Simple Summary
Durum wheat is an essential crop, particularly in regions with harsh climates, yet it faces threats from pathogens and environmental stressors that can reduce yield and quality. Understanding the genetic basis of stress resistance is a critical for developing more resilient varieties. Among defense-related genes, defensins play a crucial role in plant immunity by targeting fungal pathogens and enhancing stress tolerance. However, their role in durum wheat remains largely unexplored. In this study, we identified 28 defensin-related genes and analyzed their structure, evolutionary relationships, and predicted functions. These genes contain regulatory elements linked to stress responses and hormone signaling, suggesting their involvement in plant protection. Most defensin proteins were predicted to be secreted outside plant cells, where they may interact with pathogens. Computer-based modeling suggested that these proteins can bind to fungal membranes, which highlights their potential antimicrobial role in durum wheat. Further experimental analysis revealed that multiple defensin genes are highly expressed under stress conditions, underscoring their importance in protecting durum wheat from diseases and environmental challenges. These findings provide new insights into the natural defense mechanisms of durum wheat and open possibilities for developing stress-resistant varieties through breeding or biotechnology.
Abstract
Plant defensins (PDFs) are a group of cationic antimicrobial peptides that are distinguished by their unique tertiary structure and play significant roles in physiological metabolism, growth, and stress tolerance. Defensins are key components of plant innate immunity; they can target a wide variety of microorganisms. This study aimed to identify and investigate the role of Triticum durum PDFs (TdPDFs) in response to environmental stresses. Prior to this, in silico analyses of TdPDF genes were conducted to assess their chromosomal locations, conserved motifs, exon–intron distribution, and cis-regulatory elements in the promoter regions. Additionally, bioinformatic analyses were performed to characterize the structure of TdPDF proteins, evaluate their phylogenetic relationships, predict their subcellular localization, and estimate their physicochemical properties. Docking studies were conducted to assess the interactions between TdPDF proteins and the fungal plasma membrane. A total of 28 TdPDF genes were identified in durum wheat based on their conserved domain PF00304 (gamma-thionin). These genes are distributed across all chromosomes of the durum wheat genome, except for chromosomes 4A and 7A. Analysis of the promoters of these genes revealed numerous elements associated with development, hormone responsiveness, and environmental stress. The majority of TdPDF proteins were predicted to be located extracellular. In addition, TdPDF proteins were classified into three clusters based on sequence similarity. Phylogenetic analysis suggested that TdPDF proteins share ancestral similarities with the PDF sequences of other monocotyledonous species. Molecular docking studies revealed that TdPDF proteins interact with fungal plasma membranes, suggesting that they play a critical role in the resistance of plants to pathogen infections. Expression analysis underlined the crucial role of nine TdPDF genes in the defense responses of durum wheat against both pathogenic and environmental stressors. Overall, our findings underscore the potential of TdPDF genes in host-plant resistance and highlight opportunities for their application in crop improvement toward stress tolerance.
1. Introduction
Durum wheat (Triticum durum) is an allotetraploid species within the monocotyledonous Triticeae tribe, originating from the hybridization and subsequent chromosomal doubling of Triticum urartu and Aegilops speltoides Tausch [1].
Durum wheat (Triticum turgidum L. spp. durum) ranks among the world’s most widely cultivated crops [2]. However, in the Mediterranean region, its productivity is significantly constrained by environmental factors, particularly drought and salinity [3], as well as biotic stress from pathogenic fungi such as Fusarium oxysporum [4,5]. Under biotic stress triggered by fungi, bacteria, or viruses, plants initiate a complex molecular dialogue involving reactive oxygen species (ROS), salicylic acid (SA), jasmonic acid (JA), ethylene (ET), and nitric oxide [6]. These signaling molecules orchestrate defense responses, with SA primarily activated in response to biotrophic pathogen attacks [7] and ET modulating responses to necrotrophic pathogens and abiotic stresses [8]. The JA pathway is particularly active during herbivorous insect attacks [9,10]. The cross-talk between SA, JA, and ET synergistically enhances defensin gene expression and boosts plant immunity [7]. Plant defensins (PDFs), a key class of cysteine-rich antimicrobial peptides (AMPs), play a central role in innate plant defense [11]. Structurally, defensins adopt a conserved βαββ “knottin fold” stabilized by disulfide bonds, comprising one α-helix and three antiparallel β-sheets [12,13,14]. This structural arrangement provides the stability necessary for their effective functioning [14]. PDFs are predominantly located in the cell wall and extracellular space [15], which allows them to participate in defense responses against pathogens and pests [16]. Most plant PDFs have a biologically conserved domain known as the gamma-thonin domain (PF00304) [17]. PDF proteins serve numerous biological functions, including roles in plant development and seed growth [18,19,20]. They exhibit antifungal [21] and antibacterial activities [22], contribute to resistance against zinc [23], act as α-amylase inhibitors [24], and possess antioxidant properties with low cytotoxic effects [25]. In addition, several studies have shown that PDF proteins are responsible for stress tolerance to salinity, drought, and cold [23,26,27]. The first PDF gene was identified in seeds of monocot and dicot species [15]. Their diversification has evolved in response to environmental challenges and microbial threats [28]. The protein from chickpeas induces resistance against F. oxysporum f. sp. ciceris and R. bataticola [29]. The ZmD32 defensin protein showed a similar response against Candida albicans, C. auris, C. glabrata, C. krusei, C. parapsilosis, C. tropicalis, and F. graminearum [30]. Both γ1-zeathionins and γ2-zeathionins (PDC-1) act as sodium channel blockers [31,32]. In addition, the BcDef1, isolated from Brugmansia candida (Bc), exhibited antioxidant activity [25]. Several studies have illustrated that the constitutive expression of a radish defensin improved tomato resistance to Alternaria solani [33]. Moreover, certain PDFs demonstrate crucial activities, such as protease inhibitors [34] and anti-cancer agents [35].
Although PDFs have been studied in other species, their presence and function in durum wheat remain largely unexplored. In this context, advances in genomic technologies and databases offer new opportunities to identify novel genes, investigate their regulatory mechanisms, and explore potential interactions with other stress-response genes. This study aimed to leverage genomic data to identify and characterize PDF genes and proteins in durum wheat, including their chromosomal localization, conserved motifs, cis-regulatory elements, phylogenetic relationships, structural properties, and protein interactions. Additionally, it explored their potential roles and regulatory mechanisms under various biotic and abiotic stress conditions through bioinformatics analyses and qPCR-based expression profiling. This comprehensive analysis of TdPDFs across the T. durum genome aimed to enhance our understanding of plant disease and stress-resistance genes, ultimately supporting the development of improved wheat varieties.
2. Materials and Methods
2.1. Plant Material and Stress Treatments
The experiment was carried out using the widely cultivated Tunisian durum wheat variety ‘Karim’ (Triticum turgidum L. subsp. durum (Desf.) Husn.), with seeds sourced from Centre d’AppuiChebika-CRDA Kairouan, Tunisia. The seeds were sterilized in 70% ethanol for 1–2 min, followed by rinsing with sterile water, and then allowed to germinate in Petri dishes for 7 days to produce seedlings [36]. To examine the response of 9 TdPDF genes to stress (Table S1), ten-day-old seedlings grown hydroponically in a nutrient solution were subjected to different stress conditions, including salinity, osmotic, cold, abscisic acid (ABA), salicylic acid (SA), and methyl jasmonate (MeJA). Salt (150 mM NaCl) and osmotic stress (20% (w/v) PEG 6000) were applied as described by Ben Romdhane et al. [37]. For cold treatment, the seedlings were exposed to 4 °C. The plants were maintained in a controlled environment chamber (phytotron) with the following settings: temperature (25 ± 5 °C), light intensity (280 µmol m−2 s−1), photoperiod (16 h light/8 h dark), and relative humidity (60 ± 10%). The leaves were sprayed with the phytohormones ABA (100 µM), SA (100 µM), and MeJA (100 µM), while a control group received a water spray.
The F. graminearum strain was grown on potato dextrose agar (PDA) plates at 30 °C for seven days. Afterward, fifteen-day-old plants were inoculated with a freshly prepared spore suspension (2 × 107 spores/mL) in Hoagland’s solution. Following inoculation, plants were kept in high humidity at 25 °C under a 16 h photoperiod. Harvests were made at 1, 3, 24, and 48 h post-inoculation to evaluate infection development and the plant’s defense responses. To analyze tissue-specific gene expression, samples were collected from various plant parts (leaves, stems, roots, spikes, anthers, and developing seeds at 21 days post-anthesis) from greenhouse-grown plants. Each tissue was collected separately, frozen in liquid nitrogen immediately, and stored at −80 °C to preserve RNA integrity.
2.2. Identification and Chromosomal Mapping of TdPDF Genes in Durum Wheat
The TdPDF protein sequences were retrieved from the Ensembl Plants database (https://plants.ensembl.org/index.html; accessed on 22 January 2025), while the TaPDF protein sequences [17] were sourced from the Grain Genes database (https://wheat.pw.usda.gov/GG3; accessed on 22 January 2025) and served as query sequences for searching TdPDF proteins via the BLAST tool version 1.4.0, employing an expected E-value threshold of E−50. Then, the full sequences of TdPDF proteins were screened using SMART (https://smart.embl-heidelberg.de/, accessed on 22 January 2025) and INTER PRO to identify the PF00304 gamma-thionin domain. The chromosomal positions of all TdPDF genes were plotted using the PhenoGram Plot tool (https://visualization.ritchielab.org/phenograms/plot, accessed on 22 January 2025).
