Investigating the Antimicrobial Efficacy of Cannabinoids and Their Derivatives Against Neisseria Gonorrhoeae by Computational Analysis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
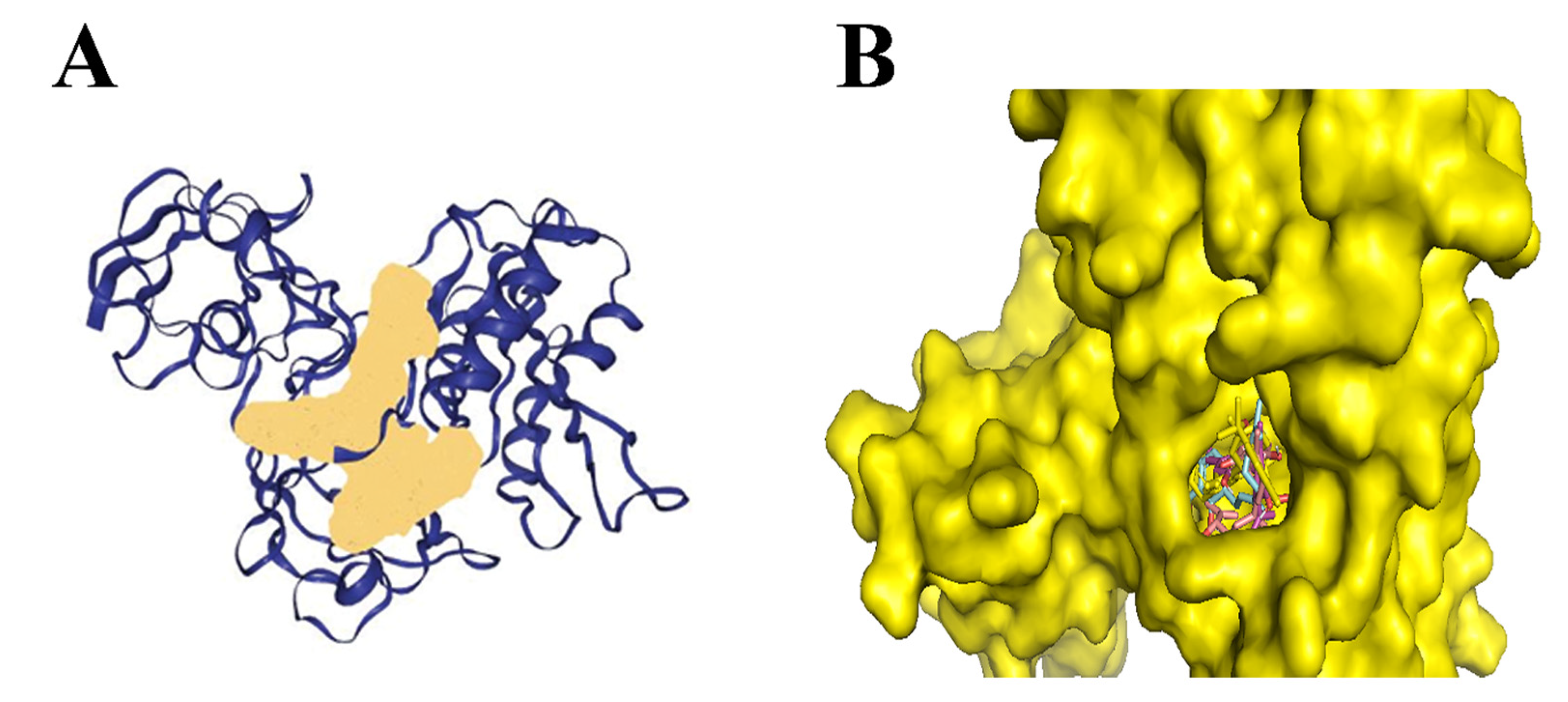
2.1. Binding Site Prediction of the Protein
2.2. Fingerprint Search
2.3. Molecular Docking
2.4. Evaluation of Protein-Ligand Interaction
2.5. Comparative Drug-likeness Assessment of Identified Cannabinoids and Its Derivatives Against Antibiotics Used in Treating N. gonorrhoeae Infection
3. Results and Discussion
3.1. Ligand Selection and Identification of Natural Products Through Fingerprint Searches
3.2. Molecular Docking Analysis of Cannabinoids and Its Derivates Against N. gonorrhoeae; 2Fe-2s Iron–Sulfur Cluster Binding Domain-Containing Protein
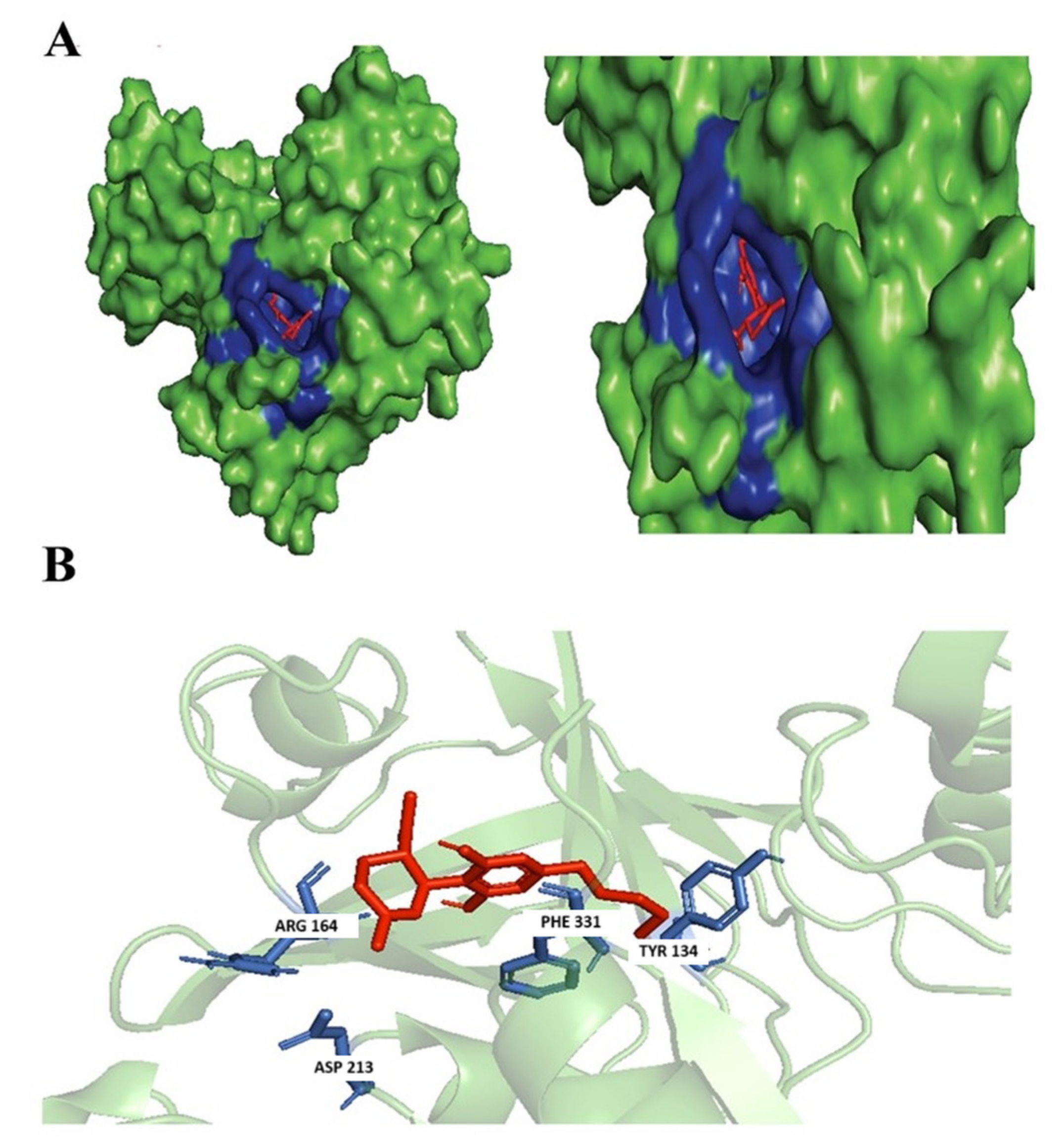
3.3. Evaluation of Protein Ligand Interaction of the Identified Cannabinoids
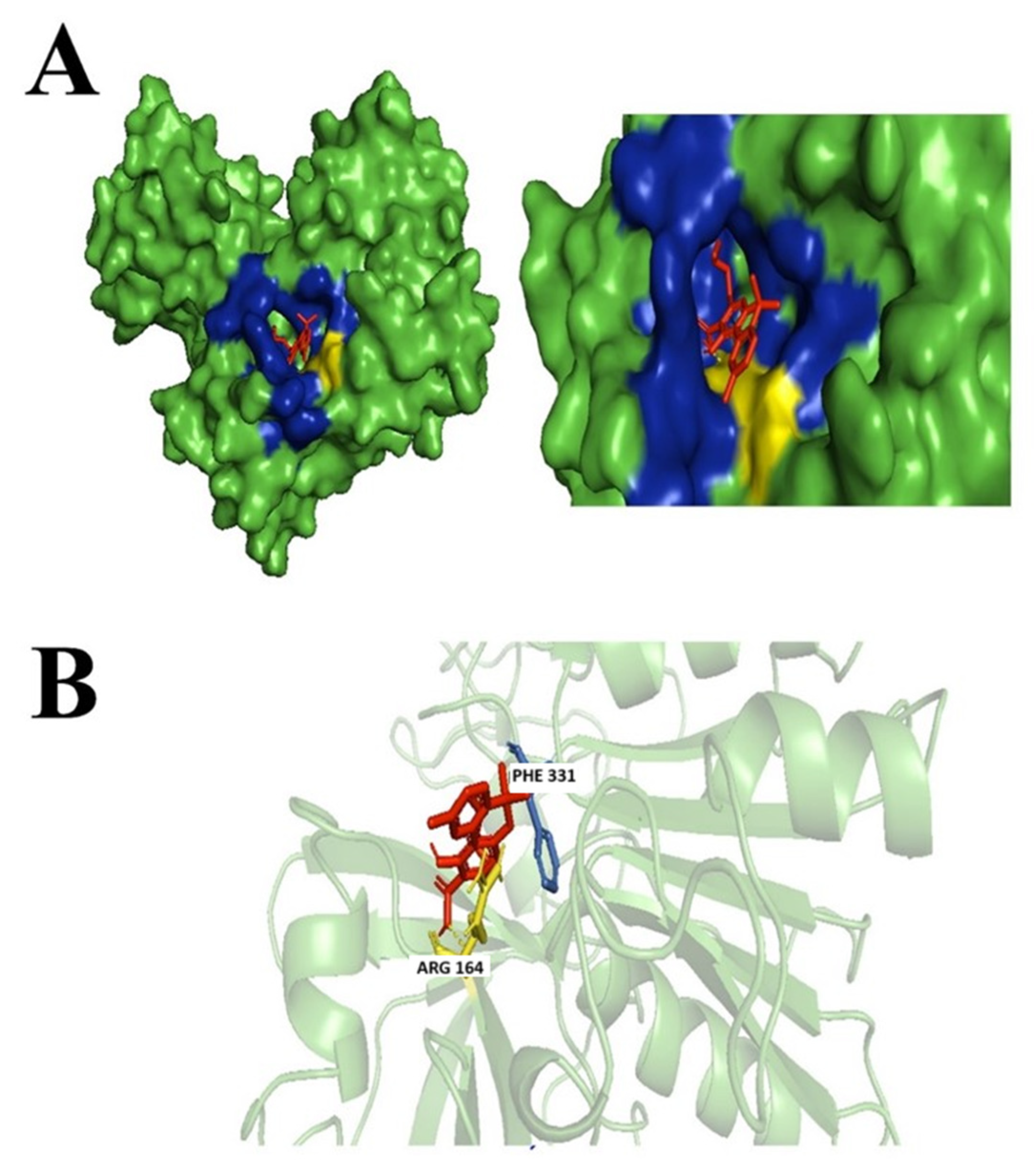
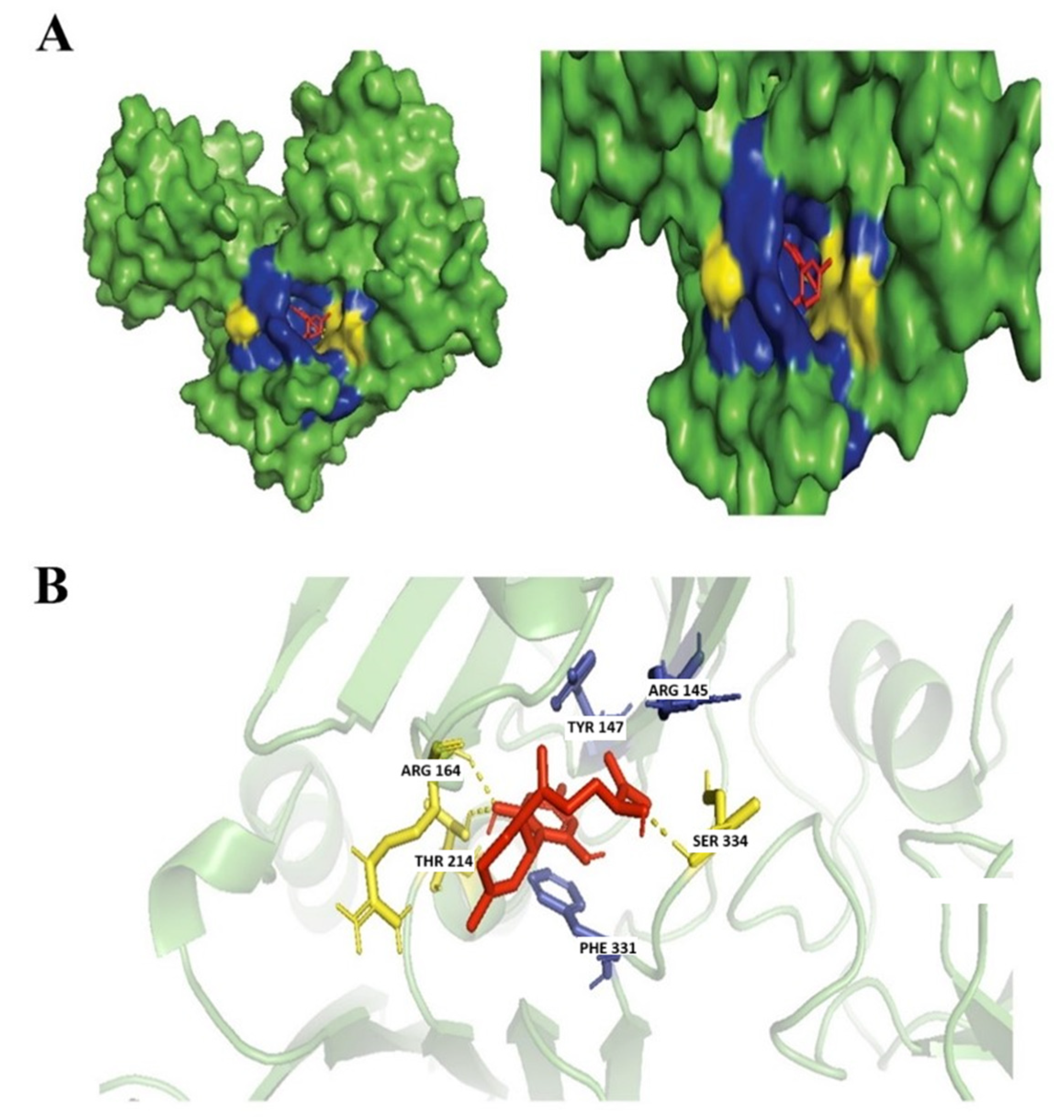
3.3.1. (+)-Cannabidiol: 1,3-Benzenediol, 2-[3-methyl-6-(1-methylethenyl)-2-cyclohexen-1-yl]-5-pentyl-, (1R-Trans)-
3.3.2. Dronabinol
3.3.3. Cannabinolic Acid A (CBNA)
3.3.4. Cannabigerolic Acid (CBGA)
