains, B. luteolum 26C and B. casei 13A, isolated from Red Sea sponges. Whole-genome sequencing and functional annotation revealed their secondary metabolite gene clusters and genomic traits associated with adaptation to the region’s challenging physicochemical conditions. These include genes for osmotic regulation, hypersalinity tolerance, survival under nutrient limitation, and resistance to toxic compounds. Importantly, both strains exhibited antimicrobial activity against different pathogenic microorganisms, further emphasizing their ecological role. Collectively, these findings underscore the adaptive capacity of Brevibacterium in challenging marine environments and its potential biotechnological value.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology14091271/s1, Table S1: Functional annotation of B. luteolum strain 26C-specific singletons identified by orthologous cluster analysis using OrthoVenn3. Table S2: Functional annotation of B. casei strain 13A-specific singletons identified by orthologous cluster analysis using OrthoVenn3.
Author Contributions
All authors contributed to the study’s conception and design. Y.S.M.: Conceptualization, Grant application, Investigation, and Writing—review the final draft. M.E.S.: Conceptualization, Investigation, Methodology, and Writing—original draft. A.M.S.: Conceptualization, Formal analysis, Data curation, Investigation, Methodology, Validation, Visualization, and Writing—original draft. H.L.K.: Conceptualization, Investigation, Methodology, and Writing—original draft. S.M.S.: Conceptualization, Investigation, Validation, Supervision, Project administration, and Writing—review and editing the final draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deanship of Research and Graduate Studies (DRG), Ajman University’s grant AY 2024/2025 ref. number 2024-IRG-MED-1.
Institutional Review Board Statement
The study protocol was approved by the Ethics Committee of the Faculty of Pharmacy, Suez Canal University (protocol code 201810PHD2 on 17 October 2018) and by the Ajman University’s Research Ethics Committee (ref. number M-F-H-3-Oct/2024).
Data Availability Statement
The original data presented in the study are openly available in the National Center for Biotechnology Information (NCBI) GenBank repository. For Brevibacterium casei strain 13A, the sequence data are accessible via BioProject accession PRJNA1280801, Biosample accession SAMN49535178, SRA accession SRR34103316, and Genome accession GCA_051216575.1. For Brevibacterium luteolum strain 26C, the sequence data are accessible via BioProject accession PRJNA1280809, Biosample accession SAMN49535213, SRA accession SRR34103435, and Genome accession GCA_051216595.1.
Acknowledgments
We would like to thank the Gulf Medical University colleagues and the following Ajman University medical students for their technical assistance: Najmah Al-Hamadi, Abdul Ilah Dakak, Abdullattif Al-Atta, and Layal Odeh.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AAI | Average Amino Acid Identity |
| ANI | Average Nucleotide Identity |
| ANIm | ANI using MUMmer alignment |
| BGC(s) | Biosynthetic Gene Cluster(s) |
| CDs/CDSs | Coding Sequences |
| dDDH | Digital DNA–DNA Hybridization |
| DMSO | Dimethyl Sulfoxide |
| GI(s) | Genomic Island(s) |
| GTDB | Genome Taxonomy Database |
| GTDB-Tk | GTDB Toolkit |
| MGE(s) | Mobile Genetic Element(s) |
| NSW | Natural Seawater |
| Prokka | Rapid Prokaryotic Genome Annotation |
| TYGS | Type Strain Genome Server |
| WGS | Whole-Genome Sequencing |
References
- Rasul, N.M.A.; Stewart, I.C.F. The Red Sea: The Formation, Morphology, Oceanography and Environment of a Young Ocean Basin; Springer: Berlin/Heidelberg, Germany, 2015; ISBN 978-3-662-45200-4. [Google Scholar]
- Pearman, J.K.; Kürten, S.; Sarma, Y.V.B.; Jones, B.H.; Carvalho, S. Biodiversity Patterns of Plankton Assemblages at the Extremes of the Red Sea. FEMS Microbiol. Ecol. 2016, 92, fiw002. [Google Scholar] [CrossRef]