2.3. Characterization of TdPDF Proteins
The physicochemical properties of the TdPDF proteins, including the molecular weight (MW), instability index (II), isoelectric point (pI), and grand average of the hydropathicity (GRAVY) of TdPDF proteins, were determined by the ProtParam tool within the ExPASy bioinformatics web tool (https://web.expasy.org/protparam/, accessed on 22 January 2025, [38]). In addition, to identify the subcellular localization of the TdPDF proteins, the BUSCA web tool (https://busca.biocomp.unibo.it/, accessed on 22 January 2025) was employed using the “Eukarya—plant—16 compartments” taxonomic origin option [39]. The signal peptide of TdPDF was predicted using SignalP5.1 (https://www.cbs.dtu.dk/services/SignalP/, accessed on 22 January 2025, [40]) by selecting “Eukarya” as the organism group.
2.4. Phylogenetic Analysis
The protein sequences of PDFs from Arabidopsis thaliana (TAIR database) (https://www.Arabidopsis.org/index.jsp, accessed on 22 January 2025), Triticum aestivum, and Oryza sativa were retrieved from Ensembl Plants. Multiple alignments of the PDF sequences were performed using the Clustal W algorithm for protein sequences using MEGA11 [41]. Maximum likelihood phylogenetic analysis of the TdPDF sequences was conducted using 1000 bootstrap replicates in MEGA11, incorporating the PDF sequences of Arabidopsis thaliana, Oryza sativa, and Triticum aestivum as outgroups.
The resulting protein trees were visualized using the Interactive Tree of Life tool (iTOL, available at https://itol.embl.de/, accessed on 22 January 2025, [42]). Additionally, the conservation of the TdPDF motif was assessed using the MEME v5.4.1 tool and visualized alongside the trees (https://meme-suite.org, accessed on 22 January 2025, [43]).
2.5. Analyses of TdPDF Genes and Promoter Regions
The Plant Compara tool from the Ensembl Plants database was employed to identify homologs, paralogs, and orthologs for each gene. Additionally, duplication events impacting these genes were investigated using TBtools v1.095 software [44] to analyze the associated evolutionary pressures. To examine the exon–intron structures of TdPDF genes, the Gene Structure Display Server (GSDS) 2.0 (https://gsds.gao-lab.org, accessed on 22 January 2025) was employed [45].
To analyze the putative cis-acting regulatory elements in the promoter regions, the 2 kb sequences upstream of each gene, obtained from the Ensembl Plants database, were examined using the PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html, accessed on 22 January 2025) and New PLACE (https://www.dna.affrc.go.jp/PLACE/?action=newplace, accessed on 22 January 2025) databases.
2.6. Quantitative RT PCR Analyses
Total RNA was extracted from plant tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), followed by treatment with DNase I (MBI Fermentas, Hanover, MD, USA) at 37 °C for 15 min to eliminate residual genomic DNA. Two micrograms of RNA were reverse-transcribed into cDNA using M-MLV reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). The resulting cDNA was diluted 1:5 and subjected to amplification with gene-specific primers designed using Primer3 (https://primer3plus.com/cgi-bin/dev/primer3plus.cgi, accessed on 22 January 2025) and SYBR Green RT-PCR master mix (Roche Diagnostics, Mannheim, Germany). Quantitative RT-qPCR was performed using the LightCycler 480 system (Roche Diagnostics International Ltd., Rotkreuz, Switzerland) in triplicate, following the protocol described by Ben Saad et al. [46]. The thermal cycling conditions included an initial denaturation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 20 s, 60 °C for 30 s, and 72 °C for 1 min. To confirm primer specificity, a melting curve analysis was conducted after the 40 cycles.
The relative expression levels of the nine TdPDF genes were quantified using the 2−ΔΔCT method, where CT denotes the cycle threshold, as outlined by Livak and Schmittgen [47]. Expression levels were normalized to the CDC gene (Ta54227), a cell division control protein from the AAA-superfamily of ATPases, as reported by Giménez et al. [48]. Results represent the mean relative expression ratios obtained from three independent experiments, each with three biological replicates.
2.7. Docking Studies and Structural Modeling
The SWISS-MODEL server (https://swissmodel.expasy.org/interactive, accessed on 22 January 2025, [49]) was employed to predict the three-dimensional structures of TdPDFs. The full protein was modeled using Alpha Fold [50] generated templates due to the lack of experimental structures. Structures with additional domains compared to the other models were characterized using the pfam server [51]. The defensin domain was modeled using the crystal structures of the rice defensin OsAFP1 (PDB ID: 6lcq) and grapevine defensin VvK1 (PDB ID: 7c31) as templates. The resulting structures were visualized with Schrödinger Maestro (Academic version 13). Defensin domain homology structures were clustered based on TM scores using the TM-align algorithm [52]. Disulfide bond predictions were conducted using the DI pro tool (available at https://scratch.proteomics.ics.uci.edu/, accessed on 22 January 2025).
Molecular docking studies were conducted on a subset of modeled TdPDF proteins, using phosphatidylinositol 4,5-bisphosphate (PIP2) as the ligand, extracted from the crystal structure of the plant defensin NaD1 in complex with PIP2 (PDB ID: 4CQK). The docking analysis was performed using Auto Dock Vina [53]. The binding site was inferred from the experimental structure of NaD1 with PIP2 (4CQK). For TdPDF9, the docking grid was centered at (9.444, 19.556, 28.139) with dimensions of 28 × 34 × 26, while for TdPDF20, the grid was centered at (9.444, 20.528, 24.472) with dimensions of 22 × 26 × 26. In both cases, an energy range of 4 and an exhaustiveness value of 8 were applied. Visualization and analysis of 2D molecular interactions were carried out using Schrödinger Maestro (Academic version 13).
2.8. Assessments of Protein-Protein Interaction Network
Protein-protein interaction analysis of TdPDF proteins was performed using the STRING database (https://string-db.org/, accessed on 22 January 2025, [54]). The interaction networks were predicted based on the closest orthologs of TdPDF proteins in Triticum aestivum due to the lack of Triticum durum data in the database. The analysis was performed using the full STRING network type with a medium confidence threshold of 0.400 and a maximum of 10 interactions to identify and visualize both direct and indirect protein associations.
3. Results
3.1. Screening and Identification of TdPDF Genes in Durum Wheat
In the present study, we identified 28 PDF genes containing the conserved domain PF00304 (gamma-thionin). These genes were named according to their chromosomal locations (TdPDF1–28). TdPDF genes are distributed in all chromosomes in the durum wheat genome except for chromosomes 4A and 7A. Each chromosome harbors multiple TdPDF genes, as illustrated in Figure 1. Notably, our findings demonstrate that many TdPDF genes located on the same chromosome appear to be in close proximity to one another. Additional details regarding the positions and characteristic features of these. genes can be found in Table 1.
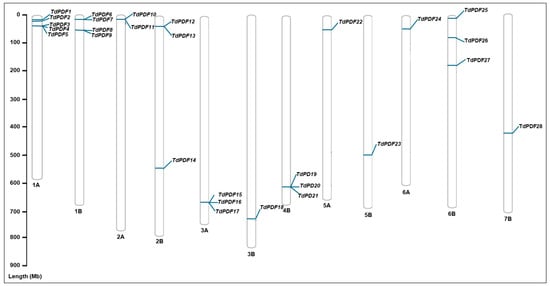
Figure 1.
Locations of the 28 TdPDF genes on durum wheat chromosomes. The scale on the left represents the length of the chromosomes. Mb = million base pair.

Table 1.
Overview of chromosomal position and characteristics of TdPDF genes in durum wheat.
3.2. TdPDF Protein Characterization and Subcellular Localization Prediction
The in silico analysis indicated that the length of TdPDF proteins ranges from 74 (TdPDF14and TdPDF9) to 123 (TdPDF17) amino acids. Their molecular weights vary from 7.942 kDa (TdPDF9) to 13.426 kDa (TdPDF17). The isoelectric points (pI) of these proteins also show considerable variation, with values between 5.55 (TdPDF27) and 9.79 (TdPDF4). Moreover, a value of pI below 9 (19 of the total proteins analyzed) indicates that these proteins are acidic, while PDF proteins with pI above 9 (9 of the total proteins) are basic (Table 2). Regarding protein stability, 15 out of the 28 TdPDF proteins were predicted to be stable in a test tube, as their instability index was below 40. Based on the grand average of hydropathicity (GRAVY) index, 18 of the proteins were classified as hydrophobic, while 10 were identified as hydrophilic. Most TdPDF proteins were predicted to be located in the extracellular space, with the exception of TdPDF17, which was localized in the mitochondrion (Table 2).

Table 2.
Physiochemical characteristics and predicted subcellular localization of TdPDF proteins.
3.3. Conserved Motif and Phylogenetic Analysis of TdPDF Proteins
To examine the evolutionary relationships between TdPDF proteins in durum wheat and other species, a phylogenetic tree was constructed with PDF protein sequences from T. aestivum (n = 73), T. turgidum (n = 28), A. thaliana (n = 15), and O. sativa (n = 11). The results suggest that TdPDF proteins share ancestral similarities with PDF sequences from other monocotyledonous species (Figure 2). The TdPDF proteins were classified into three clusters (C1, C2, and C3) based on genetic distance, and the three clusters were subdivided into six sub-clusters.
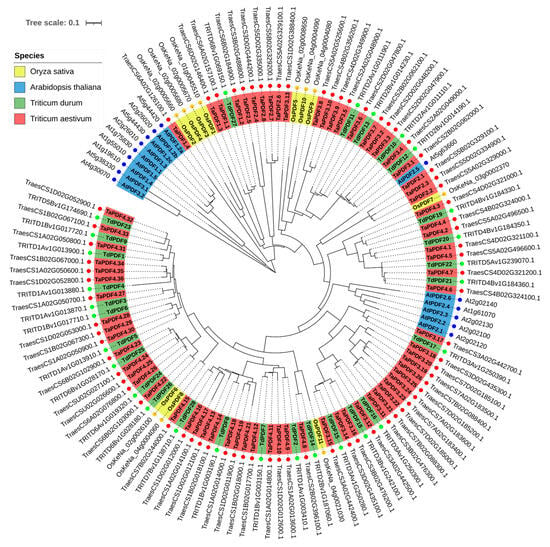
Figure 2.
