Exploring Novel Therapeutic Targets in Breast Cancer via Comprehensive Omics Profiling and Experimental Verification
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Detection of DEGs
2.3. eQTL Analysis of Exposure Data
2.4. Ascertainment of Outcome Data
2.5. MR Analysis
2.6. GO/KEGG Enrichment Analysis
2.7. Immune Cell Analysis
2.8. Differential Analysis of the TCGA Database
2.9. Establishing Protein–Protein Interaction Networks and Nomogram Models
2.10. Gene Set Enrichment Analysis (GSEA) Enrichment Analysis
2.11. Cells and Culture Conditions
2.12. Cell Transfection
| ATOH8 siRNA | 5′-GGUGCCGUGCUACUCAUAUTT-3′ |
| DNase2 siRNA | 5′-CAAGAACCCUGGAACAGCAGCAUCA-3′ |
2.13. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
| ATOH8 forward: 5′-CAGGTGCCGTGCTACTCATA-3′; |
| ATOH8 reverse: 5′-AGTCACTCCTTGCGCTTCTT-3′; |
| DNase2 forward: 5′-TCGCCTTCCTGCTCTACAAT-3′; |
| DNase2 reverse: 5′-CCCATCTTCGAGAACTGAGC-3′; |
| β-Tubulin forward: 5′-CTCTGAAGCTGACCACACCA-3′; |
| β-Tubulin reverse: 5′-GCCAGGCATAAAGAAATGGA-3′. |
2.14. Cell Proliferation Assay
2.15. Cell Wound Healing Assay
2.16. Transwell Migration and Invasion Assays
2.17. Analysis of Drug Sensitivity in Risk Subtypes
2.18. Statistical Analysis
3. Results
3.1. Overview of the Four GEO Datasets
3.2. DEG Identification
3.3. MR Analysis
3.4. GO and KEGG Enrichment Analysis
3.5. Evaluation of Immune Cell Infiltration in Breast Cancer
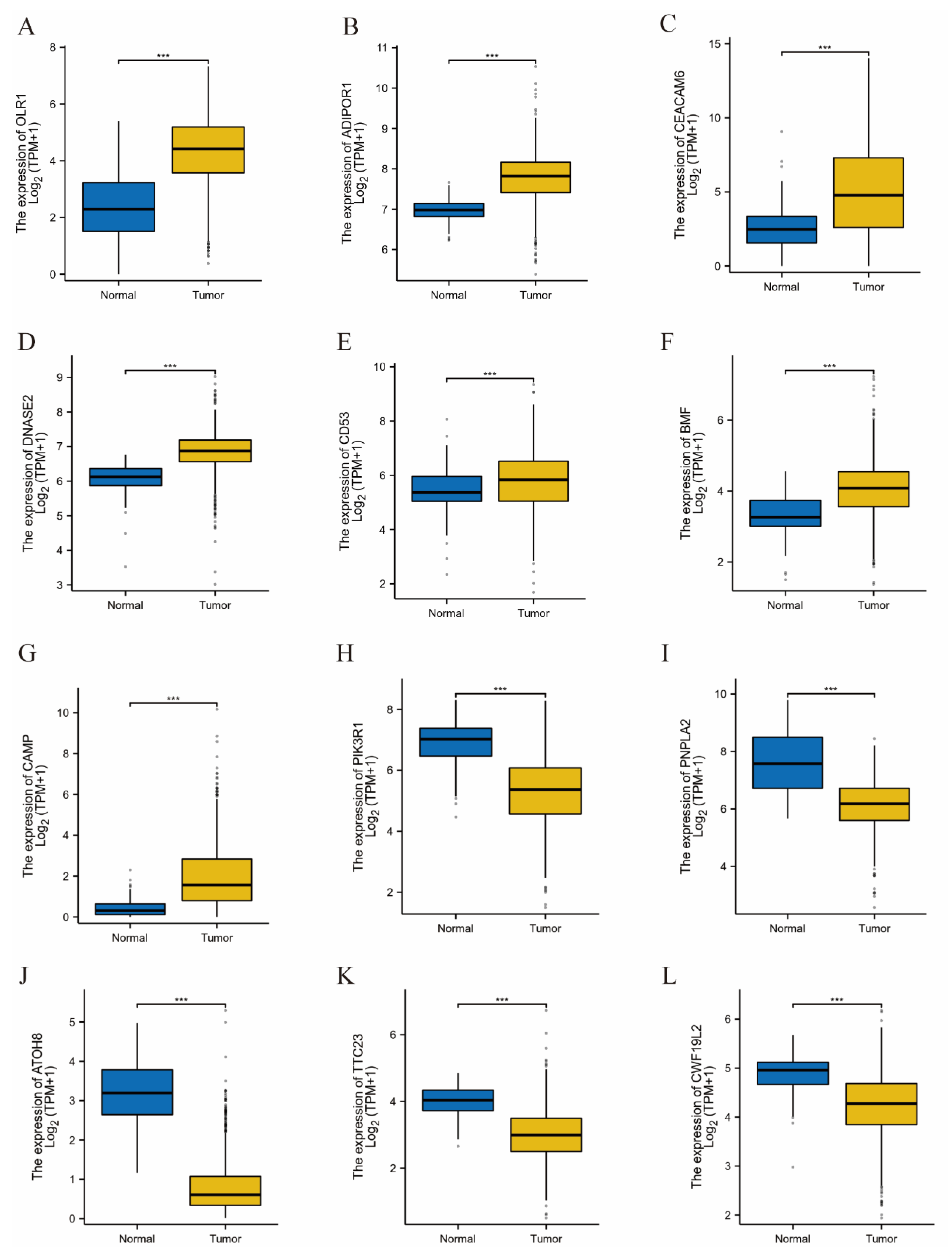
3.6. Validation of Results Using TCGA Data
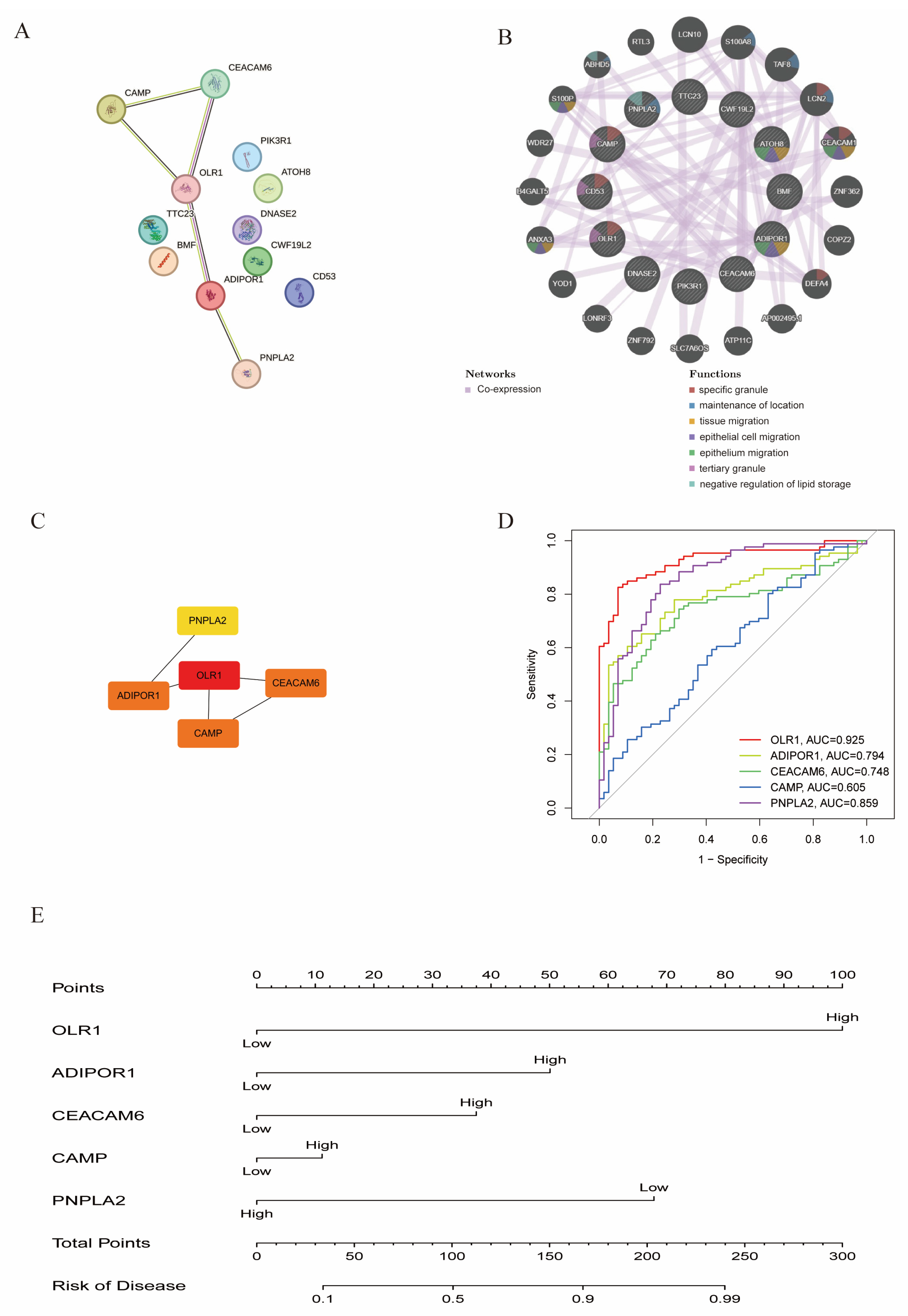
3.7. Construction of PPI Networks and Nomogram Model
3.8. In Vitro Functional Investigation into the Roles of DNASE2 and ATOH8 in Breast Cancer Development
