Three Circulating miRNAs Related to Non-Small-Cell Lung Cancer Progression: An Integrative Analysis of Their Biological Roles
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. miRNAs Microarray Data
2.2. Survival Analysis
2.3. Prediction of miRNA Target Genes
2.4. Expression and Function of Target Genes
2.5. Cell Proliferation and Migration
2.6. Data Analysis
3. Results
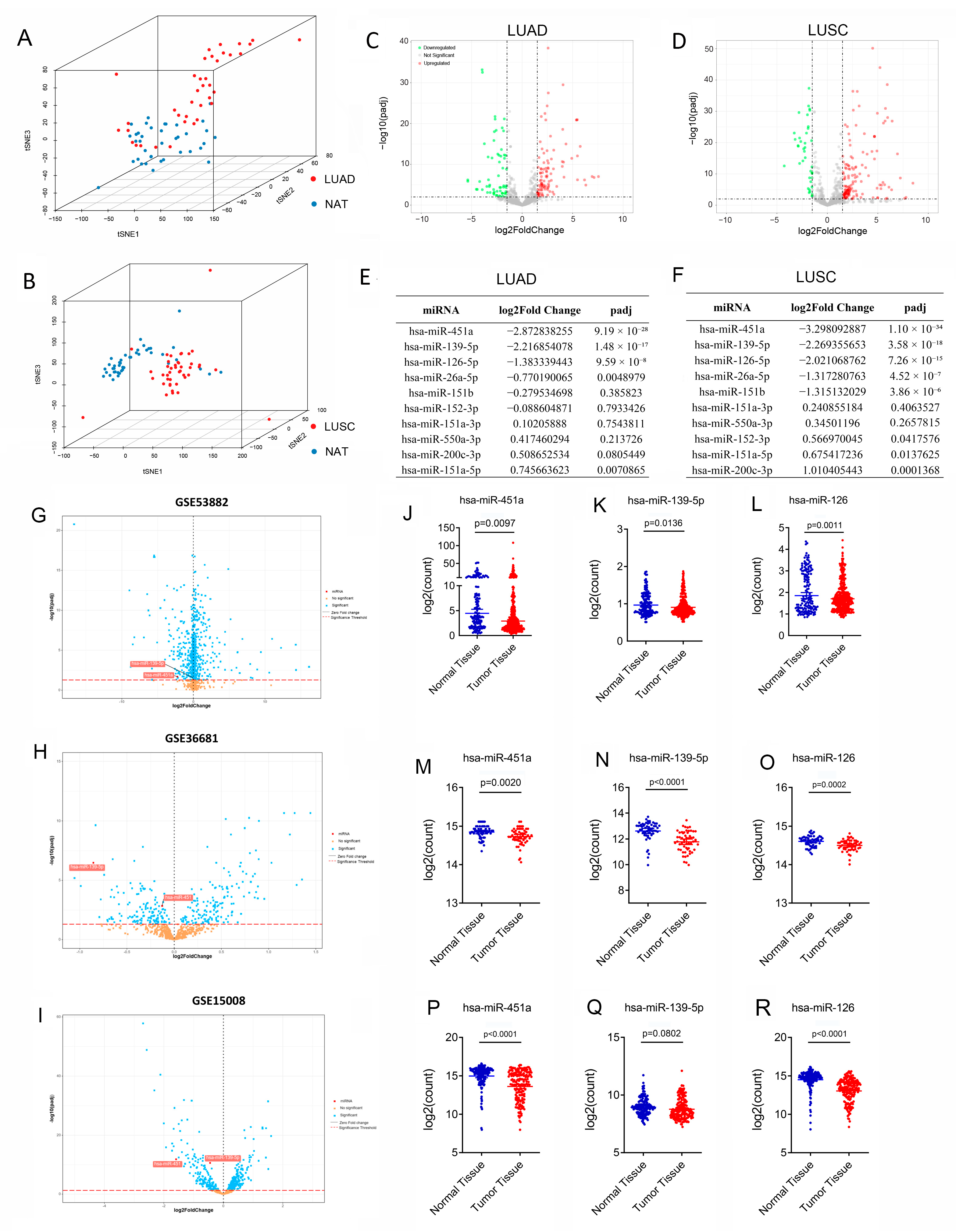
3.1. Ten Circulating miRNAs Validated in Lung Cancer Tissues
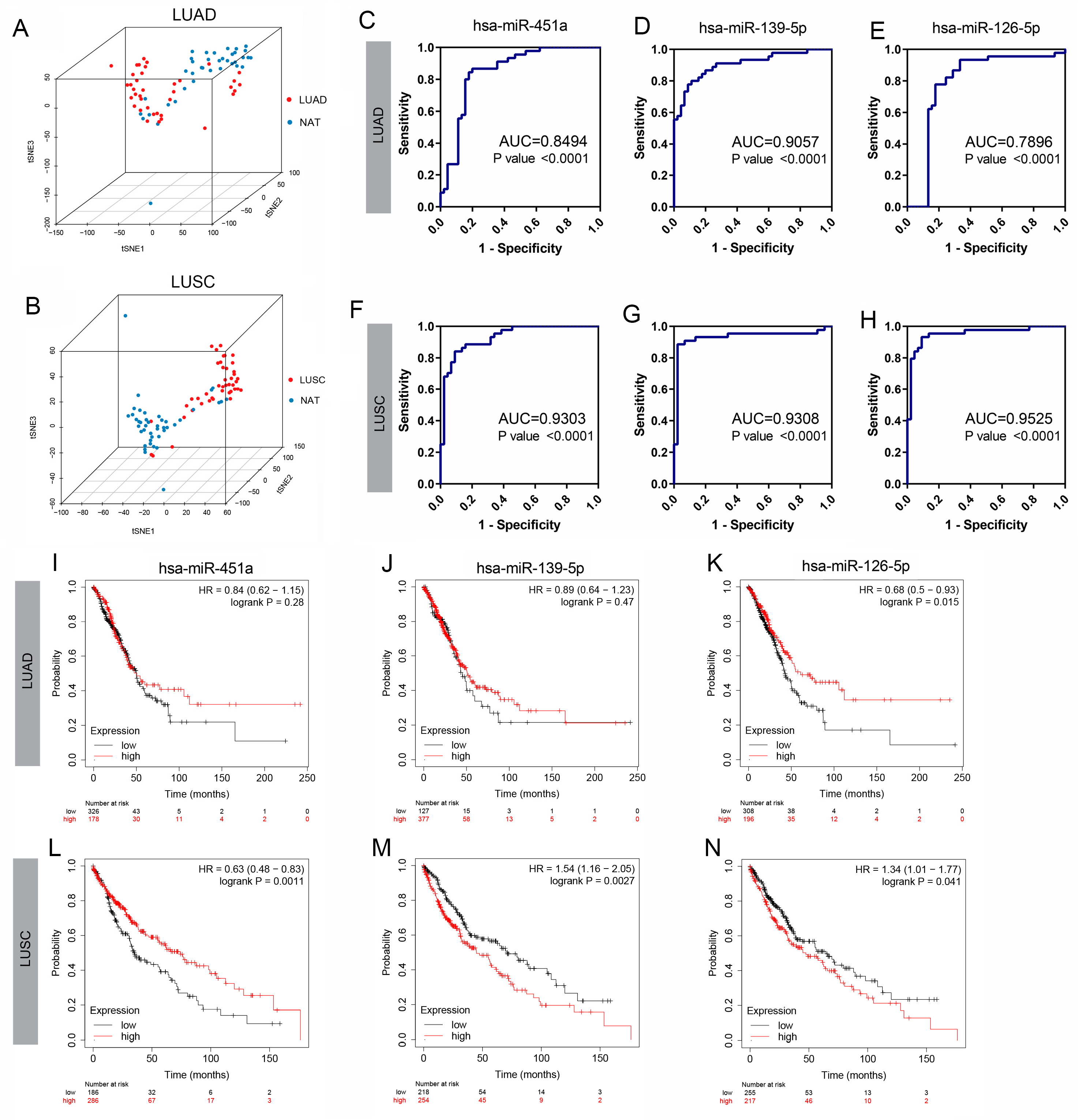
3.2. Confirmation of Three DE-miRNAs’ Clinical Significance