Phylogenetic tree analysis of A. thaliana, T. aestivum, O. sativa, and T. turgidum PDF proteins. The four subgroups of PDFs are presented with different colors. Red squares refer to T. aestivum (TaPDF) proteins, red circles for A. thaliana (AtPDF) proteins, red stars for T. turgidum (TdPDF) proteins, and red triangles for O. sativa (OsPDF).
The analysis of the motif organization of TdPDF proteins revealed that they share Motif 2 (blue, Figure 3). Motif 3 was conserved across all TdPDF proteins, except for three sequences (TdPDF4, TdPDF16, and TdPDF17). In contrast, Motif 1 (red) was present in all sequences, except for one sequenceTdPDF23, while Motif 4 is conserved in 22 of the detected proteins.
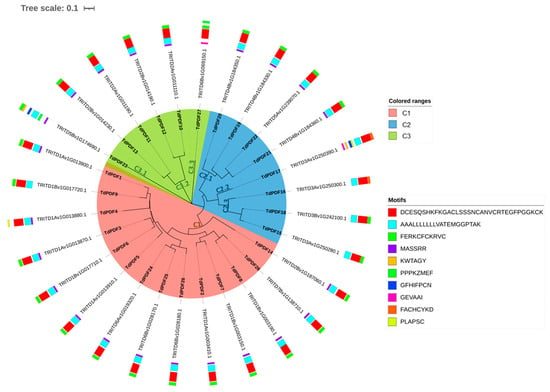
Figure 3.
Analysis of Triticum durum TdPDF protein structures. The circular tree was generated using TdPDF protein sequences.
3.4. TdPDF Gene Duplication and Ka/Ks Analysis
Gene duplication (GD) shaped the evolution of genomes in plants. This phenomenon might occur via several mechanisms, including whole genome duplication (WGD), single-gene duplications, and segmental duplications [55]. WGD serves as a significant factor in biological complexity, evolutionary innovation, and adaptation to diverse environmental conditions [56]. In T. durum, we identified eight pairs of TdPDF homologs that resulted from the whole-genome duplication (WGD) event, as well as 12 pairs of paralogs that most probably originated from segmental duplication (Table 3). The analysis of Ka/Ks ratios indicated that only the pair TdPDF7/TdPDF28 exhibited a value greater than 1 (Table 4), suggesting these genes have evolved during a period of positive selection, which accelerates evolution [57]. Conversely, all other gene pairs displayed Ka/Ks values below 1, with the highest ratios recorded at 0.7835 for TdPDF20/TdPDF21 (Table 4). These lower ratios imply that these gene pairs have evolved under strong purifying selection [57].

Table 3.
TdPDF gene duplication and their orthologs in T. aestivum.

Table 4.
Ka/Ks analysis for the duplicated TdPDF genes in durum wheat.
3.5. Gene Structure and Cis-Regulatory Element Analyses
The genomic information of TdPDF genes was used to map gene structure using GSDS2.0. The analysis revealed that 24 out of 28 TdPDF genes consist of two exons separated by an intron, with coding regions located at their 5′ ends (Figure 4). The remaining genes, TdPDF20, TdPDF21, TdPDF22, and TdPDF24, contain only an encoding sequence (exon). Additionally, all TdPDF genes contained the same number of exons as their Triticum aestivum orthologs, except for the orthologous pairs TdPDF21–TaPDF14, which have one and four exons, respectively, and TdPDF15–TaPDF10, which have two and three exons, respectively (Table 3).
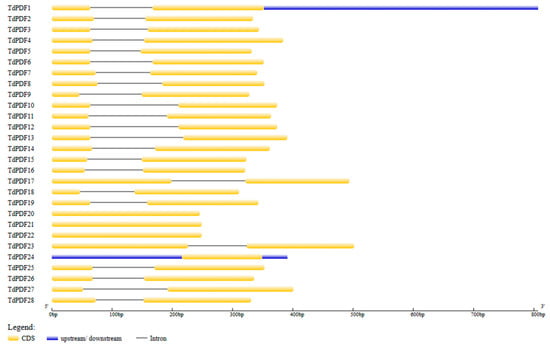
Figure 4.
Exon–intron structure analysis of TdPDF genes.
Cis-acting regulatory elements play a critical role in the regulation of gene expression under biotic and abiotic stresses [58] and the induction of related genes by supervising promoter efficiency [59]. The analysis of sequence 2 kb upstream of TdPDF genes revealed multiple cis-elements, which were classified into three categories. Regarding the category of hormone-responsive elements, abscisic acid (ABRE, ABRE3a, ABRE4a, and ABRE2) and methyl jasmonate (MeJA, CGTCA-motifs, and TGACG-motifs) cis-elements were frequently detected across TdPDF genes. Auxin (TGA element, Aux core), salicylic acid (TCA element), gibberellic acid (Pbox, Gare motif), and ethylene (ERE) cis-elements were also detected, although less frequently.
Additionally, cis-acting regulatory elements associated with environmental stress were identified, including those related to drought (MBS), low temperature (LTR), anaerobic induction (ARE), and biotic stress (box-S, W-box). Furthermore, developmental-related cis-elements were detected, such as those involved in meristem function (CAT-box and CCGTCC-box), endosperm regulation (GCN4 motif), and metabolic regulation (o2-site) (Figure 5).
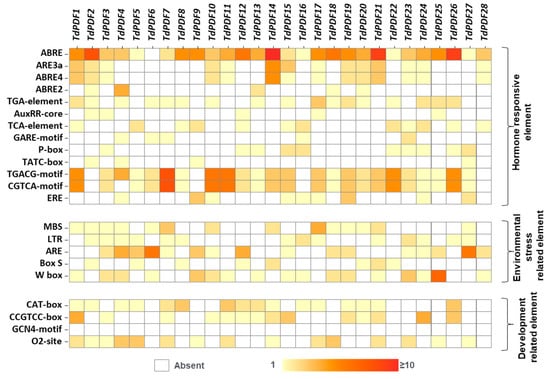
Figure 5.
Heatmap of the cis-acting elements in the promoter regions of TdPDF genes.
3.6. Docking Analysis and Structural Modeling
The structural modeling of the full TdPDF proteins revealed that the most common conformation consisted of an α-helix representing the signal peptide, which had a relatively consistent length (around 20–35 amino acids), followed by the defensin domain, which included one α-helix and three antiparallel β-sheets (Figure 6). This conformation was observed in 25 out of 28 models, with only three deviating from this structure.
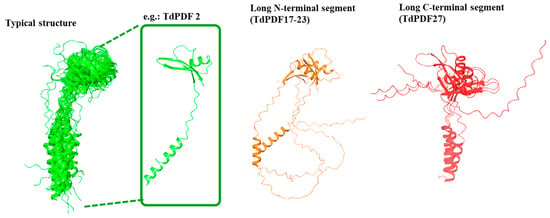
Figure 6.
Structural characterization of the full protein models of durum wheat (Triticum durum) plant defensins.
Specifically, for two models (TdPDF17 and TdPDF23), the N-terminal α-helix segment was long, exceeding 70 amino acids. Additionally, the TdPDF27 structure featured a long segment extending toward the C-terminal region of the protein, located after the β-sheets. Regarding the defensin domains, structural clustering based on TM score identified two major clusters, C1 and C2, along with a singleton, TdPDF4 (Figure 7A).
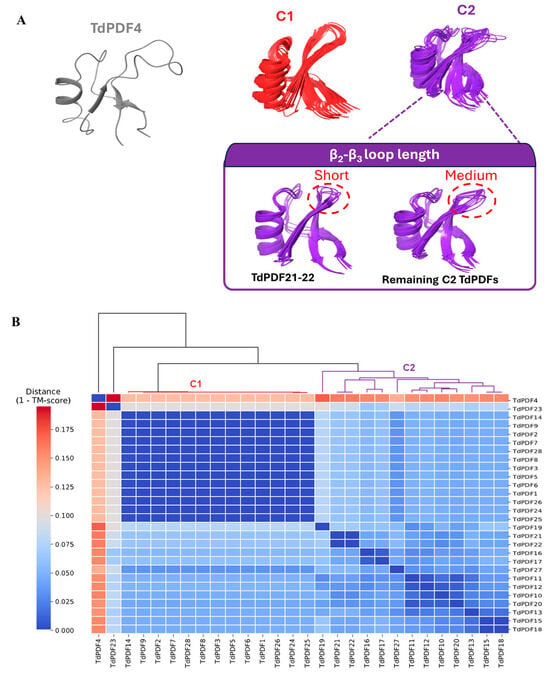
Figure 7.
Structural clustering (A) and 3D representation (B) of the defensin domain in modeled structures of durum wheat (Triticum durum) plant defensin proteins.
From a structural perspective, cluster C1 exhibited high structural similarity, characterized by a protruding β2-β3 loop due to relatively long β2 and β3 sheets. TdPDF models within cluster C2 were more structurally diverse, with the β2-β3 loop length varying considerably, ranging from short in TdPDF16 and TdPDF17 to medium (Figure 7B).