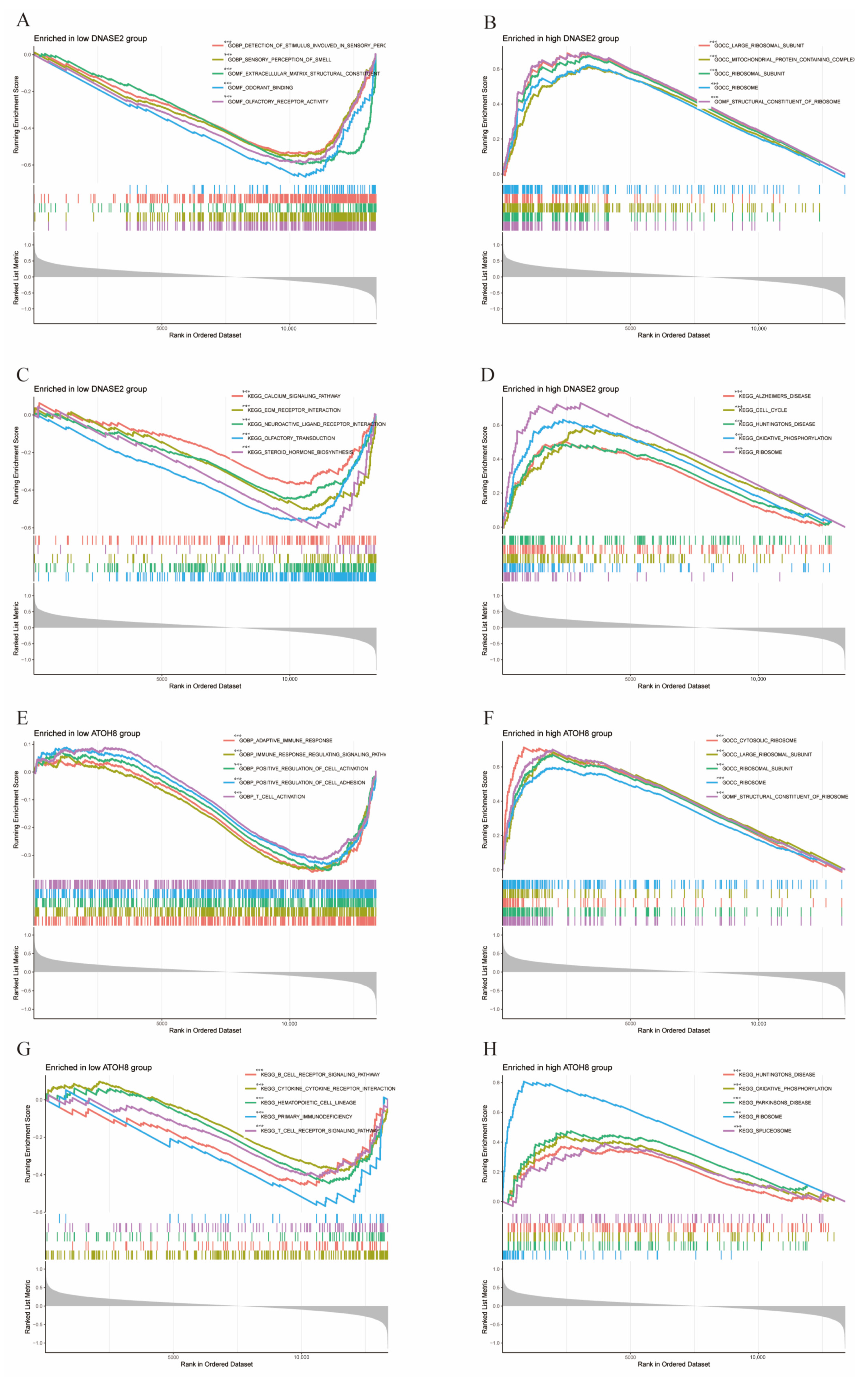
3.9. GSEA Enrichment Analysis
3.10. Drug Sensitivity Between High and Low Expression Groups of ATOH8 and DNASE2 Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEGs | Differentially expressed genes |
| eQTL | Expression quantitative trait loci |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MR | Mendelian randomization |
| PPI | Protein–protein interaction |
| LogFC | LogFoldChange |
| PCA | Principal component analysis |
| IVW | Inverse variance weighted |
| ROC | Receiver operator characteristic |
References
- Britt, K.L.; Cuzick, J.; Phillips, K.A. Key steps for effective breast cancer prevention. Nat. Rev. Cancer 2020, 20, 417–436. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, H.; Song, X.; Yang, Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin. Cancer Biol. 2020, 60, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Trapani, D.; Ginsburg, O.; Fadelu, T.; Lin, N.U.; Hassett, M.; Ilbawi, A.M.; Anderson, B.O.; Curigliano, G. Global challenges and policy solutions in breast cancer control. Cancer Treat. Rev. 2022, 104, 102339. [Google Scholar] [CrossRef] [PubMed]
- Taurin, S.; Alkhalifa, H. Breast cancers, mammary stem cells, and cancer stem cells, characteristics, and hypotheses. Neoplasia 2020, 22, 663–678. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, W.; Liu, S.; Chen, C. Targeting Breast Cancer Stem Cells. Int. J. Biol. Sci. 2023, 19, 552–570. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, M.; Xu, F.; Jiang, S. Wnt signaling in breast cancer: Biological mechanisms, challenges and opportunities. Mol. Cancer 2020, 19, 165. [Google Scholar] [CrossRef]