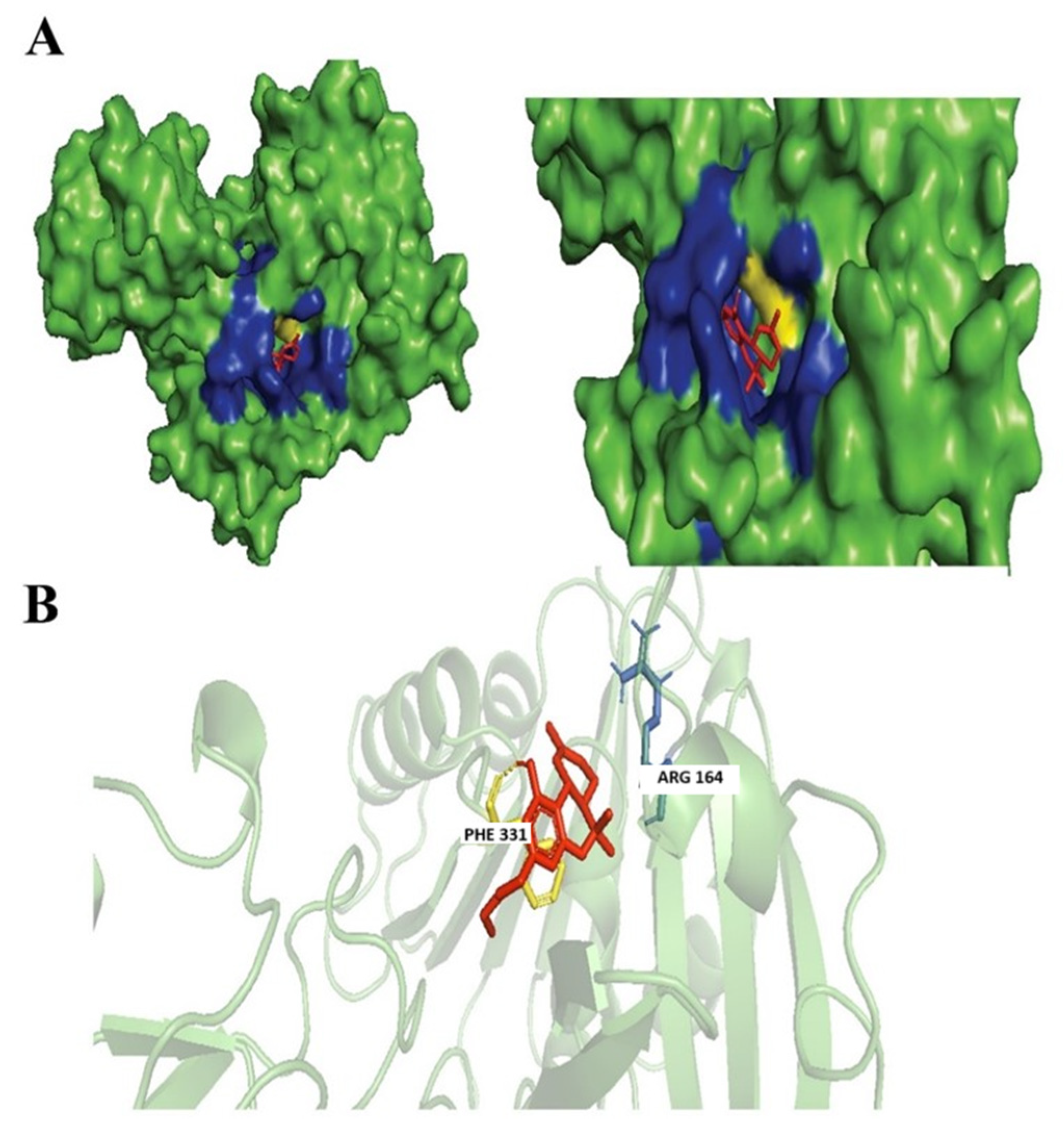
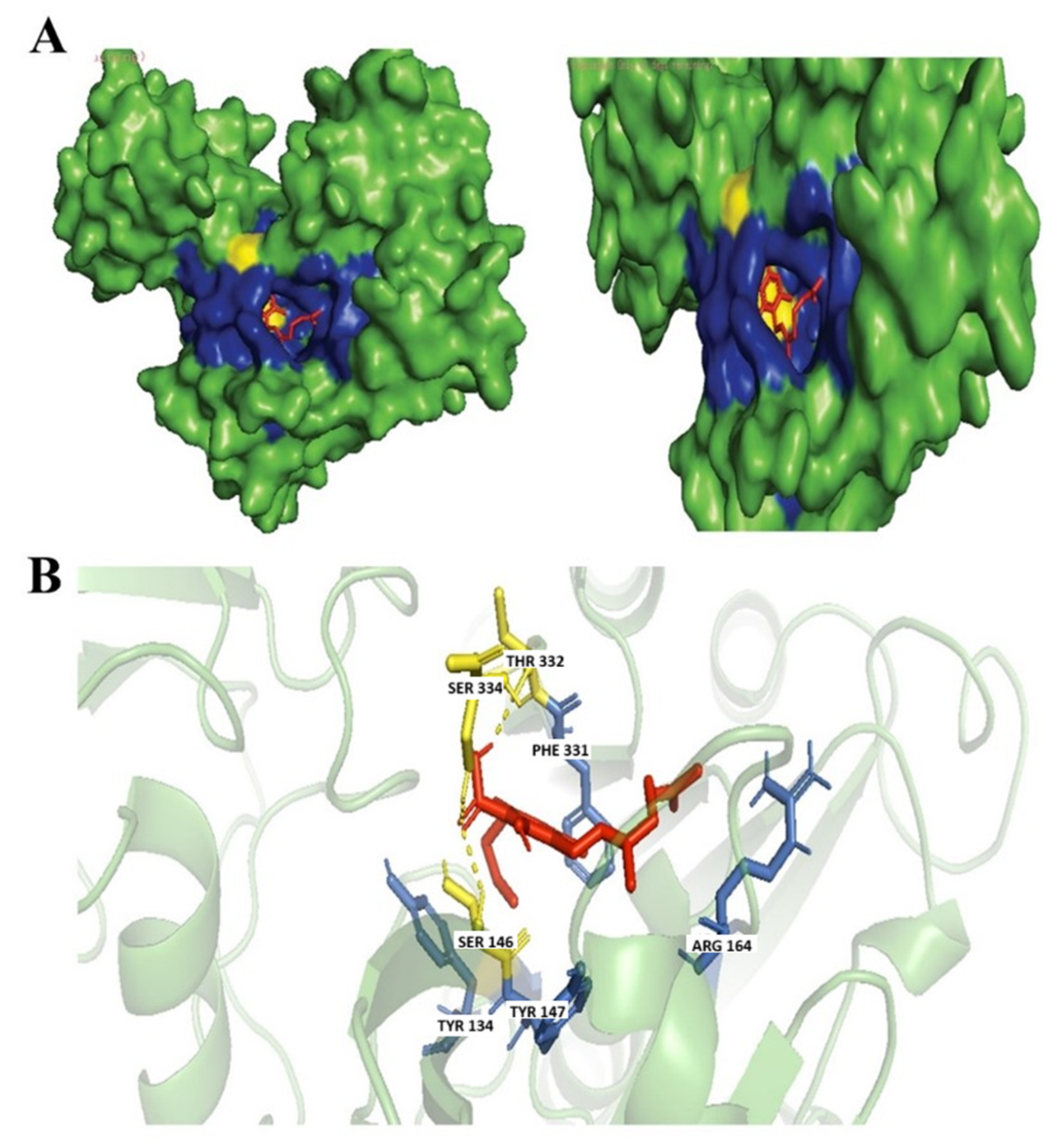
3.4. Evaluation of Protein Ligand Interaction of the Cannabinoid’s Derivatives
Ferruginene C
3.5. Drug-likeness and Pharmacokinetics Profile Assessment of the Identified Cannabinoids and Derivatives
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette |
| ADT | AutoDock Tools |
| ADME | Absorption, Distribution, Metabolism, Excretion |
| AIDS | Acquired Immunodeficiency Syndrome |
| AMR | Antimicrobial Resistance |
| BBB | Blood–Brain Barrier |
| BOILED-Egg | Brain or Intestinal EstimateD permeation method |
| BSTIs | Bacterial Sexually Transmitted Infections |
| CB | Cannabinoid |
| CBN | Cannabinol |
| CBNA | Cannabinolic Acid A |
| CBGA | Cannabigerolic Acid |
| CNS | Central Nervous System |
| CYP | Cytochrome P450 |
| ETC | Electron Transport Chain |
| ESC | Extended-Spectrum Cephalosporins |
| FDA | Food and Drug Administration |
| Fe-S | Iron–sulfur Proteins |
| FeS | Iron–sulfur |
| FUR | Ferric Uptake Regulator |
| GI | Gastrointestinal |
| hERG | Human Ether-à-Go-Go-Related Gene |
| HIA | Human Intestinal Absorption |
| MIC | Lowest Inhibitory Concentration |