Disulfide bond prediction revealed that all TdPDF structures were stabilized via two to five disulfide bonds, with a median count of four bonds. The docking analysis was performed on TdPDF9 (representative of C1) and TdPDF15 (representative of C2) with PIP2. As shown in Figure 8, the docking results suggested that PIP2 can interact with TdPDF dimers, occupying a binding pocket situated between the two monomers. The interaction between PIP2 and TdPDF is primarily mediated through hydrogen bonds and pi-cation interactions between the phosphate groups and the inositol ring of PIP2 and residues located on the β2-β3 loop of the TdPDF protein.
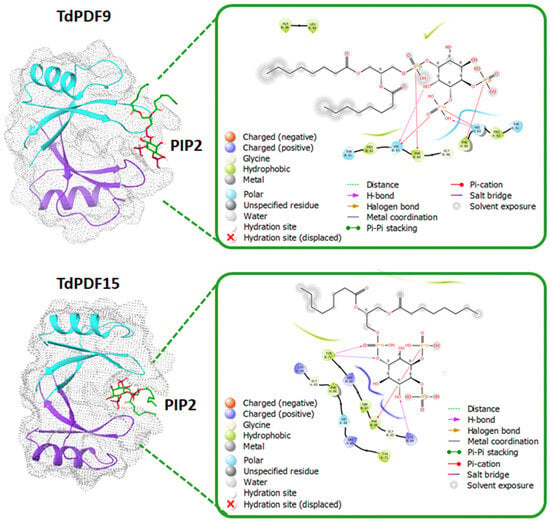
Figure 8.
Docking analysis of TdPDF9 (representative of subcluster C2b) and TdPDF20 (representative of subcluster C2c) with phosphatidylinositol 4,5-bisphosphate (PIP2). The estimated binding affinity was −4.8 kcal/mol for TdPDF9 and −5.4 kcal/mol for TdPDF15.
3.7. Protein-Protein Interaction Network Analysis
Protein-protein interaction analysis identified interaction networks for 28 TdPDF orthologs in Triticum aestivum. Among these, 27 TdPDFs shared a similar interaction network (Figure 9A). This network included two proteins involved in photosynthesis (photosystem II reaction center protein T and chlorophyll a–b binding protein), a protein essential for stabilizing the manganese cluster (oxygen-evolving enhancer protein 1), a member of the universal ribosomal protein uL5 family, and six uncharacterized proteins. In contrast, only TdPDF27 orthologs exhibited distinct interaction patterns (Figure 9B), engaging with three RNA-binding proteins containing the ASCH domain, five proteins possessing the ZnMc domain (zinc-dependent metalloprotease), and one protein with a kinase domain.
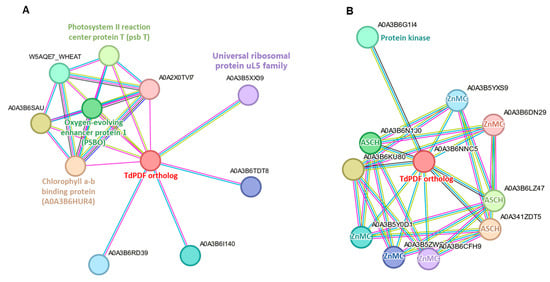
Figure 9.
Protein-protein interaction (PPI) analysis of TdPDF protein orthologs in Triticum aestivum. (A) Shared protein-protein interaction network of 27 TdPDFs orthologs. (B) Pro-tein-protein interaction network of TdPDF27 orthologs in Triticum aestivum.
3.8. TdPDF Genes Are Highly Expressed in Developing Seeds and Induced by Multiple Stresses and Phytohormones Treatments
To investigate the functions of the TdPDF gene family in Triticum durum, we analyzed the expression levels of these genes in different tissues—leaves, stems, roots, spikes, seeds, embryos, and anthers—using RT-qPCR (Figure 10A). The results revealed that TdPDF13 showed significantly higher expression in leaves compared to the other tissues, suggesting it may be involved in leaf-specific functions such as photosynthesis, leaf development, or response to environmental factors. Moreover, TdPDF27 has the highest expression in anthers, which indicates it could control pollen development. In contrast, TdPDF25 was highly expressed in embryos, suggesting its role in early developmental processes, including embryo formation and seed maturation. TdPDF25 and TdPDF23 were highly expressed in seeds (10.78- and 10.13-fold, respectively), suggesting that these genes may play a role in seed development. TdPDF15 was predominantly expressed in roots, which might indicate its potential role in root growth and root-related processes, such as nutrient uptake or stress responses in the root system. TdPDF11 was accumulated in stems, which suggests its involvement in stem development or vascular tissue function. Overall, the differential expression of TdPDF genes across various tissues and organs points to their diverse roles in plant growth, development, and reproduction (Figure 10B). It highlights how these genes may be adapted to perform specific functions in distinct parts of the plant. For the analysis of the expression levels of nine TdPDF genes in the defense responses of durum wheat against both pathogenic and environmental stressors, we computed the average expression level among three replicates with the standard error of the mean (SEM) to help compare the expression profiles.
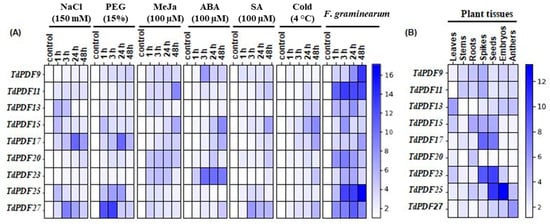
Figure 10.
Expression pattern of durum wheat PDF genes. (A) Heatmap showing the expression pattern of TdPDF genes in T. durum plants subjected to 150 mM NaCl, 15% PEG−6000, 100 μM ABA, 100 μM SA, 100 µM MeJa, and F. garminearum for 1, 3, 24, and 48 h. (B) Heatmap of the expression pattern of TdPDF genes in roots, stems, leaves, spikes, seeds, embryos, and anthers. The specific reference CDC gene was used as an internal control for qPCR normalization. The relative expression values were calculated using the 2−ΔΔCt method [47]. Three plants were used per treatment per replicate.
Next, to investigate the impact of phytohormones, as well as abiotic or biotic stress factors, on the transcript accumulation of the nine TdPDF genes, ten-day-old wheat seedlings were subjected to NaCl, PEG, MeJa, ABA, SA, or cold treatments and F. garminearum infection. Tissues were collected after 1, 3, 24, and 48 h. As illustrated in Figure 10A, the results of RT-qPCR showed that all these stresses induced the expression of most of the nine TdPDF genes. Salt stress enhanced the expression of TdPDF11 and TdPDF27 after 3 h and 24 h, respectively, of stress treatment. At this time of stress exposure, this induction reached the maximum, and it was about nine- and ten-fold, respectively, compared to the control condition. The highest increase in PEG-induced TdPDF expression was observed for TdPDF27, TdPDF17, and TdPDF25. Cold treatments significantly increased the expression level of TdPDF17 and TdPDF15 five- and six-fold, respectively, after 48 h. Under MeJa treatment, the expression levels of TdPDF11 and TdPDF15 increased approximately eight-and six-fold, respectively, after 48 h of stress application. The upregulation of TdPDF15 and TdPDF23 were observed after 48 h of ABA treatment, whereas it was only slightly induced by SA treatment.
In contrast, during the biotic stress treatment, all TdPDF genes were pronouncedly expressed in tissues inoculated by F. graminearum at 1, 3, 24, and 48 h (Figure 10B). These results confirm that the TdPDF genes may be associated with response to pathogen attack, consistent with the prediction of the PDF gene function. The results from this experiment demonstrated that analyzed TdPDF genes play distinctive roles in durum wheat responses to exogenous phytohormone treatments, which resemble phytohormonal cascades activated upon several abiotic or biotic stressors. Direct treatments with abiotic and biotic stress factors confirmed the involvement of TdPDF genes in plant reactions to a wide range of environmental stimuli.
4. Discussion
Defensins are proteins that may have either overlapping or distinct roles in plant defense and development. Although PDF genes have been identified in various plants, they have not been previously explored in durum wheat. Wheat defensins are particularly crucial due to wheat’s vulnerability to diseases like Fusarium head blight, which can severely impact yield and quality [60]. These defensins are distinct from those found in other crops, where the immune response to such pathogens is less pronounced [61,62]. Therefore, in this study, we identified 28 TdPDF genes in durum wheat using bioinformatics tools. This number is significantly lower than in common wheat (n = 73). However, the number of PDF genes varies considerably across species. This variation scan impacts their resistance to pathogens, stress adaptation, and overall resilience, contributing to evolutionary advantages and potential for crop improvement. One defensin (Tm-AMP-D1) was found in T. monococcum seeds, two in barley seeds (γ-hordothionin (γ-H) and ω-hordothionin (ω-H), six (Sd1–6) in sugarcane (Saccharum spp.), eight in T. kiharae, 11 in rice, and 15 in maize [17,29,63,64,65,66,67,68,69,70,71]. Model dicot A. thaliana possesses 15 PDF genes, while in legumes, the number usually ranges from several to over a dozen (for instance, from four in Vigna angularis to 22 in chickpea) [17,29,63,64,65,66,67,68,69,70,71]. Much higher numbers of sequences was found in the genomes of Brassica napus-37, and Vitis vinifera-79 [33,69].