- Hanker, A.B.; Sudhan, D.R.; Arteaga, C.L. Overcoming Endocrine Resistance in Breast Cancer. Cancer Cell 2020, 37, 496–513. [Google Scholar] [CrossRef]
- Liu, T.; Hooda, J.; Atkinson, J.M.; Whiteside, T.L.; Oesterreich, S.; Lee, A.V. Exosomes in Breast Cancer—Mechanisms of Action and Clinical Potential. Mol. Cancer Res. 2021, 19, 935–945. [Google Scholar] [CrossRef]
- Ye, F.; Dewanjee, S.; Li, Y.; Jha, N.K.; Chen, Z.S.; Kumar, A.; Vishakha Behl, T.; Jha, S.K.; Tang, H. Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer. Mol. Cancer 2023, 22, 105. [Google Scholar] [CrossRef]
- Lau, K.H.; Tan, A.M.; Shi, Y. New and Emerging Targeted Therapies for Advanced Breast Cancer. Int. J. Mol. Sci. 2022, 23, 2288. [Google Scholar] [CrossRef]
- Jacobs, A.T.; Martinez Castaneda-Cruz, D.; Rose, M.M.; Connelly, L. Targeted therapy for breast cancer: An overview of drug classes and outcomes. Biochem. Pharmacol. 2022, 204, 115209. [Google Scholar] [CrossRef] [PubMed]
- The Lancet. Breast cancer targeted therapy: Successes and challenges. Lancet 2017, 389, 2350. [Google Scholar] [CrossRef]
- Harbeck, N.; Ewer, M.S.; De Laurentiis, M.; Suter, T.M.; Ewer, S.M. Cardiovascular complications of conventional and targeted adjuvant breast cancer therapy. Ann. Oncol. 2011, 22, 1250–1258. [Google Scholar] [CrossRef]
- Tsuchida, J.; Rothman, J.; McDonald, K.A.; Nagahashi, M.; Takabe, K.; Wakai, T. Clinical target sequencing for precision medicine of breast cancer. Int. J. Clin. Oncol. 2019, 24, 131–140. [Google Scholar] [CrossRef]