- Thomas, T.; Moitinho-Silva, L.; Lurgi, M.; Björk, J.R.; Easson, C.; Astudillo-García, C.; Olson, J.B.; Erwin, P.M.; López-Legentil, S.; Luter, H.; et al. Diversity, Structure and Convergent Evolution of the Global Sponge Microbiome. Nat. Commun. 2016, 7, 11870. [Google Scholar] [CrossRef]
- Hentschel, U.; Piel, J.; Degnan, S.M.; Taylor, M.W. Genomic Insights into the Marine Sponge Microbiome. Nat. Rev. Microbiol. 2012, 10, 641–654. [Google Scholar] [CrossRef]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The Sponge Holobiont in a Changing Ocean: From Microbes to Ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- Laport, M.; Santos, O.; Muricy, G. Marine Sponges: Potential Sources of New Antimicrobial Drugs. Curr. Pharm. Biotechnol. 2009, 10, 86–105. [Google Scholar] [CrossRef]
- Sedeek, A.M.; Elfeky, H.; Hanora, A.S.; Solyman, S.M. Genomic Insights into Biosynthesis and Adaptation in the Bioactive Marine Bacterium Streptomyces albidoflavus VIP-1 from the Red Sea. BMC Microbiol. 2025, 25, 372. [Google Scholar] [CrossRef] [PubMed]
- Kamel, H.L.; Hanora, A.; Solyman, S.M. Metataxonomic, Bioactivity and Microbiome Analysis of Red Sea Marine Sponges from Egypt. Mar. Genom. 2022, 61, 100920. [Google Scholar] [CrossRef]
- El Samak, M.; Lotfy, H.; Sedeek, A.M.; Mohamed, Y.S.; Solyman, S.M. Genomic Characterization of Marine Staphylococcus shinii strain SC-M1C: Potential Genetic Adaptations and Ecological Role. Microorganisms 2025, 13, 1866. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; She, J.; Fu, J.; Wang, J.; Ye, Y.; Yang, B.; Liu, Y.; Zhou, X.; Tao, H. Advances in Natural Products from the Marine-Sponge-Associated Microorganisms with Antimicrobial Activity in the Last Decade. Mar. Drugs 2023, 21, 236. [Google Scholar] [CrossRef]
- Ibrahim, J.A.A.; Botcha, S.; Prattipati, S.D. Marine Actinomycetes: A Promising Source of Novel Therapeutics and Pharmaceutical Bioactive Compounds—A Review. Microbe 2025, 7, 100383. [Google Scholar] [CrossRef]
- Lee, S.D. Brevibacterium marinum sp. Nov., Isolated from Seawater. Int. J. Syst. Evol. Microbiol. 2008, 58, 500–504. [Google Scholar] [CrossRef]
- Bhadra, B.; Raghukumar, C.; Pindi, P.K.; Shivaji, S. Brevibacterium oceani sp. Nov., Isolated from Deep-Sea Sediment of the Chagos Trench, Indian Ocean. Int. J. Syst. Evol. Microbiol. 2008, 58, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Huang, P.; Dong, C.; Deng, X. Nickel Bioaccumulation by a Marine Bacterium Brevibacterium sp. (X6) Isolated from Shenzhen Bay, China. Mar. Pollut. Bull. 2021, 170, 112656. [Google Scholar] [CrossRef] [PubMed]
- Olender, A.; Rutyna, P.; Niemcewicz, M.; Bogut, A.; Ciesielka, M.; Teresiński, G. Draft Whole-Genome Sequence of Brevibacterium casei strain Isolated from a Bloodstream Infection. Braz. J. Microbiol. 2020, 51, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Cumsille, A.; Serna-Cardona, N.; González, V.; Claverías, F.; Undabarrena, A.; Molina, V.; Salvà-Serra, F.; Moore, E.R.B.; Cámara, B. Exploring the Biosynthetic Gene Clusters in Brevibacterium: A Comparative Genomic Analysis of Diversity and Distribution. BMC Genom. 2023, 24, 622. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: Memory friendly classification with the genome taxonomy database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A Web Server for Prokaryotic Species Circumscription Based on Pairwise Genome Comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The Enveomics Collection: A Toolbox for Specialized Analyses of Microbial Genomes and Metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar] [CrossRef]
- Alonge, M.; Soyk, S.; Ramakrishnan, S.; Wang, X.; Goodwin, S.; Sedlazeck, F.J.; Lippman, Z.B.; Schatz, M.C. RaGOO: Fast and Accurate Reference-Guided Scaffolding of Draft Genomes. Genome Biol. 2019, 20, 224. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-Db: A Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022, 88, e0099122. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S. IslandViewer 4: Expanded Prediction of Genomic Islands for Larger-Scale Datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Vader, L.; Szenei, J.; Reitz, Z.L.; Augustijn, H.E.; Cediel-Becerra, J.D.D.; de Crécy-Lagard, V.; Koetsier, R.A.; Williams, S.E.; et al. antiSMASH 8.0: Extended Gene Cluster Detection Capabilities and Analyses of Chemistry, Enzymology, and Regulation. Nucleic Acids Res. 2025, 53, W32–W38. [Google Scholar] [CrossRef]