In durum wheat, all 28 TdPDF proteins contain a gamma-thionin domain (PF00304), which is highly conserved across species and has been identified in PDF proteins from various plants [72]. Gene-family evolution is strongly influenced by tandem, segmental, and whole-genome duplications [73]. Our results indicate that both tandem and segmental duplications play a role in the evolution of TdPDF genes. Notably, many TdPDF genes located on the same chromosome are positioned near each other, suggesting that they likely originate from segmental duplication events and, therefore, may share similar functions or be involved in related biochemical pathways [55]. Duplications may also contribute to the evolution of distinct PDF paralogs with specialized roles in stress tolerance [74]. Such duplications could lead to functional differentiation, with some copies evolving to better defend against biotic stresses, such as fungal pathogens, while others may play a role in mitigating abiotic stresses like drought or salinity [75]. Recent studies have highlighted that gene duplication events allow for the accumulation of beneficial mutations, enhancing the adaptability of plants to changing environments [76]. Consistent with previous findings [56,57], our analysis confirmed that all TdPDF proteins contain a signal peptide. This signal peptide mediates the transport of defensins to the extracellular space, where they likely perform their main biological functions [58]. In silico analysis predicted extracellular localization for most TdPDF proteins, while one is localized in the mitochondrion. This variation in the localization of proteins in the cell may be related to factors such as protein-protein interaction. This result aligns with previous studies, which report extracellular localization for PDF proteins in rice [59], Arabidopsis thaliana [77], peanut [78], lentil [79], and Brassica napus [34]. Defensin genes are marked by a typical exon–intron–exon structure, with some exceptions previously reported [70,80]. All identified TdPDF genes contain a single intron with variable length, while the sizes of the two exons remain relatively conserved. These findings are consistent with other reports that indicated that most of the plant defensins feature a single intron [81,82]. Gene structure and physicochemical property analyses showed that PDF genes exhibit highly conserved architecture [83]. Molecular weight analysis classified durum wheat PDFs as a short peptide with a relatively low molecular weight. Their three-dimensional structure consists of one α-helix and three antiparallel β-sheets, stabilized by disulfide bonds. This is in agreement with previous studies [26,69,84]. The structural homologies observed in defensins highlight phylogenetic relationships between plant species, with PDFs serving as effective phylogenetic markers [20].
All TdPDF genes are implicated in the plant’s responses to a range of abiotic stresses, as revealed through promoter region analysis. Examining the promoter regions of genes is crucial for uncovering their potential functions [85]. The identification of GA-responsive cis-elements (such as P box, GARE motifs, and TATC box) suggests that these genes are likely regulated by GA and may play a role in GA signaling pathways and seed development. Similarly, the presence of drought-related cis-elements (DREs and MBS) indicates their potential involvement in drought resistance. Additionally, other cis-acting elements, including endosperm-related elements (AAGAA motifs and GCN4 motifs) and meristem-related motifs (CAT box and CCGTCC box), were found, implying that these genes may be important for plant development and tissue-specific expression. The abscisic acid-responsive cis-acting element ABRE and the core promoter elements TATA box and CAAT box in the promoter of the wheat PDF gene are all growth- and development-related elements. This suggests that TdPDF is associated with plant growth and development. A comparison of the cis-acting elements in the PDF genes of common and durum wheat revealed that some genes share similar elements in their promoter regions. The diversity of cis-regulatory elements in the promoter regions of PDF genes plays a fundamental role in the regulation of various pathways. In addition, identifying stress-responsive TdPDF genes offers significant potential for enhancing durum wheat productivity in climate-stressed regions. These genes are integral in plant defense mechanisms under abiotic stress conditions such as drought, heat, and salinity, which are exacerbated by climate change. Research has shown that PDF genes (chickpea defensin) help maintain plant stability by regulating stress-induced responses, reducing oxidative damage, and improving water-use efficiency [86]. Additionally, PtDefensin can increase disease resistance, as stressed plants are more susceptible to pathogens [87]. Understanding the molecular pathways governing these stress-responsive genes provides insights into how durum wheat can be adapted to thrive in harsher environments, enhancing yield stability. By integrating PDF genes into breeding programs, wheat varieties can be developed that are more resilient to climate change, reducing the need for external inputs such as irrigation and pesticides [88].
Plant defensins are generally recognized for their antimicrobial activity, primarily targeting fungi [89]. Defensins are known for their ability to bind and interact with phospholipids, key components of the fungal cell membrane, leading to pore formation and membrane disruption [29,90]. The primary mechanism of action for plant defensins is thought to involve the disruption of the pathogen’s cell membrane and wall integrity through the interactions between the positively charged regions of defensins and the negatively charged components of fungal membranes [91]. Additionally, defensins may bind to fungal wall components, inhibiting cell wall synthesis or enzyme activity, further impairing fungal growth [92].
PDFs disrupt microbial cell membranes by forming pores [93]. In contrast, chitinases target the cell walls of fungi by breaking down chitin and protease inhibitors block proteases, preventing pathogens from accessing plant proteins [94]. While thionins, another class of AMPs, are larger and more complex [95], TdPDFs are smaller and diffuse more rapidly through plant tissues, allowing them to quickly target pathogens and reduce pathogen load.
To verify the potential functions of TdPDF genes, we conducted an analysis of their expression profiles across various tissues and observed a wide range of expression patterns. Most TdPDF genes showed tissue-specific upregulation, including, specifically, TdPDF13 in the leaf, TdPDF15 in the root, TdPDF25 in the embryo, TdPDF27 in the anther, and TdPDF25 and TdPDF23 in the seeds. In Medicago sativa, defensin genes are expressed in leaves, flowers, and seeds but not in roots, whereas in Medicago truncatula, their expression is limited to seeds and absent in other organs, corroborating that defensin expression is tissue-specific [82]. In Populus trichocarpa, the PtDef gene is expressed in petioles, roots, stems, and leaves, with the highest expression observed in petioles, followed by roots, stems, and leaves [96]. ArabidopsisPDF2.2 is expressed in all organs of healthy plants except in the stems and seeds, and the expression of PDF2.2 in the leaves can change upon infection by the pathogen Alternaria brassicicola [97]. Plant defensins have also been found in leaves, floral organs, tubers, fruit, and roots, and their expression in various plant organs provides a first line of defense against pathogen attack [19,56,83,84,98,99,100]. According to the literature, seeds are the most abundant source of defensins [19]. Seed defensins suppress fungal growth, enhancing seedling survival rate [101]. Similarly, in durum wheat, TdPDFs are substantially abundant in seedlings and storage tissues, suggesting their role in plant growth and development.
Abiotic stressors like heat, drought, salinity, and nutrient deficiencies are major contributors to global crop losses [102]. Recently, the role of antimicrobial peptides (AMPs) in plant responses to abiotic stress is becoming increasingly acknowledged [103], along with the involvement of certain plant defensins [23,86,104]. Nevertheless, our understanding of the function of defensin’s abiotic stress responses remains incomplete. In the present study, we found that most TdPDF genes were regulated by abiotic stress, as well as the stress-related phytohormones ABA, MeJa, and SA. The diverse expression patterns of various TdPDF genes in response to phytohormone treatments suggest their involvement in a range of physiological reactions to stress factors. Our findings are in line with those of previous studies on other species [17,29], where heat and drought stress could trigger the expression of chickpea defensins in Arabidopsis thaliana [86], and the Dhn8 (dehydrin protein) from Hordeum vulgare L. was highly expressed in leaves under drought treatment [26]. In addition, the expression levels of OsDEF7 and OsDEF8 were affected by several stressful conditions [59]. Considering biotic stress response, TdPDF expression was upregulated during F. graminearum infection. Similarly, the expression levels of TaPDFs were differentially up-regulated by this pathogen [17]. Studies have demonstrated that PsDef1 levels rise significantly in Scots pine seedlings during germination and in response to pathogenic infection by Heterobasidion annosum [105]. Also, when Medicago truncatula is infected by Xanthomonas campestris, the expressions of the MtDef5 gene are upregulated, suggesting their antibacterial role [22]. Upregulation of PDFs under pathogen infection was also reported in pea, tobacco, Arabidopsis, and spruce [26]. Our results demonstrated that TdPDFs bind to phospholipids, suggesting that a similar mechanism may occur during contact with the fungal cell membrane. Such binding triggers pore formation and destabilizes membrane integrity, which may likely contribute to the inhibition of fungal growth, as suggested by Thevissen et al. [106] and observed in previous studies [29,90]. Additionally, research by Shahzad et al. [104] on representatives of Arabidposis genes revealed that apart from being involved in zinc tolerance, AhPDF1s in A. halerii exhibits antifungal properties. To validate the bioinformatics predictions made in the study, future studies should incorporate experimental approaches such as gene editing (CRISPR-Cas9) for knockout or overexpression studies in durum wheat, heterologous expression in Arabidopsis or E. coli, and integration with omics technologies like proteomics and metabolomics to validate gene function. Additionally, gene co-expression analysis and phenotypic assessment can further confirm the predicted roles of these genes.
5. Conclusions
In conclusion, this study provides the first comprehensive genome-wide analysis of the defensin gene family in durum wheat. A total of 28 TdPDF genes were identified and analyzed in terms of their structure, localization, phylogenetic relationships, conserved domain organization, protein interaction, docking analysis, and tissue-specific expression patterns. It allowed for comprehensive characterization of this gene family in durum wheat, as well as for gaining an insight into the structure, functions, and regulation of their encoded proteins. Due to a thorough examination of nine TdPDF gene expression profiles under abiotic and biotic stress conditions, novel information was gathered concerning the role of defensins in durum wheat’s response to environmental stimuli. These findings could serve as valuable resources for improving stress tolerance in wheat and other crop plants.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology14040404/s1, Table S1: Sequences of primers used in RT-qPCR analysis.