- Bedard, P.L.; Hyman, D.M.; Davids, M.S.; Siu, L.L. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 2020, 395, 1078–1088. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Hamilton, E.; Shastry, M.; Shiller, S.M.; Ren, R. Targeting HER2 heterogeneity in breast cancer. Cancer Treat. Rev. 2021, 100, 102286. [Google Scholar] [CrossRef]
- Nagini, S. Breast Cancer: Current Molecular Therapeutic Targets and New Players. Anticancer. Agents Med. Chem. 2017, 17, 152–163. [Google Scholar] [CrossRef]
- Li, Z.; Wei, H.; Li, S.; Wu, P.; Mao, X. The Role of Progesterone Receptors in Breast Cancer. Drug Des. Devel Ther. 2022, 16, 305–314. [Google Scholar] [CrossRef]
- Swain, S.M.; Shastry, M.; Hamilton, E. Targeting HER2-positive breast cancer: Advances and future directions. Nat. Rev. Drug Discov. 2023, 22, 101–126. [Google Scholar] [CrossRef]
- Goldvaser, H.; AlGorashi, I.; Ribnikar, D.; Seruga, B.; Templeton, A.J.; Ocana, A.; Amir, E. Efficacy of extended adjuvant therapy with aromatase inhibitors in early breast cancer among common clinicopathologically-defined subgroups: A systematic review and meta-analysis. Cancer Treat. Rev. 2017, 60, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.; Piccart-Gebhart, M.J.; Gelber, R.D.; Procter, M.; Goldhirsch, A.; de Azambuja, E.; Castro, G., Jr.; Untch, M.; Smith, I.; Gianni, L.; et al. 11 years’ follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive early breast cancer: Final analysis of the HERceptin Adjuvant (HERA) trial. Lancet 2017, 389, 1195–1205. [Google Scholar] [CrossRef]
- Sporikova, Z.; Koudelakova, V.; Trojanec, R.; Hajduch, M. Genetic Markers in Triple-Negative Breast Cancer. Clin. Breast Cancer 2018, 18, e841–e850. [Google Scholar] [CrossRef]