| NO | Nitric Oxide |
| NPs | Natural Products |
| NPASS | Natural Product Activity and Species Source |
| PLIP | Protein-Ligand Interaction Profiler |
| Ro5 | Rule-of-Five |
| ROS | Reactive Oxygen Species |
| SPMs | Secondary Plant Metabolites |
| STD | Sexually Transmitted Disease |
| SWISSADME | Swiss Institute of Bioinformatics ADME prediction tool |
| THC | Δ-9-Tetrahydrocannabinol |
| THCA | Tetrahydrocannabinol Acid |
| VDss | Volume of Distribution |
| 3D | 3-Dimensional |
References
- World Health Organization (WHO). WHO Guidelines for the Treatment of Neisseria gonorrhoeae; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- Whittles, L.K.; White, P.J.; Paul, J.; Didelot, X. Epidemiological trends of antibiotic resistant Gonorrhoea in the United Kingdom. Antibiotics 2018, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Calado, J.; Castro, R.; Lopes, Â.; Campos, M.J.; Rocha, M.; Pereira, F. Antimicrobial resistance and molecular characteristics of Neisseria gonorrhoeae isolates from men who have sex with men. Int. J. Infect. Dis. 2019, 79, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Derbie, A.; Mekonnen, D.; Woldeamanuel, Y.; Abebe, T. Azithromycin resistant gonococci: A literature review. Antimicrob. Resist. Infect. Control. 2020, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019. [Google Scholar]
- Farha, M.A.; El-Halfawy, O.M.; Gale, R.T.; Macnair, C.R.; Carfrae, L.A.; Zhang, X.; Jentsch, N.G.; Magolan, J.; Brown, E.D. Uncovering the Hidden Antibiotic Potential of Cannabis. ACS Infect. Dis. 2020, 6, 338–346. [Google Scholar] [CrossRef]
- Paritala, H.; Carroll, K. New Targets and Inhibitors of Mycobacterial Sulfur Metabolism. Infect. Disord—Drug Targets 2013, 13, 85–115. [Google Scholar] [CrossRef]
- Hernández-Cervantes, R.; Méndez-DÍaz, M.; Prospéro-García, Ó.; Morales-Montor, J. Immunoregulatory Role of Cannabinoids during Infectious Disease. Neuroimmunomodulation 2018, 24, 183–199. [Google Scholar] [CrossRef]
- Justino, M.C.; Almeida, C.C.; Gonçalves, V.L.; Teixeira, M.; Saraiva, L.M. Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron–sulphur clusters. FEMS Microbiol. Lett. 2006, 257, 278–284. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Multi-Drug Resistant Gonorrhoea. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/multi-drug-resistant-gonorrhoea (accessed on 3 February 2022).