Author Contributions
Conceptualization, N.G., A.B.H. and R.B.S.; methodology, N.G., F.S., M.T.B., W.B.R., A.W., N.B., Y.C., A.B.H., M.K., M.I.K., S.G. and R.B.S.; software, N.G. and F.S.; validation, R.B.S., W.B.R. and A.B.H.; formal analysis, N.G., F.S., W.B.R., M.T.B., A.W., N.B., Y.C., A.B.H., M.K., S.G. and R.B.S.; investigation, N.G. and S.G.; resources, M.K. and A.B.H.; data curation, N.G., F.S., M.T.B., W.B.R., A.W., N.B., Y.C., A.B.H., M.K., M.I.K., S.G. and R.B.S.; writing—original draft preparation, N.G., W.B.R., A.W., M.T.B., N.B., Y.C., A.B.H., M.K., S.G. and R.B.S.; writing—review and editing; M.K., A.W., S.G. and A.B.H.; Supervision, R.B.S. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through the large research group projects of Narjes Baazaoui (Project under grant number (RGP. 2/88/45)). This research was partially funded by the Tunisian Ministry of Higher Education and Scientific Research (Program contract 2023–2027).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are contained within the article or Supplementary Materials.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through the large research group projects of Narjes Baazaoui (Project under grant number (RGP. 2/88/45).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Vendramin, V.; Ormanbekova, D.; Scalabrin, S.; Scaglione, D.; Maccaferri, M.; Martelli, P.; Salvi, S.; Jurman, I.; Casadio, R.; Cattonaro, F.; et al. Genomic Tools for Durum Wheat Breeding: De Novo Assembly of Svevo Transcriptome and SNP Discovery in Elite Germplasm. BMC Genom. 2019, 20, 278. [Google Scholar] [CrossRef]
- Marcotuli, I.; Colasuonno, P.; Hsieh, Y.S.Y.; Fincher, G.B.; Gadaleta, A. Non-Starch Polysaccharides in Durum Wheat: A Review. Int. J. Mol. Sci. 2020, 21, 2933. [Google Scholar] [CrossRef]
- Maggio, A.; De Pascale, S.; Fagnano, M.; Barbieri, G. Saline Agriculture in Mediterranean Environments. Ital. J. Agron. 2011, 6, e7. [Google Scholar] [CrossRef]
- Avery, S.V.; Singleton, I.; Magan, N.; Goldman, G.H. The Fungal Threat to Global Food Security. Fungal Biol. 2019, 123, 555–557. [Google Scholar] [CrossRef]
- Ntui, V.O.; Azadi, P.; Thirukkumaran, G.; Khan, R.S.; Chin, D.P.; Nakamura, I.; Mii, M. Increased Resistance to Fusarium Wilt in Transgenic Tobacco Lines Co-Expressing Chitinase and Wasabi Defensin Genes. Plant Pathol. 2011, 60, 221–231. [Google Scholar] [CrossRef]
- Pappas, M.; Baptista, P.; Broufas, G.; Dalakouras, A.; Djobbi, W.; Flors, V.; Msaad Guerfali, M.; Khayi, S.; Mentag, R.; Pastor, V.; et al. Biological and Molecular Control Tools in Plant Defense; Springer: Berlin/Heidelberg, Germany, 2020; pp. 3–43. ISBN 978-3-030-51034-3. [Google Scholar]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C.M. Networking by Small-Molecule Hormones in Plant Immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef]
- Wu, J.; Baldwin, I.T. New Insights into Plant Responses to the Attack from Insect Herbivores. Annu. Rev. Genet. 2010, 44, 1–24. [Google Scholar] [CrossRef]
- Thaler, J.S.; Farag, M.A.; Paré, P.W.; Dicke, M. Jasmonate-Deficient Plants Have Reduced Direct and Indirect Defences against Herbivores. Ecol. Lett. 2002, 5, 764–774. [Google Scholar] [CrossRef]
- Bodenhausen, N.; Reymond, P. Signaling Pathways Controlling Induced Resistance to Insect Herbivores in Arabidopsis. Mol. Plant-Microbe Interact. 2007, 20, 1406–1420. [Google Scholar] [CrossRef]
- Nawrot, R.; Barylski, J.; Nowicki, G.; Broniarczyk, J.; Buchwald, W.; Goździcka-Józefiak, A. Plant Antimicrobial Peptides. Folia Microbiol. 2014, 59, 181–196. [Google Scholar] [CrossRef]
- Caldwell, J.E.; Abildgaard, F.; Dzakula, Z.; Ming, D.; Hellekant, G.; Markley, J.L. Solution Structure of the Thermostable Sweet-Tasting Protein Brazzein. Nat. Struct. Biol. 1998, 5, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Gracy, J.; Le-Nguyen, D.; Gelly, J.-C.; Kaas, Q.; Heitz, A.; Chiche, L. KNOTTIN: The Knottin or Inhibitor Cystine Knot Scaffold in 2007. Nucleic Acids Res. 2008, 36, D314–D319. [Google Scholar] [CrossRef]
- Swathi Anuradha, T.; Divya, K.; Jami, S.K.; Kirti, P.B. Transgenic Tobacco and Peanut Plants Expressing a Mustard Defensin Show Resistance to Fungal Pathogens. Plant Cell Rep. 2008, 27, 1777–1786. [Google Scholar] [CrossRef]
- Terras, F.R.; Eggermont, K.; Kovaleva, V.; Raikhel, N.V.; Osborn, R.W.; Kester, A.; Rees, S.B.; Torrekens, S.; Van Leuven, F.; Vanderleyden, J. Small Cysteine-Rich Antifungal Proteins from Radish: Their Role in Host Defense. Plant Cell 1995, 7, 573–588. [Google Scholar]
- Jha, S.; Tank, H.; Prasad, B.; Chattoo, B. Expression of Dm-AMP1 in Rice Confers Resistance to Magnaporthe oryzae and Rhizoctonia Solani. Transgenic Res. 2008, 18, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wang, Y.; Tang, M.; Chen, W.; Chai, Y.; Wang, W. Bioinformatic Analysis of Wheat Defensin Gene Family and Function Verification of Candidate Genes. Front. Plant Sci. 2023, 14, 1279502. [Google Scholar] [CrossRef]
- Franco, O.L. Peptide Promiscuity: An Evolutionary Concept for Plant Defense. FEBS Lett. 2011, 585, 995–1000. [Google Scholar] [CrossRef]
- Stotz, H.U.; Thomson, J.; Wang, Y. Plant Defensins: Defense, Development and Application. Plant Signal. Behav. 2009, 4, 1010–1012. [Google Scholar] [CrossRef]
- van der Weerden, N.; Anderson, M. Plant Defensins: Common Fold, Multiple Functions. Fungal Biol. Rev. 2013, 26, 121–131. [Google Scholar] [CrossRef]
- Khan, R.S.; Darwish, N.A.; Khattak, B.; Ntui, V.O.; Kong, K.; Shimomae, K.; Nakamura, I.; Mii, M. Retransformation of Marker-Free Potato for Enhanced Resistance against Fungal Pathogens by Pyramiding Chitinase and Wasabi Defensin Genes. Mol. Biotechnol. 2014, 56, 814–823. [Google Scholar] [CrossRef]
- Velivelli, S.L.S.; Islam, K.T.; Hobson, E.; Shah, D.M. Modes of Action of a Bi-Domain Plant Defensin MtDef5 Against a Bacterial Pathogen Xanthomonas campestris. Front. Microbiol. 2018, 9, 934. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, M.; Sels, J.; Richard, O.; Czernic, P.; Loubet, S.; Jacquier, A.; François, I.E.J.A.; Cammue, B.P.A.; Lebrun, M.; Berthomieu, P.; et al. A Putative Novel Role for Plant Defensins: A Defensin from the Zinc Hyper-Accumulating Plant, Arabidopsis halleri, Confers Zinc Tolerance. Plant J. 2006, 47, 329–342. [Google Scholar] [CrossRef]
- Lin, K.-F.; Lee, T.-R.; Tsai, P.-H.; Hsu, M.-P.; Chen, C.-S.; Lyu, P.-C. Structure-Based Protein Engineering for Alpha-Amylase Inhibitory Activity of Plant Defensin. Proteins 2007, 68, 530–540. [Google Scholar] [CrossRef]
- Kaewklom, S.; Wongchai, M.; Petvises, S.; Hanpithakphong, W.; Aunpad, R. Structural and Biological Features of a Novel Plant Defensin from Brugmansia x Candida. PLoS ONE 2018, 13, e0201668. [Google Scholar] [CrossRef]
- Lay, F.T.; Anderson, M.A. Defensins--Components of the Innate Immune System in Plants. Curr. Protein Pept. Sci. 2005, 6, 85–101. [Google Scholar] [CrossRef]
- Parisi, K.; Doyle, S.R.; Lee, E.; Lowe, R.G.T.; van der Weerden, N.L.; Anderson, M.A.; Bleackley, M.R. Screening the Saccharomyces cerevisiae Nonessential Gene Deletion Library Reveals Diverse Mechanisms of Action for Antifungal Plant Defensins. Antimicrob. Agents Chemother. 2019, 63, e01097-19. [Google Scholar] [CrossRef] [PubMed]
- Farvardin, A.; González-Hernández, A.I.; Llorens, E.; Camañes, G.; Scalschi, L.; Vicedo, B. The Dual Role of Antimicrobial Proteins and Peptides: Exploring Their Direct Impact and Plant Defense-Enhancing Abilities. Plants 2024, 13, 2059. [Google Scholar] [CrossRef]
- Nitnavare, R.B.; Pothana, A.; Yeshvekar, R.K.; Bhattacharya, J.; Sapara, V.; Reddy, P.S.; Ramtirtha, Y.; Tarafdar, A.; Sharma, M.; Bhatnagar-Mathur, P. Chickpea Defensin Gene Family: Promising Candidates for Resistance Against Soil-Borne Chickpea Fungal Pathogens. J. Plant Growth Regul. 2022, 42, 6244–6260. [Google Scholar] [CrossRef]