- Chapdelaine, A.G.; Sun, G. Challenges and Opportunities in Developing Targeted Therapies for Triple Negative Breast Cancer. Biomolecules 2023, 13, 1207. [Google Scholar] [CrossRef]
- Westra, H.J.; Peters, M.J.; Esko, T.; Yaghootkar, H.; Schurmann, C.; Kettunen, J.; Christiansen, M.W.; Fairfax, B.P.; Schramm, K.; Powell, J.E.; et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013, 45, 1238–1243. [Google Scholar] [CrossRef] [PubMed]
- Dudbridge, F. Polygenic Mendelian Randomization. Cold Spring Harb. Perspect. Med. 2021, 11, a039586. [Google Scholar] [CrossRef]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian Randomization. JAMA 2017, 318, 1925–1926. [Google Scholar] [CrossRef]
- Wu, G.; Wang, D.; Xiong, F.; Wang, Q.; Liu, W.; Chen, J.; Chen, Y. The emerging roles of CEACAM6 in human cancer (Review). Int. J. Oncol. 2024, 64, 27. [Google Scholar] [CrossRef]
- Oh, M.; Batty, S.; Banerjee, N.; Kim, T.H. High extracellular glucose promotes cell motility by modulating cell deformability and contractility via the cAMP-RhoA-ROCK axis in human breast cancer cells. Mol. Biol. Cell 2023, 34, ar79. [Google Scholar] [CrossRef]
- Rossi, T.; Zamponi, R.; Chirico, M.; Pisanu, M.E.; Iorio, E.; Torricelli, F.; Gugnoni, M.; Ciarrocchi, A.; Pistoni, M. BETi enhance ATGL expression and its lipase activity to exert their antitumoral effects in triple-negative breast cancer (TNBC) cells. J. Exp. Clin. Cancer Res. 2023, 42, 7. [Google Scholar] [CrossRef]