- Sun, J.; Lu, F.; Luo, Y.; Bie, L.; Xu, L.; Wang, Y. OrthoVenn3: An Integrated Platform for Exploring and Visualizing Orthologous Data across Genomes. Nucleic Acids Res. 2023, 51, W397–W403. [Google Scholar] [CrossRef]
- Ngugi, D.K.; Antunes, A.; Brune, A.; Stingl, U. Biogeography of Pelagic Bacterioplankton Across an Antagonistic Temperature—Salinity Gradient in the Red Sea. Mol. Ecol. 2012, 21, 388–405. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A Communal Catalogue Reveals Earth’s Multiscale Microbial Diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Onraedt, A.; Soetaert, W.; Vandamme, E. Industrial Importance of the Genus Brevibacterium. Biotechnol. Lett. 2005, 27, 527–533. [Google Scholar] [CrossRef]
- Al-Zereini, W.A. Bioactive Crude Extracts from Four Bacterial Isolates of Marine Sediments from Red Sea, Gulf of Aqaba, Jordan. Jordan J. Biol. Sci. 2014, 7, 133–173. [Google Scholar] [CrossRef]
- Abdel-Razik, M.A.; Azmy, A.F.; Dishisha, T.; El-Gendy, A.O.; Afzan, A.; Kamal, N.; Tawfike, A.; Sebak, M. Screening of Red Sea- and Mediterranean Sea-Derived Actinomycetes for Antimicrobial and Antitumor Activities: LC-ESI-HRMS-Based Metabolomics Study. Microb. Cell Fact. 2025, 24, 136. [Google Scholar] [CrossRef]
- El Samak, M.; Zakeer, S.; Hanora, A.; Solyman, S.M. Metagenomic and Metatranscriptomic Exploration of the Egyptian Red Sea Sponge Theonella sp. Associated Microbial Community. Mar. Genom. 2023, 70, 101032. [Google Scholar] [CrossRef]
- Sedeek, A.M.; Ismail, M.M.; Elsayed, T.R.; Ramadan, M.A. Recent Methods for Discovering Novel Bioactive Metabolites, Specifically Antimicrobial Agents, from Marine-Associated Micro-Organisms. Lett. Appl. Microbiol. 2022, 75, 511–525. [Google Scholar] [CrossRef]
- Juhas, M.; Van Der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic Islands: Tools of Bacterial Horizontal Gene Transfer and Evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef] [PubMed]
- Audrey, B.; Cellier, N.; White, F.; Jacques, P.-É.; Burrus, V. A Systematic Approach to Classify and Characterize Genomic Islands Driven by Conjugative Mobility Using Protein Signatures. Nucleic Acids Res. 2023, 51, 8402–8412. [Google Scholar] [CrossRef]
- Ravcheev, D.A.; Best, A.A.; Sernova, N.V.; Kazanov, M.D.; Novichkov, P.S.; Rodionov, D.A. Genomic Reconstruction of Transcriptional Regulatory Networks in Lactic Acid Bacteria. BMC Genom. 2013, 14, 94. [Google Scholar] [CrossRef]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, Function, and Evolution of Bacterial ATP-Binding Cassette Systems. Microbiol. Mol. Biol. Rev. MMBR 2008, 72, 317–364. [Google Scholar] [CrossRef]
- Blumer, C.; Haas, D. Mechanism, Regulation, and Ecological Role of Bacterial Cyanide Biosynthesis. Arch. Microbiol. 2000, 173, 170–177. [Google Scholar] [CrossRef]
- Newton, G.L.; Buchmeier, N.; Fahey, R.C. Biosynthesis and Functions of Mycothiol, the Unique Protective Thiol of Actinobacteria. Microbiol. Mol. Biol. Rev. MMBR 2008, 72, 471. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Efflux-Mediated Heavy Metal Resistance in Prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Crow, A.; Acheson, R.M.; Le Brun, N.E.; Oubrie, A. Structural basis of Redox-coupled protein substrate selection by the cytochrome c biosynthesis protein ResA. J. Biol. Chem. 2004, 279, 23654–23660. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, K.A.; Gangloff, S.; Rothstein, R. The RecQ DNA Helicases in DNA Repair. Annu. Rev. Genet. 2010, 44, 393. [Google Scholar] [CrossRef]
- Hickson, I.D. RecQ Helicases: Caretakers of the Genome. Nat. Rev. Cancer 2003, 3, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Anjou, C.; Lotoux, A.; Morvan, C.; Martin-Verstraete, I. From Ubiquity to Specificity: The Diverse Functions of Bacterial Thioredoxin Systems. Environ. Microbiol. 2024, 26, e16668. [Google Scholar] [CrossRef]