- Kerenga, B.K.; McKenna, J.A.; Harvey, P.J.; Quimbar, P.; Garcia-Ceron, D.; Lay, F.T.; Phan, T.K.; Veneer, P.K.; Vasa, S.; Parisi, K.; et al. Salt-Tolerant Antifungal and Antibacterial Activities of the Corn Defensin ZmD32. Front. Microbiol. 2019, 10, 795. [Google Scholar] [CrossRef]
- Kant, P.; Liu, W.-Z.; Pauls, K.P. PDC1, a Corn Defensin Peptide Expressed in Escherichia coli and Pichia pastoris Inhibits Growth of Fusarium Graminearum. Peptides 2009, 30, 1593–1599. [Google Scholar] [CrossRef]
- Kushmerick, C.; de Souza Castro, M.; Santos Cruz, J.; Bloch, C.; Beirão, P.S. Functional and Structural Features of Gamma-Zeathionins, a New Class of Sodium Channel Blockers. FEBS Lett. 1998, 440, 302–306. [Google Scholar] [CrossRef]
- Parashina, E.; Shadenkov, A.; Lavrova, N.; Avetisov, V. The Use of the Defensive Peptide (Defensin) Gene from Radish Seeds in Order to Improve the Resistance of Tomatoes towards Diseases Caused by Fungi. Biotekhnologiya 1999, 15, 35–41. [Google Scholar]
- de Beer, A.; Vivier, M.A. Four Plant Defensins from an Indigenous South African Brassicaceae Species Display Divergent Activities against Two Test Pathogens despite High Sequence Similarity in the Encoding Genes. BMC Res. Notes 2011, 4, 459. [Google Scholar] [CrossRef]
- Lin, P.; Wong, J.H.; Ng, T.B. A Defensin with Highly Potent Antipathogenic Activities from the Seeds of Purple Pole Bean. Biosci. Rep. 2009, 30, 101–109. [Google Scholar] [CrossRef]
- Bouteraa, M.T.; Garzoli, S.; Romdhane, W.B.; Baazaoui, N.; Chouaibi, Y.; Hsouna, A.B.; Saad, R.B. Foliar Application of Lobularia Maritima-Derived Polysaccharides Modulates Chemical Composition and Enhances Salt Tolerance in Greenhouse-Cultivated Durum Wheat. J. Soil. Sci. Plant Nutr. 2025, 25, 589–602. [Google Scholar] [CrossRef]
- Ben Romdhane, W.; Ben Saad, R.; Meynard, D.; Zouari, N.; Tarroum, M.; Ali, A.; Droc, G.; Périn, C.; Morel, J.-B.; Fki, L.; et al. Expression of an A20/AN1 Stress-Associated Protein from Aeluropus littoralis in Rice Deregulates Stress-Related Genes. J. Plant Growth Regul. 2022, 41, 848–862. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Springer Protocols Handbooks; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-59259-890-8. [Google Scholar]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An Integrative Web Server to Predict Subcellular Localization of Proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Ben Saad, R.; Safi, H.; Ben Hsouna, A.; Brini, F.; Ben Romdhane, W. Functional Domain Analysis of LmSAP Protein Reveals the Crucial Role of the Zinc-Finger A20 Domain in Abiotic Stress Tolerance. Protoplasma 2019, 256, 1333–1344. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Giménez, M.J.; Pistón, F.; Atienza, S.G. Identification of Suitable Reference Genes for Normalization of qPCR Data in Comparative Transcriptomics Analyses in the Triticeae. Planta 2011, 233, 163–173. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-Align: A Protein Structure Alignment Algorithm Based on the TM-Score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010, 31, 455. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis Thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Lay, F.T.; Brugliera, F.; Anderson, M.A. Isolation and Properties of Floral Defensins from Ornamental Tobacco and Petunia. Plant Physiol. 2003, 131, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Oomen, R.J.F.J.; Séveno-Carpentier, E.; Ricodeau, N.; Bournaud, C.; Conéjéro, G.; Paris, N.; Berthomieu, P.; Marquès, L. Plant Defensin AhPDF1.1 Is Not Secreted in Leaves but It Accumulates in Intracellular Compartments. New Phytol. 2011, 192, 140–150. [Google Scholar] [CrossRef]
- Cools, T.L.; Struyfs, C.; Cammue, B.P.; Thevissen, K. Antifungal Plant Defensins: Increased Insight in Their Mode of Action as a Basis for Their Use to Combat Fungal Infections. Future Microbiol. 2017, 12, 441–454. [Google Scholar] [CrossRef]
- Weerawanich, K.; Webster, G.; Ma, J.K.-C.; Phoolcharoen, W.; Sirikantaramas, S. Gene Expression Analysis, Subcellular Localization, and in Planta Antimicrobial Activity of Rice (Oryza sativa L.) Defensin 7 and 8. Plant Physiol. Biochem. 2018, 124, 160–166. [Google Scholar] [CrossRef]
- Xu, M.; Wang, Q.; Wang, G.; Zhang, X.; Liu, H.; Jiang, C. Combatting Fusarium Head Blight: Advances in Molecular Interactions between Fusarium graminearum and Wheat. Phytopathol. Res. 2022, 4, 37. [Google Scholar] [CrossRef]
- Odintsova, T.I.; Slezina, M.P.; Istomina, E.A. Defensins of Grasses: A Systematic Review. Biomolecules 2020, 10, 1029. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, M.; Zhang, Z.; Ren, L.; Du, L.; Zhang, B.; Xu, H.; Xin, Z. Expression of a Radish Defensin in Transgenic Wheat Confers Increased Resistance to Fusarium graminearum and Rhizoctonia cerealis. Funct. Integr. Genom. 2011, 11, 63–70. [Google Scholar] [CrossRef]
- Odintsova, T.I.; Rogozhin, E.A.; Baranov, Y.; Musolyamov, A.K.; Yalpani, N.; Egorov, T.A.; Grishin, E.V. Seed Defensins of Barnyard Grass Echinochloa crusgalli (L.) Beauv. Biochimie 2008, 90, 1667–1673. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Jiang, N.; Meng, J.; Hou, X.; Yang, G.; Luan, Y. Identification and Characterization of Defensin Genes Conferring Phytophthora infestans Resistance in Tomato. Physiol. Mol. Plant Pathol. 2018, 103, 28–35. [Google Scholar] [CrossRef]
- Graham, M.; Silverstein, K.; Vandenbosch, K. Defensin-like Genes: Genomic Perspectives on a Diverse Superfamily in Plants. Crop Sci. 2008, 48, S-3–S-11. [Google Scholar] [CrossRef]
- Nanni, V.; Schumacher, J.; Giacomelli, L.; Brazzale, D.; Sbolci, L.; Moser, C.; Tudzynski, P.; Baraldi, E. VvAMP2, a Grapevine Flower-Specific Defensin Capable of Inhibiting Otrytis Cinerea Growth: Insights into Its Mode of Action. Plant Pathol. 2014, 63, 899–910. [Google Scholar] [CrossRef]
- De-Paula, V.S.; Razzera, G.; Medeiros, L.; Miyamoto, C.A.; Almeida, M.S.; Kurtenbach, E.; Almeida, F.C.L.; Valente, A.P. Evolutionary Relationship between Defensins in the Poaceae Family Strengthened by the Characterization of New Sugarcane Defensins. Plant Mol. Biol. 2008, 68, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Mendez, E.; Moreno, A.; Colilla, F.; Pelaez, F.; Limas, G.G.; Mendez, R.; Soriano, F.; Salinas, M.; de Haro, C. Primary Structure and Inhibition of Protein Synthesis in Eukaryotic Cell-Free System of a Novel Thionin, Gamma-Hordothionin, from Barley Endosperm. Eur. J. Biochem. 1990, 194, 533–539. [Google Scholar] [CrossRef]
- Liu, Y.; Hua, Y.-P.; Chen, H.; Zhou, T.; Yue, C.-P.; Huang, J.-Y. Genome-Scale Identification of Plant Defensin (PDF) Family Genes and Molecular Characterization of Their Responses to Diverse Nutrient Stresses in Allotetraploid Rapeseed. PeerJ 2021, 9, e12007. [Google Scholar] [CrossRef]
- Giacomelli, L.; Nanni, V.; Lenzi, L.; Zhuang, J.; Dalla Serra, M.; Banfield, M.J.; Town, C.D.; Silverstein, K.A.T.; Baraldi, E.; Moser, C. Identification and Characterization of the Defensin-like Gene Family of Grapevine. Mol. Plant Microbe Interact. 2012, 25, 1118–1131. [Google Scholar] [CrossRef]
- Padovan, L.; Segat, L.; Tossi, A.; Calsa, T.; Ederson, A.K.; Brandao, L.; Guimarães, R.L.; Pandolfi, V.; Pestana-Calsa, M.C.; Belarmino, L.C.; et al. Characterization of a New Defensin from Cowpea (Vigna unguiculata (L.) Walp.). Protein Pept. Lett. 2010, 17, 297–304. [Google Scholar] [CrossRef]
- Clavijo, B.J.; Venturini, L.; Schudoma, C.; Accinelli, G.G.; Kaithakottil, G.; Wright, J.; Borrill, P.; Kettleborough, G.; Heavens, D.; Chapman, H.; et al. An Improved Assembly and Annotation of the Allohexaploid Wheat Genome Identifies Complete Families of Agronomic Genes and Provides Genomic Evidence for Chromosomal Translocations. Genome Res. 2017, 27, 885–896. [Google Scholar] [CrossRef]
- Paterson, A.H.; Freeling, M.; Tang, H.; Wang, X. Insights from the Comparison of Plant Genome Sequences. Annu. Rev. Plant Biol. 2010, 61, 349–372. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Park, C. Adaptive evolution of β-defensin gene clusters by gene duplication and loss during environmental changes in mammals. J. Anim. Breed. Genom. 2022, 6, 143–154. [Google Scholar]