- Wang, B.; Zhao, H.; Zhao, L.; Zhang, Y.; Wan, Q.; Shen, Y.; Bu, X.; Wan, M.; Shen, C. Up-regulation of OLR1 expression by TBC1D3 through activation of TNFalpha/NF-kappaB pathway promotes the migration of human breast cancer cells. Cancer Lett. 2017, 408, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Q.; Chen, D.Y.; Li, B.; Gao, Z.J.; Feng, H.F.; Yu, X.; Liu, Z.; Wang, Y.; Li, W.G.; Sun, S.; et al. Single-cell analysis of white adipose tissue reveals the tumor-promoting adipocyte subtypes. J. Transl. Med. 2023, 21, 470. [Google Scholar] [CrossRef] [PubMed]
- Hornsveld, M.; Tenhagen, M.; van de Ven, R.A.; Smits, A.M.; van Triest, M.H.; van Amersfoort, M.; Kloet, D.E.; Dansen, T.B.; Burgering, B.M.; Derksen, P.W. Restraining FOXO3-dependent transcriptional BMF activation underpins tumour growth and metastasis of E-cadherin-negative breast cancer. Cell Death Differ. 2016, 23, 1483–1492. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, P.; Antonov, A.; Anemona, L.; Vangapandou, C.; Montanaro, M.; Botticelli, A.; Mauriello, A.; Melino, G.; Catani, M.V. New immunological potential markers for triple negative breast cancer: IL18R1, CD53, TRIM, Jaw1, LTB, PTPRCAP. Discov. Oncol. 2021, 12, 6. [Google Scholar] [CrossRef]
- Liu, H.X.; Lian, L.; Hou, L.L.; Liu, C.X.; Ren, J.H.; Qiao, Y.B.; Wen, S.Y.; Li, Q.S. Herb pair of Huangqi-Danggui exerts anti-tumor immunity to breast cancer by upregulating PIK3R1. Anim. Model. Exp. Med. 2024, 7, 234–258. [Google Scholar] [CrossRef]
- Nordgard, S.H.; Johansen, F.E.; Alnaes, G.I.; Bucher, E.; Syvanen, A.C.; Naume, B.; Borresen-Dale, A.L.; Kristensen, V.N. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosomes Cancer 2008, 47, 680–696. [Google Scholar] [CrossRef]
- Breast Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 1 July 2024).