- Spellberg, B.; Powers, J.H.; Brass, E.P.; Miller, L.G.; Edwards, J.E. Trends in antimicrobial drug development: Implications for the future. Clin. Infect. Dis. 2004, 38, 1279–1286. [Google Scholar] [CrossRef]
- Sholler, D.J.; Huestis, M.A.; Amendolara, B.; Vandrey, R.; Cooper, Z.D. Therapeutic potential and safety considerations for the clinical use of synthetic cannabinoids. Pharmacol. Biochem. Behav. 2020, 199, 173059. [Google Scholar] [CrossRef]
- Blaskovich, M.A.T.; Kavanagh, A.M.; Elliott, A.G.; Zhang, B.; Ramu, S.; Amado, M.; Lowe, G.J.; Hinton, A.O.; Pham, D.M.T.; Zuegg, J.; et al. The antimicrobial potential of cannabidiol. Commun. Biol. 2021, 4, 7. [Google Scholar] [CrossRef]
- Omeershffudin, U.N.M.; Kumar, S. Antibiotic Resistance in Neisseria gonnorhoeae: A Broad-spectrum Drug Target Identification by Using Subtractive Genomics. Genom. Inform. 2023; in press. [Google Scholar]
- Fontenot, C.R.; Ding, H. Ferric uptake regulator (Fur) binds a [2Fe-2S] cluster to regulate intracellular iron homeostasis in Escherichia coli. J. Biol. Chem. 2023, 299, 104748. [Google Scholar] [CrossRef]
- Volkamer, A.; Kuhn, D.; Grombacher, T.; Rippmann, F.; Rarey, M. Combining Global and Local Measures for Structure-Based Druggability Predictions. J. Chem. Inf. Model. 2012, 52, 360–372. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel. J. Cheminform. 2011, 3, 33. Available online: https://jcheminf.biomedcentral.com/track/pdf/10.1186/1758-2946-3-33 (accessed on 1 January 2025). [CrossRef]
- Schrödinger, L.L.C.; DeLano, W. PyMOL. Available online: https://www.schrodinger.com/platform/products/pymol (accessed on 1 January 2025).
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the Scope of the Protein–Ligand Interaction Profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.A. ADMET Properties: Overview and Current Topics; Springer: Singapore, 2017; pp. 113–133. [Google Scholar]
- Walters, W.P. Going further than Lipinski’s rule in drug design. Expert. Opin. Drug Discov. 2012, 7, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Mortenson, L.E.; Valentine, R.C.; Carnahan, J.E. Ferredoxin in the Phosphoroclastic Reaction of Pyruvic Acid and Its Relation to Nitrogen Fixation in Clostridium pasteurianum. J. Biol. Chem. 1963, 238, 794–800. [Google Scholar] [CrossRef]
- Miller, H.K.; Auerbuch, V. Bacterial iron-sulfur cluster sensors in mammalian pathogens. Metallomics 2015, 7, 943–956. [Google Scholar] [CrossRef]
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005, 74, 247–281. [Google Scholar] [CrossRef] [PubMed]
- Catalano-Dupuy, D.L.; López-Rivero, A.; Soldano, A.; Ceccarelli, E.A. Redox proteins as targets for drugs development against pathogens. Curr. Pharm. Des. 2013, 19, 2594–2605. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.M.; Krupp, J.; Clark, S.; Isberg, R.R. Iron-Sulfur Cluster Repair Contributes to Yersinia pseudotuberculosis Survival within Deep Tissues. Infect. Immun. 2019, 87, e00533-19. [Google Scholar] [CrossRef] [PubMed]
- Walford, G.; Loscalzo, J. Nitric oxide in vascular biology. J. Thromb. Haemost. 2003, 1, 2112–2118. [Google Scholar] [CrossRef]
- Isabella, V.M.; Lapek, J.D.; Kennedy, E.M.; Clark, V.L. Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol. Microbiol. 2009, 71, 227–239. [Google Scholar] [CrossRef]
- Gurung, A.B.; Bhattacharjee, A.; Ali, M.A. Exploring the Physicochemical Profile and the Binding Patterns of Selected Novel Anticancer Himalayan Plant Derived Active Compounds with Macromolecular Targets. Inform. Med. Unlocked 2016, 5, 1–14. [Google Scholar] [CrossRef]
- Nunn, A.V.W.; Guy, G.W.; Bell, J.D. Informing the Cannabis Conjecture: From Life’s Beginnings to Mitochondria, Membranes and the Electrome—A Review. Int. J. Mol. Sci. 2023, 24, 13070. [Google Scholar] [CrossRef]
- Hadacek, F.; Bachmann, G.; Engelmeier, D.; Chobot, V. Hormesis and a chemical raison d’ětre for secondary plant metabolites. Dose-Response 2011, 9, 79–116. [Google Scholar] [CrossRef]
- Wang, X.; Auwerx, J. Systems phytohormone responses to mitochondrial proteotoxic stress. Mol. Cell 2017, 68, 540–551. [Google Scholar] [CrossRef]
- Sunil, B.; Talla, S.K.; Aswani, V.; Raghavendra, A.S. Optimization of photosynthesis by multiple metabolic pathways involving interorganelle interactions: Resource sharing and ROS maintenance as the bases. Photosynth. Res. 2013, 117, 61–71. [Google Scholar] [CrossRef]
- Janecki, M.; Graczyk, M.; Lewandowska, A.A.; Pawlak, Ł. Anti-Inflammatory and Antiviral Effects of Cannabinoids in Inhibiting and Preventing SARS-CoV-2 Infection. Int. J. Mol. Sci. 2022, 23, 4170. [Google Scholar] [CrossRef]