- Du, L.; Ma, Z.; Mao, H. Duplicate Genes Contribute to Variability in Abiotic Stress Resistance in Allopolyploid Wheat. Plants 2023, 12, 2465. [Google Scholar] [CrossRef]
- Kondrashov, F.A. Gene Duplication as a Mechanism of Genomic Adaptation to a Changing Environment. Proc. Biol. Sci. 2012, 279, 5048–5057. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.S.M.; Pires, Á.S.; Damaceno, N.B.; Rigueiras, P.O.; Maximiano, M.R.; Franco, O.L.; Porto, W.F. In Silico Characterization of Class II Plant Defensins from Arabidopsis thaliana. Phytochemistry 2020, 179, 112511. [Google Scholar] [CrossRef]
- Zhao, K.; Ren, R.; Ma, X.; Zhao, K.; Qu, C.; Cao, D.; Ma, Q.; Ma, Y.; Gong, F.; Li, Z.; et al. Genome-Wide Investigation of Defensin Genes in Peanut (Arachis hypogaea L.) Reveals AhDef2.2 Conferring Resistance to Bacterial Wilt. Crop J. 2022, 10, 809–819. [Google Scholar] [CrossRef]
- Mir Drikvand, R.; Sohrabi, S.M.; Samiei, K. Molecular Cloning and Characterization of Six Defensin Genes from Lentil Plant (Lens culinaris L.). 3 Biotech. 2019, 9, 104. [Google Scholar] [CrossRef]
- Houlné, G.; Meyer, B.; Schantz, R. Alteration of the Expression of a Plant Defensin Gene by Exon Shuffling in Bell Pepper (Capsicum annuum L.). Mol. Gen. Genet. 1998, 259, 504–510. [Google Scholar] [CrossRef]
- Spelbrink, R.G.; Dilmac, N.; Allen, A.; Smith, T.J.; Shah, D.M.; Hockerman, G.H. Differential Antifungal and Calcium Channel-Blocking Activity among Structurally Related Plant Defensins. Plant Physiol. 2004, 135, 2055–2067. [Google Scholar] [CrossRef]
- Hanks, J.N.; Snyder, A.K.; Graham, M.A.; Shah, R.K.; Blaylock, L.A.; Harrison, M.J.; Shah, D.M. Defensin Gene Family in Medicago Truncatula: Structure, Expression and Induction by Signal Molecules. Plant Mol. Biol. 2005, 58, 385–399. [Google Scholar] [CrossRef]
- Thomma, B.P.H.J.; Cammue, B.P.A.; Thevissen, K. Plant Defensins. Planta 2002, 216, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, K.A.T.; Graham, M.A.; Paape, T.D.; VandenBosch, K.A. Genome Organization of More than 300 Defensin-like Genes in Arabidopsis. Plant Physiol. 2005, 138, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Ben Saad, R.; Ben Romdhane, W.; Zouari, N.; Ben Hsouna, A.; Harbaoui, M.; Brini, F.; Ghneim-Herrera, T. Characterization of a Novel LmSAP Gene Promoter from Lobularia maritima: Tissue Specificity and Environmental Stress Responsiveness. PLoS ONE 2020, 15, e0236943. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Yusuf, M.A.; Yadav, P.; Narayan, S.; Kumar, M. Overexpression of Chickpea Defensin Gene Confers Tolerance to Water-Deficit Stress in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 290. [Google Scholar] [CrossRef]
- Wei, H.; Movahedi, A.; Xu, C.; Sun, W.; Wang, X.; Li, D.; Zhuge, Q. Overexpression of PtDefensin Enhances Resistance to Septotis populiperda in Transgenic Poplar. Plant Sci. 2020, 292, 110379. [Google Scholar] [CrossRef]
- Gao, L.; Kantar, M.B.; Moxley, D.; Ortiz-Barrientos, D.; Rieseberg, L.H. Crop Adaptation to Climate Change: An Evolutionary Perspective. Mol. Plant 2023, 16, 1518–1546. [Google Scholar] [CrossRef]
- Sathoff, A.E.; Velivelli, S.; Shah, D.M.; Samac, D.A. Plant Defensin Peptides Have Antifungal and Antibacterial Activity Against Human and Plant Pathogens. Phytopathology 2019, 109, 402–408. [Google Scholar] [CrossRef]
- Poon, I.K.; Baxter, A.A.; Lay, F.T.; Mills, G.D.; Adda, C.G.; Payne, J.A.; Phan, T.K.; Ryan, G.F.; White, J.A.; Veneer, P.K.; et al. Phosphoinositide-Mediated Oligomerization of a Defensin Induces Cell Lysis. Elife 2014, 3, e01808. [Google Scholar] [CrossRef]
- Vriens, K.; Cammue, B.P.A.; Thevissen, K. Antifungal Plant Defensins: Mechanisms of Action and Production. Molecules 2014, 19, 12280–12303. [Google Scholar] [CrossRef]
- Oeemig, J.S.; Lynggaard, C.; Knudsen, D.H.; Hansen, F.T.; Nørgaard, K.D.; Schneider, T.; Vad, B.S.; Sandvang, D.H.; Nielsen, L.A.; Neve, S.; et al. Eurocin, a New Fungal Defensin: Structure, Lipid Binding, and Its Mode of Action. J. Biol. Chem. 2012, 287, 42361–42372. [Google Scholar] [CrossRef]
- Tanvir, A. Antimicrobial Peptides and Their Interaction Cell Membrane and Microbial Surface Interactions. J. Med. Org. Chem. 2022, 5, 86–92. [Google Scholar]
- Bakhat, N.; Vielba-Fernández, A.; Padilla-Roji, I.; Martínez-Cruz, J.; Polonio, Á.; Fernández-Ortuño, D.; Pérez-García, A. Suppression of Chitin-Triggered Immunity by Plant Fungal Pathogens: A Case Study of the Cucurbit Powdery Mildew Fungus Podosphaera xanthii. J. Fungi 2023, 9, 771. [Google Scholar] [CrossRef] [PubMed]
- Odintsova, T.I.; Slezina, M.P.; Istomina, E.A. Plant thionins: Structure, biological functions and potential use in biotechnology. Vavilov J. Genet. Breed. 2018, 22, 667–675. [Google Scholar] [CrossRef]
- Wei, H.; Movahedi, A.; Xu, C.; Sun, W.; Wang, P.; Li, D.; Yin, T.; Zhuge, Q. Characterization, Expression Profiling, and Functional Analysis of PtDef, a Defensin-Encoding Gene from Populus trichocarpa. Front. Microbiol. 2020, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J.; Broekaert, W.F. Tissue-Specific Expression of Plant Defensin Genes PDF2.1 and PDF2.2 in Arabidopsis thaliana. Plant Physiol. Biochem. 1998, 36, 533–537. [Google Scholar] [CrossRef]
- Carvalho, A.d.O.; Gomes, V.M. Plant Defensins--Prospects for the Biological Functions and Biotechnological Properties. Peptides 2009, 30, 1007–1020. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, L.; Yang, C.-L.; Han, S.; Yang, S.; Liu, G.; Zeng, Q.; Liu, Y. Molecular Identification and Antifungal Activity of a Defensin (PaDef) from Spruce. J. Plant Growth Regul. 2021, 41, 494–506. [Google Scholar] [CrossRef]
- Domingo, G.; Locato, V.; Cimini, S.; Ciceri, L.; Marsoni, M.; De Gara, L.; Bracale, M.; Vannini, C. A Comprehensive Characterization and Expression Profiling of Defensin Family Peptides in Arabidopsis thaliana with a Focus on Their Abiotic Stress-Specific Transcriptional Modulation. Curr. Plant Biol. 2024, 39, 100376. [Google Scholar] [CrossRef]
- Osborn, R.W.; De Samblanx, G.W.; Thevissen, K.; Goderis, I.; Torrekens, S.; Van Leuven, F.; Attenborough, S.; Rees, S.B.; Broekaert, W.F. Isolation and Characterisation of Plant Defensins from Seeds of Asteraceae, Fabaceae, Hippocastanaceae and Saxifragaceae. FEBS Lett. 1995, 368, 257–262. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Modulation of Antioxidant Defense System Is Associated with Combined Drought and Heat Stress Tolerance in Citrus. Front. Plant Sci. 2017, 8, 953. [Google Scholar] [CrossRef]
- Kulaeva, O.; Kliukova, M.; Afonin, A.; Sulima, A.; Zhukov, V.; Tikhonovich, I. The Role of Plant Antimicrobial Peptides (AMPs) in Response to Biotic and Abiotic Environmental Factors. Biol. Commun. 2020, 65, 187–199. [Google Scholar] [CrossRef]
- Shahzad, Z.; Ranwez, V.; Fizames, C.; Marquès, L.; Le Martret, B.; Alassimone, J.; Godé, C.; Lacombe, E.; Castillo, T.; Saumitou-Laprade, P.; et al. Plant Defensin Type 1 (PDF1): Protein Promiscuity and Expression Variation within the Arabidopsis Genus Shed Light on Zinc Tolerance Acquisition in Arabidopsis Halleri. New Phytol. 2013, 200, 820–833. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, V. Recombinant Expression, Affinity Purification and Functional Characterization of Scots Pine Defensin 1. Appl. Microbiol. Biotechnol. 2011, 89, 1093–1101. [Google Scholar] [CrossRef]
- Thevissen, K.; de Mello Tavares, P.; Xu, D.; Blankenship, J.; Vandenbosch, D.; Idkowiak-Baldys, J.; Govaert, G.; Bink, A.; Rozental, S.; de Groot, P.W.J.; et al. The Plant Defensin RsAFP2 Induces Cell Wall Stress, Septin Mislocalization and Accumulation of Ceramides in Candida albicans. Mol. Microbiol. 2012, 84, 166–180. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).