- Liu, X.; Li, X.; Wang, S.; Liu, Q.; Feng, X.; Wang, W.; Huang, Z.; Huang, Y.; Wu, J.; Cai, M.; et al. ATOH8 binds SMAD3 to induce cellular senescence and prevent Ras-driven malignant transformation. Proc. Natl. Acad. Sci. USA 2023, 120, e2208927120. [Google Scholar] [CrossRef]
- Huang, Q.; Li, S.; Hu, X.; Sun, M.; Wu, Q.; Dai, H.; Tan, Y.; Sun, F.; Wang, C.; Rong, X.; et al. Shear stress activates ATOH8 via autocrine VEGF promoting glycolysis dependent-survival of colorectal cancer cells in the circulation. J. Exp. Clin. Cancer Res. 2020, 39, 25. [Google Scholar] [CrossRef]
- Xu, M.; Huang, S.; Dong, X.; Chen, Y.; Li, M.; Shi, W.; Wang, G.; Huang, C.; Wang, Q.; Liu, Y.; et al. A novel isoform of ATOH8 promotes the metastasis of breast cancer by regulating RhoC. J. Mol. Cell Biol. 2021, 13, 59–71. [Google Scholar] [CrossRef]
- Yi, M.; Wu, Y.; Niu, M.; Zhu, S.; Zhang, J.; Yan, Y.; Zhou, P.; Dai, Z.; Wu, K. Anti-TGF-beta/PD-L1 bispecific antibody promotes T cell infiltration and exhibits enhanced antitumor activity in triple-negative breast cancer. J. Immunother. Cancer 2022, 10, e005543. [Google Scholar] [CrossRef]
- Anindya, R. Cytoplasmic DNA in cancer cells: Several pathways that potentially limit DNase2 and TREX1 activities. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119278. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Yu, Z.; Li, Q.; Hu, X.; Liu, X.; Li, F.; Li, S.; Chen, Y. Circ_0073228 serves as a competitive endogenous ribonucleic acid to facilitate proliferation and inhibit apoptosis of hepatocellular carcinoma cells by the miR-139-5p/deoxyribonuclease II axis. J. Gene Med. 2023, 25, e3507. [Google Scholar] [CrossRef] [PubMed]
- Das, C.; Adhikari, S.; Bhattacharya, A.; Chakraborty, S.; Mondal, P.; Yadav, S.S.; Adhikary, S.; Hunt, C.R.; Yadav, K.K.; Pandita, S.; et al. Epigenetic-Metabolic Interplay in the DNA Damage Response and Therapeutic Resistance of Breast Cancer. Cancer Res. 2023, 83, 657–666. [Google Scholar] [CrossRef] [PubMed]





| GSE ID | Samples | Tissues | Platform | Experiment Type |
|---|---|---|---|---|
| GSE109169 | 25 cases and 25 controls | Breast cancer | GPL5175 | 13 January 2018 |
| GSE113865 | 3 cases and 3 controls | Breast cancer | GPL10558 | 30 April 2023 |
| GSE139038 | 41 cases and 24 controls | Breast cancer | GPL27630 | 18 October 2019 |
| GSE205185 | 17 cases and 5 controls | Breast cancer | GPL21158 | 1 June 2022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, S.; Cui, J.; Sun, Y.; Wang, X.; Cai, C. Exploring Novel Therapeutic Targets in Breast Cancer via Comprehensive Omics Profiling and Experimental Verification. Biology 2025, 14, 405. https://doi.org/10.3390/biology14040405
Chai S, Cui J, Sun Y, Wang X, Cai C. Exploring Novel Therapeutic Targets in Breast Cancer via Comprehensive Omics Profiling and Experimental Verification. Biology. 2025; 14(4):405. https://doi.org/10.3390/biology14040405
Chicago/Turabian StyleChai, Shengjun, Jiayong Cui, Yinuo Sun, Xiaowu Wang, and Chunmei Cai. 2025. "Exploring Novel Therapeutic Targets in Breast Cancer via Comprehensive Omics Profiling and Experimental Verification" Biology 14, no. 4: 405. https://doi.org/10.3390/biology14040405
APA StyleChai, S., Cui, J., Sun, Y., Wang, X., & Cai, C. (2025). Exploring Novel Therapeutic Targets in Breast Cancer via Comprehensive Omics Profiling and Experimental Verification. Biology, 14(4), 405. https://doi.org/10.3390/biology14040405