- Switzer, A.; Burchell, L.; McQuail, J.; Wigneshweraraj, S. The Adaptive Response to Long-Term Nitrogen Starvation in Escherichia coli Requires the Breakdown of Allantoin. J. Bacteriol. 2020, 202, e00172-20. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.P.; Thrash, J.C.; Nicora, C.D.; Lipton, M.S.; Burnum-Johnson, K.E.; Carini, P.; Smith, R.D.; Giovannoni, S.J. Proteomic and Transcriptomic Analyses of “Candidatus Pelagibacter ubique” Describe the First PII-Independent Response to Nitrogen Limitation in a Free-Living Alphaproteobacterium. mBio 2013, 4, e00133-12. [Google Scholar] [CrossRef]
- Gohil, N.; Ramírez-García, R.; Panchasara, H.; Patel, S.; Bhattacharjee, G.; Singh, V. Book Review: Quorum Sensing vs. Quorum Quenching: A Battle with no End in Sight; Kalia, V.C., Ed.; Springer: New Delhi, India, 2015; ISBN 978-81-322-1981-1. [Google Scholar]
- Fanelli, G.; Pasqua, M.; Colonna, B.; Prosseda, G.; Grossi, M. Expression Profile of Multidrug Resistance Efflux Pumps During Intracellular Life of Adherent-Invasive Escherichia coli strain LF82. Front. Microbiol. 2020, 11, 1935. [Google Scholar] [CrossRef] [PubMed]
- Sehrawat, A.; Sindhu, S.S.; Glick, B.R. Hydrogen Cyanide Production by Soil Bacteria: Biological Control of Pests and Promotion of Plant Growth in Sustainable Agriculture. Pedosphere 2022, 32, 15–38. [Google Scholar] [CrossRef]
- Flury, P.; Vesga, P.; Péchy-Tarr, M.; Aellen, N.; Dennert, F.; Hofer, N.; Kupferschmied, K.P.; Kupferschmied, P.; Metla, Z.; Ma, Z.; et al. Antimicrobial and Insecticidal: Cyclic Lipopeptides and Hydrogen Cyanide Produced by Plant-Beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 Contribute to Insect Killing. Front. Microbiol. 2017, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Michelsen, C.F.; Stougaard, P. Hydrogen Cyanide Synthesis and Antifungal Activity of the Biocontrol Strain Pseudomonas fluorescens In5 from Greenland Is Highly Dependent on Growth Medium. Can. J. Microbiol. 2012, 58, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile Genetic Elements: The Agents of Open Source Evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Hu, Y.; Qin, J.; Yu, B. Design and Optimization of ε-Poly-l-Lysine with Specific Functions for Diverse Applications. Int. J. Biol. Macromol. 2024, 262, 129513. [Google Scholar] [CrossRef]
- Hyldgaard, M.; Mygind, T.; Vad, B.S.; Stenvang, M.; Otzen, D.E.; Meyer, R.L. The Antimicrobial Mechanism of Action of Epsilon-Poly-L-Lysine. Appl. Environ. Microbiol. 2014, 80, 7758–7770. [Google Scholar] [CrossRef]
- Kuhlmann, A.U.; Bremer, E. Osmotically Regulated Synthesis of the Compatible Solute Ectoine in Bacillus Pasteurii and Related Bacillus spp. Appl. Environ. Microbiol. 2002, 68, 772–783. [Google Scholar] [CrossRef]
- Barkay, T.; Miller, S.M.; Summers, A.O. Bacterial Mercury Resistance from Atoms to Ecosystems. FEMS Microbiol. Rev. 2003, 27, 355–384. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, R.; Bhattacharyya, T.; Bhando, T.; Pathania, R. Fosfomycin Resistance in Acinetobacter baumannii Is Mediated by Efflux Through a Major Facilitator Superfamily (MFS) Transporter—AbaF. J. Antimicrob. Chemother. 2017, 72, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Nikaido, E.; Yamaguchi, A. Regulation of Multidrug Efflux Systems Involved in Multidrug and Metal Resistance of Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2007, 189, 9066–9075. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial Insertion Sequences: Their Genomic Impact and Diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef] [PubMed]
- Kabiraj, A.; Biswas, R.; Halder, U.; Bandopadhyay, R. Bacterial Arsenic Metabolism and Its Role in Arsenic Bioremediation. Curr. Microbiol. 2022, 79, 131. [Google Scholar] [CrossRef]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—A Database for DNA Restriction and Modification: Enzymes, Genes and Genomes. Nucleic Acids Res. 2015, 43, D298–D299. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).