- Baik, A.H.; Haribowo, A.G.; Chen, X.; Queliconi, B.B.; Barrios, A.M.; Garg, A.; Maishan, M.; Campos, A.R.; Matthay, M.A.; Jain, I.H. Oxygen toxicity causes cyclic damage by destabilizing specific Fe-S cluster-containing protein complexes. Mol. Cell 2023, 83, 942–960.e9. [Google Scholar] [CrossRef] [PubMed]
- Vernis, L.; El Banna, N.; Baïlle, D.; Hatem, E.; Heneman, A.; Huang, M.E. Fe-S Clusters Emerging as Targets of Therapeutic Drugs. Oxidative Med. Cell. Longev. 2017, 2017, 3647657. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Imlay, J.A. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 2007, 282, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Justino, M.C.; Almeida, C.C.; Teixeira, M.; Saraiva, L.M. Escherichia coli Di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulfur clusters. J. Biol. Chem. 2007, 282, 10352–10359. [Google Scholar] [CrossRef]
- Bulteau, A.L.; O’Neill, H.A.; Kennedy, M.C.; Ikeda-Saito, M.; Isaya, G.; Szweda, L.I. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 2004, 305, 242–245. [Google Scholar] [CrossRef]
- Borgelt, L.M.; Franson, K.L.; Nussbaum, A.M.; Wang, G.S. The pharmacologic and clinical effects of medical cannabis. Pharmacotherapy 2013, 33, 195–209. [Google Scholar] [CrossRef]
- Okwu, D.E.; Nnamdi, F.U. Cannabinoid Dronabinol alkaloid with antimicrobial activity from Cassia alata Linn. Der Chem. Sin. 2011, 2, 247–254. [Google Scholar]
- Smetanová, L.; Stĕtinová, V.; Svoboda, Z.; Kvetina, J. Caco-2 cells, biopharmaceutics classification system (BCS) and biowaiver. Acta Medica 2011, 54, 3–8. [Google Scholar]
- Stahl, V.; Vasudevan, K. Comparison of Efficacy of Cannabinoids versus Commercial Oral Care Products in Reducing Bacterial Content from Dental Plaque: A Preliminary Observation. Cureus 2020, 12, e6809. [Google Scholar] [CrossRef]
- Seephonkai, P.; Popescu, R.; Zehl, M.; Krupitza, G.; Urban, E.; Kopp, B. Ferruginenes A–C from rhododendron ferrugineum and their cytotoxic evaluation. J. Nat. Prod. 2011, 74, 712–717. [Google Scholar] [CrossRef]
- Davis, P.J.; Bosenberg, A.; Davidson, A.; Jimenez, N.; Kharasch, E.; Lynn, A.M.; Tofovic, S.P.; Woelfe, S. CHAPTER 7—Pharmacology of Pediatric Anesthesia. In Smith’s Anesthesia for Infants and Children, 8th ed.; Davis, P.J., Cladis, F.P., Motoyama, E.K., Eds.; Mosby: Philadelphia, PA, USA, 2011; pp. 179–261. [Google Scholar] [CrossRef]
- Daneman, R.; Prat, A. The blood-brain barrier. Cold Spring Harb. Perspect. Biol. 2015, 7, a020412. [Google Scholar] [CrossRef] [PubMed]
- Yee, S. In vitro permeability across Caco-2 cells (colonic) can predict in vivo (small intestinal) absorption in man–fact or myth. Pharm. Res. 1997, 14, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Yamazaki, M. Role of P-glycoprotein in pharmacokinetics: Clinical implications. Clin. Pharmacokinet. 2003, 42, 59–98. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, P.; Nagini, S. Cytochrome P450 Structure, Function and Clinical Significance: A Review. Curr. Drug Targets. 2018, 19, 38–54. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, J.; Li, M.; Zhang, Y.; Jiang, B.; Zhao, X.; Huai, C.; Shen, L.; Zhang, N.; He, L.; et al. Cytochrome P450 Enzymes and Drug Metabolism in Humans. Int. J. Mol. Sci. 2021, 22, 12808. [Google Scholar] [CrossRef]
- Föllmann, W.; Degen, G.; Oesch, F.; Hengstler, J.G. Ames Test. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 104–107. [Google Scholar] [CrossRef]
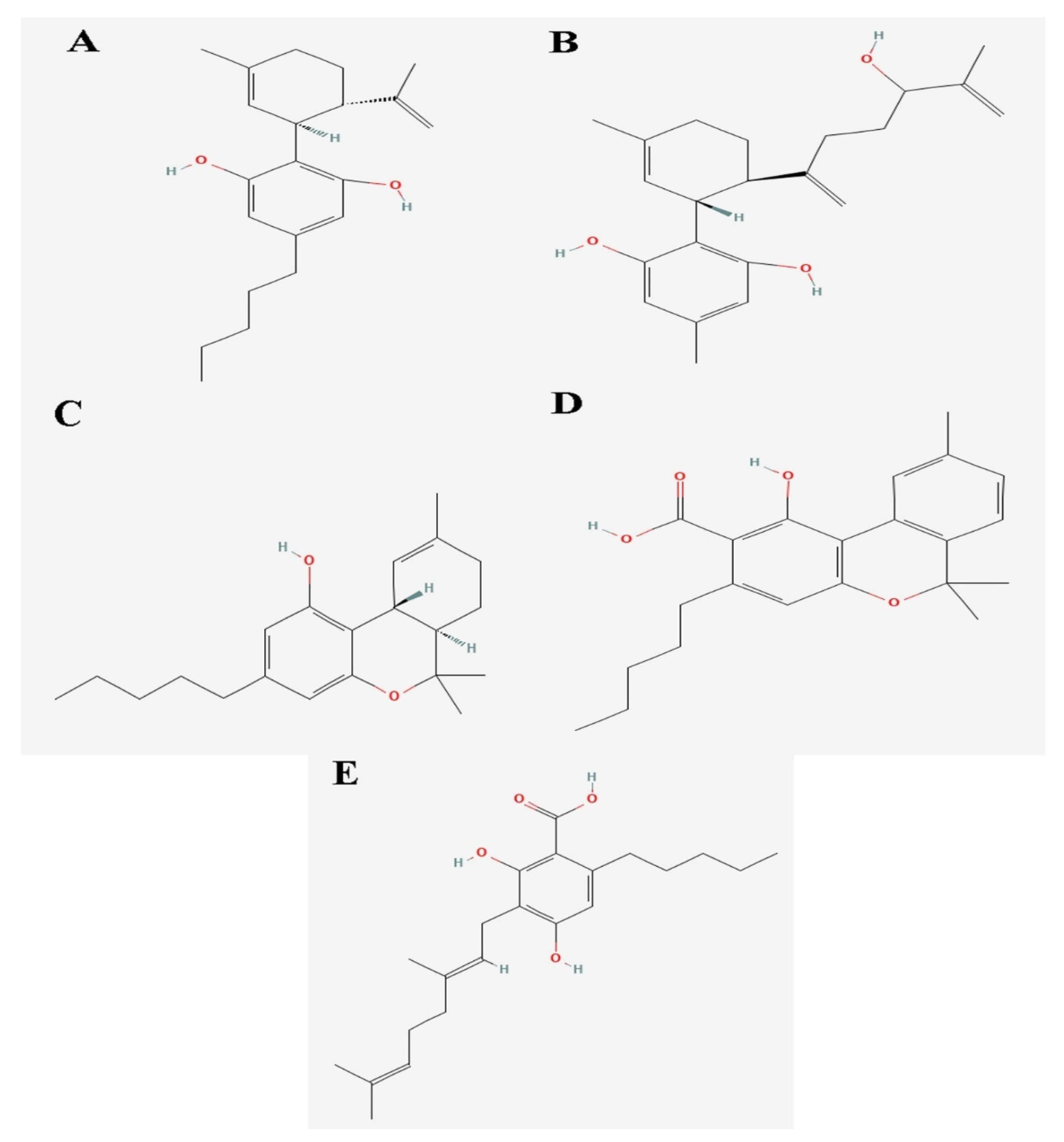
| Cannabinoids | Pubhem ID | Chemical Formula | Weight (g/mol) |
|---|---|---|---|
| Cannabidiol | 644019 | C21H30O2 | 314.46 |
| 7-Hydroxycannabidiol | 11301963 | C21H30O3 | 330.46 |
| Cannabidivarin | 11601669 | C19H26O2 | 286.41 |
| Cannabidiolic acid | 160570 | C22H30O4 | 358.4 |
| Tetrahydrocannabivarin (THCV) | 93147 | C19H26O2 | 286.41 |
| Tetrahydrocannabivarinic acid (THCVA) | 59444416 | C20H26O4 | 330.42 |
| Cannabinolic acid A (CBNA) | 3081990 | C22H26O4 | 354.44 |
| Cannabigerol (CBG) | 5315659 | C21H32O2 | 316.48 |
| Cannabigerolic acid (CBGA) | 6449999 | C22H32O4 | 360.49 |
| Searched Compound | Identified Compound | NPASS ID | PubChem ID | Chemical Formulae | Molecular Weight |
|---|---|---|---|---|---|
| Tetrahydrocannabivarin (THCV) | Dronabinol | NPC96940 | 16078 | C21H30O2 | 314.22 |
| Cannabidiol | 1,3-Benzenediol, 2-[3-methyl-6-(1-methylethenyl)-2-cyclohexen-1-yl]-5-pentyl-, (1R-trans)- | NPC186385 | 36688143 | C21H30O2 | 314.22 |
| 7-Hydroxycannabidiol | Ferruginene C | NPC319803 | 52951888 | C22H30O3 | 342.22 |
| Tetrahydrocannabivarinic acid (THCVA) | Δ9-Tetrahydrocannabinolic Acid A | NPC150928 | 98523 | C22H30O4 | 358.21 |
| Cannabigerol (CBG) | Grifolin | NPC12640 | 5372312 | C22H32O2 | 328.4 |
| Sesquicannabigerol | NPC99836 | 54669855 | C26H40O2 | 384.3 | |
| Cannabigerolic acid (CBGA) | Grifolic Acid | NPC180261 | 9976563 | C23H32O4 | 372.23 |
| Cannabidivarin | 1,3-Benzenediol, 2-[3-methyl-6-(1-methylethenyl)-2-cyclohexen-1-yl]-5-pentyl-, (1R-trans)- | NPC186385 | 36688143 | C21H30O2 | 314.22 |
| Cannabidiol | NPC299568 | 644019 | C21H30O2 | 314.46 |
| Compounds | Binding Affinity (kcal/mol) |
|---|---|
| Tetrahydrocannabivarin (THCV) | −7.9 |
| Cannabidiolic acid | −7.8 |
| Cannabidiol | −7.5 |
| Cannabinolic acid A (CBNA) | −8.1 |
| Grifolin | −7.7 |
| Ferruginene C | −8.3 |
| 1,3-Benzenediol, 2-[3-methyl-6-(1-methylethenyl)-2-cyclohexen-1-yl]-5-pentyl-, (1R-trans)- | −8.4 |
| Sesquicannabigerol | −6.6 |
| Grifolic acid | −7.9 |
| delta(9)-Tetrahydrocannabinolic acid | −7.6 |
| Dronabinol | −8.1 |
| Cannabigerol (CBG) | −6.8 |
| Cannabigerolic acid (CBGA) | −8.0 |
| 7-Hydroxycannabidiol | −7.4 |
| Tetrahydrocannabivarinic acid (THCVA) | −7.9 |
| Cannabidivarin | −7.8 |
| Cannabinoids | Molecular Weight g/mol | Lipinski Rules of 5 (ROF) | Chemical Formula | Log Po/w (WLOGP) | Log S (SILI- COS-IT) | BBB Permeation | Gastrointestinal (GI) Absorption | PGP Substrate | Suitable for Oral Bioavailability |
|---|---|---|---|---|---|---|---|---|---|
| Cannabinolic acid A (CBNA) | 354.44 | Yes; 0 violation | C22H26O4 | 5.32 | −6.84 | No | High | No | No. High lipophilicity and poorly soluble |
| Cannabigerolic acid (CBGA) | 360.49 | Yes; 1 violation: MLOGP > 4.15 | C22H32O4 | 5.76 | −5.14 | No | High | No | No. High lipophilicity, too flexible and moderately soluble |
| Dronabinol | 314.22 | Yes; 1 violation: MLOGP > 4.15 | C21H30O2 | 5.74 | −5.93 | Yes | High | No | No. High Lipophilicity |
| 1,3-Benzenediol, 2-[3-methyl-6-(1-methylethenyl)-2-cyclohexen-1-yl]-5-pentyl-, (1R-trans)- | 314.22 | Yes; 1 violation: MLOGP > 4.15 | C21H30O2 | 5.85 | −5.41 | Yes | High | No | No. High Lipophilicity |
| Ferruginene C | 342.22 | Yes; 0 violation | C22H30O3 | 5.12 | −4.51 | Yes | High | No | Yes. Moderately Soluble |
| Antibiotic | Molecular Weight g/mol | Lipinski Rules of 5 (ROF) | Chemical Formula | Log Po/w (WLOGP) | Log S (SILI- COS-IT) | BBB Permeation | Gastrointestinal (GI) Absorption | PGP Substrate | Suitable for Oral Bioavailability |
|---|---|---|---|---|---|---|---|---|---|
| Azithromycin | 748.98 | No; 2 violations: MW > 500, NorO > 10 | C38H72N2O12 | 1.52 | −2.22 | No | Yes | No | No. High molecular weight, high polarity and insoluble |
| Cefixime | 453.45 | Yes; 1 violation: NorO > 10 | C16H15N5O7S2 | −0.92 | −0.75 | No | No | No | No. Too polar |
| Ceftriaxone | 554.58 | No; 2 violations: MW > 500, NorO > 10 | C18H18N8O7S3 | −1.98 | −2.08 | No | No | No | No, high molecular weight and high polarity |
| Ciprofloxacin | 331.34 | Yes; 0 violation | C17H18FN3O3 | 1.18 | −3.5 | No | High | Yes | Yes. Highly Soluble |
| Antibiotic | Azithromycin | Cefixime | Ceftriaxone | Ciprofloxacin | |
|---|---|---|---|---|---|
| Absorption | Water solubility (log mol/L) | −3.144 | −2.73 | −2.843 | −2.692 |
| Caco2 permeability (log Papp in 10-6 cm/s) | 0.119 | −0.392 | −0.457 | 0.675 | |
| Intestinal absorption (human) (% Absorbed) | 0.119 | 14.687 | 12.93 | 98.704 | |
| Skin Permeability (log Kp) | −2.735 | −2.735 | −2.735 | −2.735 | |
| P-glycoprotein substrate | Yes | Yes | Yes | Yes | |
| P-glycoprotein I inhibitor | Yes | No | No | No | |
| P-glycoprotein II inhibitor | Yes | No | No | No | |
| Distribution | VDss (human) (log L/kg) | 1.179 | −1.647 | −2.286 | −0.395 |
| Fraction unbound (human) (Fu) | 0.71 | 0.543 | 0.365 | 0.646 | |
| BBB permeability (log BB) | −1.74 | −1.575 | −2.092 | −0.555 | |
| CNS permeability (log PS) | −4.704 | −4.038 | −4.516 | −2.957 | |
| Metabolism | CYP2D6 substrate | No | No | No | No |
| CYP3A4 substrate | No | No | No | No | |
| CYP1A2 inhibitor | No | No | No | No | |
| CYP2C19 inhibitor | No | No | No | No | |
| CYP2C9 inhibitor | No | No | No | No | |
| CYP2D6 inhibitor | No | No | No | No | |
| CYP3A4 inhibitor | No | No | No | No | |
| Excretion | Total Clearance (log mL/min/kg) | −0.404 | 0.076 | −0.188 | 0.618 |
| Renal OCT2 substrate | No | No | No | No | |
| Toxicity | AMES toxicity | No | No | No | No |
| Max. tolerated dose (human) (log mg/kg/day) | 1.088 | 1.534 | 1.099 | 0.771 | |
| hERG I inhibitor | No | No | No | No | |
| hERG II inhibitor | No | No | No | No | |
| Oral Rat Acute Toxicity (LD50) (mol/kg) | 2.352 | 1.947 | 2.319 | 2.661 | |
| Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day) | 3.013 | 2.587 | 2.614 | 0.851 | |
| Hepatotoxicity | Yes | Yes | Yes | Yes | |
| Skin Sensitisation | No | No | No | No | |
| T.Pyriformis toxicity (log μg/L) | 0.285 | 0.285 | 0.285 | 0.851 | |
| Minnow toxicity (log mM) | 0.285 | 3.613 | 3.523 | 1.71 | |
| Organ Toxicity | Hepatotoxicity | Inactive | Inactive | Inactive | Inactive |
| Neurotoxicity | Active | Inactive | Inactive | Active | |
| Nephrotoxicity | Active | Active | Inactive | Active | |
| Mutagenicity | Inactive | Active | Inactive | Active | |
| Ligand | Cannabinolic Acid A (CBNA) | 1,3-Benzenediol, 2-[3-Methyl-6-(1-methylethenyl)-2-cyclohexen-1-yl]-5-pentyl-, (1R-Trans)- | Ferruginene C | Dronabinol | Cannabigerolic Acid (CBGA) | |
|---|---|---|---|---|---|---|
| Property | Model Name | Predicted Value | ||||
| Absorption | Water solubility (log mol/L) | −3.473 | −5.609 | −4.585 | −5.042 | −3.515 |
| Caco2 permeability (log Papp in 10−6 cm/s) | 0.664 | 1.227 | 1.411 | 1.539 | 0.588 | |
| Intestinal absorption (human) (% Absorbed) | 95.906 | 89.308 | 89.599 | 91.162 | 95.782 | |
| Skin Permeability (log Kp) | −2.735 | −2.784 | −2.855 | −2.669 | −2.735 | |
| P-glycoprotein substrate | Yes | Yes | Yes | No | Yes | |
| P-glycoprotein I inhibitor | No | No | No | Yes | No | |
| P-glycoprotein II inhibitor | No | Yes | No | No | Yes | |
| Distribution | VDss (human) (log L/kg) | −1.623 | 0.771 | 0.493 | 0.977 | −1.575 |
| Fraction unbound (human) (Fu) | 0 | 0 | 0.078 | 0.005 | 0.074 | |
| BBB permeability (log BB) | 0.176 | −0.074 | −0.345 | 0.489 | −0.911 | |
| CNS permeability (log PS) | −1.853 | −1.741 | −0.345 | −1.807 | −2.269 | |
| Metabolism | CYP2D6 substrate | No | No | No | No | No |
| CYP3A4 substrate | No | Yes | No | Yes | No | |
| CYP1A2 inhibitor | No | Yes | No | Yes | No | |
| CYP2C19 inhibitor | No | Yes | No | Yes | No | |
| CYP2C9 inhibitor | Yes | No | No | No | No | |
| CYP2D6 inhibitor | No | No | No | No | No | |
| CYP3A4 inhibitor | No | Yes | Yes | No | No | |
| Excretion | Total Clearance (log ml/min/kg) | 0.663 | 1.267 | 1.198 | 0.881 | 1.278 |
| Renal OCT2 substrate | No | No | No | No | No | |
| Toxicity | AMES toxicity | No | No | No | No | No |
| Max. tolerated dose (human) (log mg/kg/day) | 0.585 | −0.18 | −0.062 | −0.207 | −0.073 | |
| hERG I inhibitor | No | No | No | No | No | |
| hERG II inhibitor | No | No | No | Yes | No | |
| Oral Rat Acute Toxicity (LD50) (mol/kg) | 2.71 | 2.55 | 2.727 | 2.62 | 2.588 | |
| Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day) | 1.971 | 2.639 | 2.443 | 1.885 | 2.057 | |
| Hepatotoxicity | No | No | No | No | No | |
| Skin Sensitisation | No | Yes | No | No | No | |
| T.Pyriformis toxicity (log µg/L) | 0.285 | 1.998 | 1.774 | 1.919 | 0.285 | |
| Minnow toxicity (log mM) | −1.328 | −0.57 | −0.198 | −0.917 | −0.825 | |
| Organ Toxicity | Hepatotoxicity | Inactive | Inactive | Active | Inactive | Inactive |
| Neurotoxicity | Inactive | Inactive | Active | Inactive | Inactive | |
| Nephrotoxicity | Active | Inactive | Inactive | Inactive | Active | |
| Mutagenicity | Inactive | Inactive | Inactive | Inactive | Inactive | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohd Omeershffudin, U.N.; Zainal Abidin, Z.; Hein, Z.M.; Che Mohd Nassir, C.M.N.; Kottakal Cheriya, E.N.; Kumar, S.; Che Ramli, M.D. Investigating the Antimicrobial Efficacy of Cannabinoids and Their Derivatives Against Neisseria Gonorrhoeae by Computational Analysis. Biology 2025, 14, 1272. https://doi.org/10.3390/biology14091272
Mohd Omeershffudin UN, Zainal Abidin Z, Hein ZM, Che Mohd Nassir CMN, Kottakal Cheriya EN, Kumar S, Che Ramli MD. Investigating the Antimicrobial Efficacy of Cannabinoids and Their Derivatives Against Neisseria Gonorrhoeae by Computational Analysis. Biology. 2025; 14(9):1272. https://doi.org/10.3390/biology14091272
Chicago/Turabian StyleMohd Omeershffudin, Umairah Natasya, Zakirah Zainal Abidin, Zaw Myo Hein, Che Mohd Nasril Che Mohd Nassir, Ebrahim Nangarath Kottakal Cheriya, Suresh Kumar, and Muhammad Danial Che Ramli. 2025. "Investigating the Antimicrobial Efficacy of Cannabinoids and Their Derivatives Against Neisseria Gonorrhoeae by Computational Analysis" Biology 14, no. 9: 1272. https://doi.org/10.3390/biology14091272
APA StyleMohd Omeershffudin, U. N., Zainal Abidin, Z., Hein, Z. M., Che Mohd Nassir, C. M. N., Kottakal Cheriya, E. N., Kumar, S., & Che Ramli, M. D. (2025). Investigating the Antimicrobial Efficacy of Cannabinoids and Their Derivatives Against Neisseria Gonorrhoeae by Computational Analysis. Biology, 14(9), 1272. https://doi.org/10.3390/biology14091272