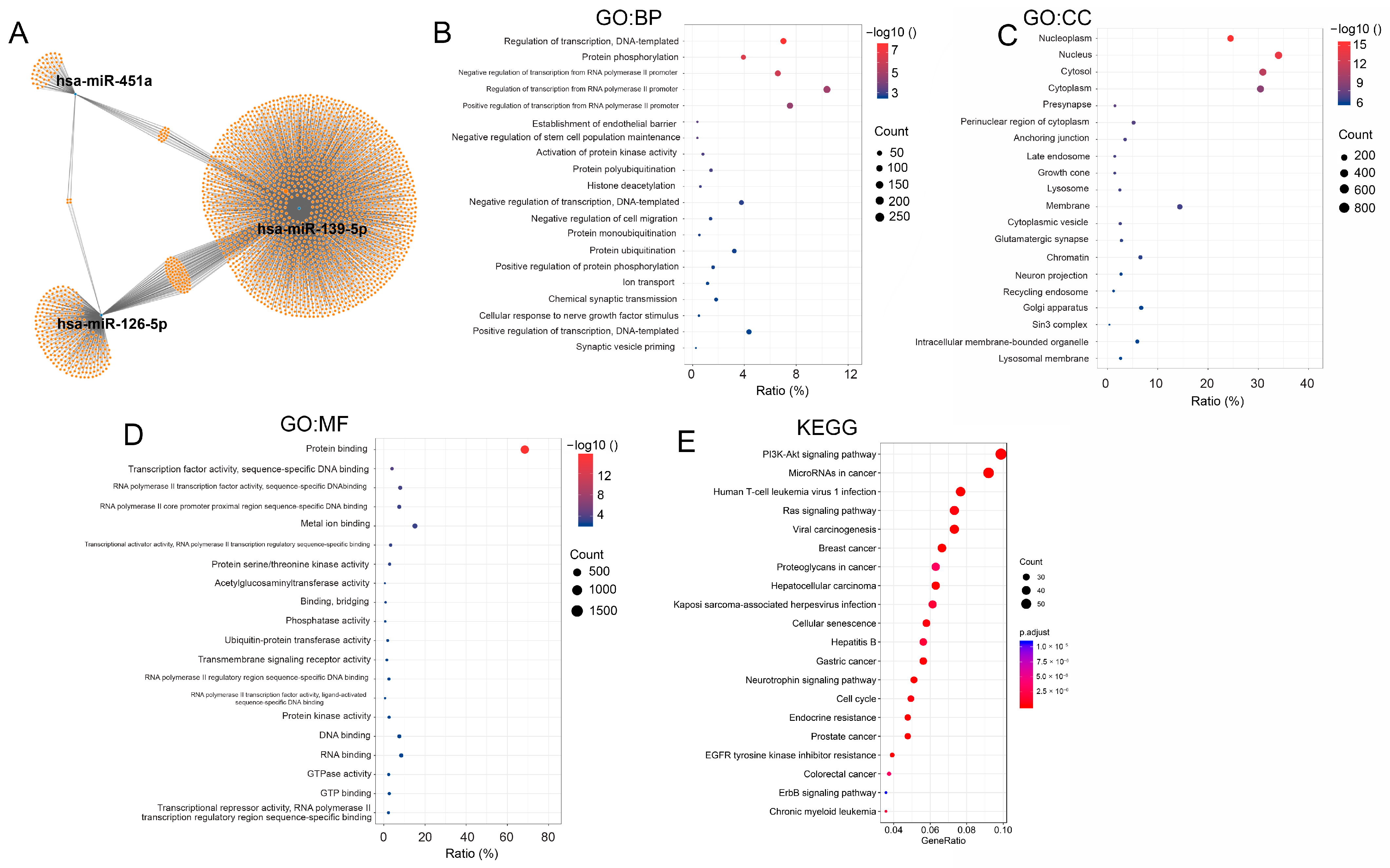
3.3. Function and Pathway Analysis of Predicted Target Genes
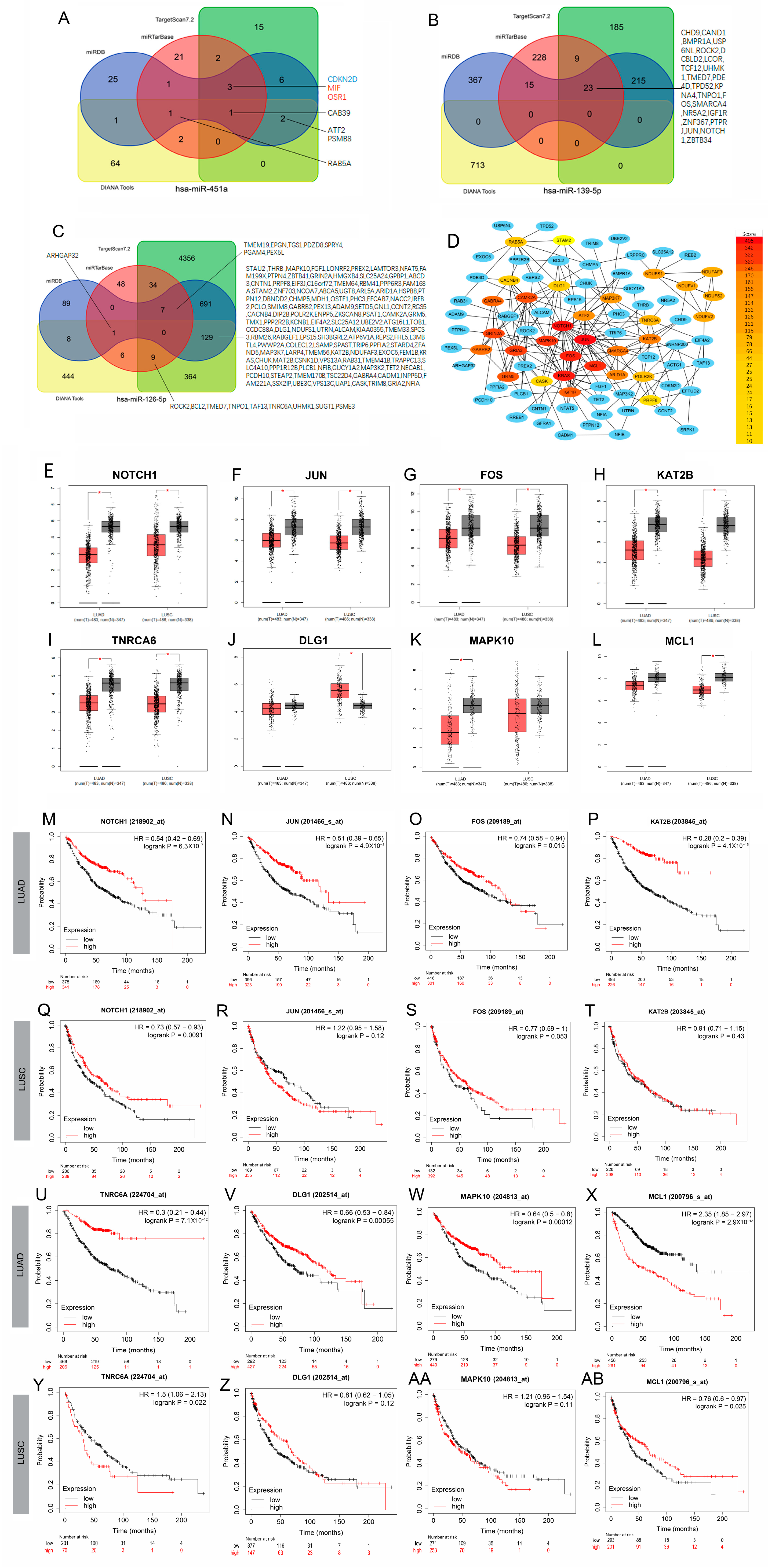
3.4. Potential Clinical Value of Predicted Target Genes of DE-miRNAs
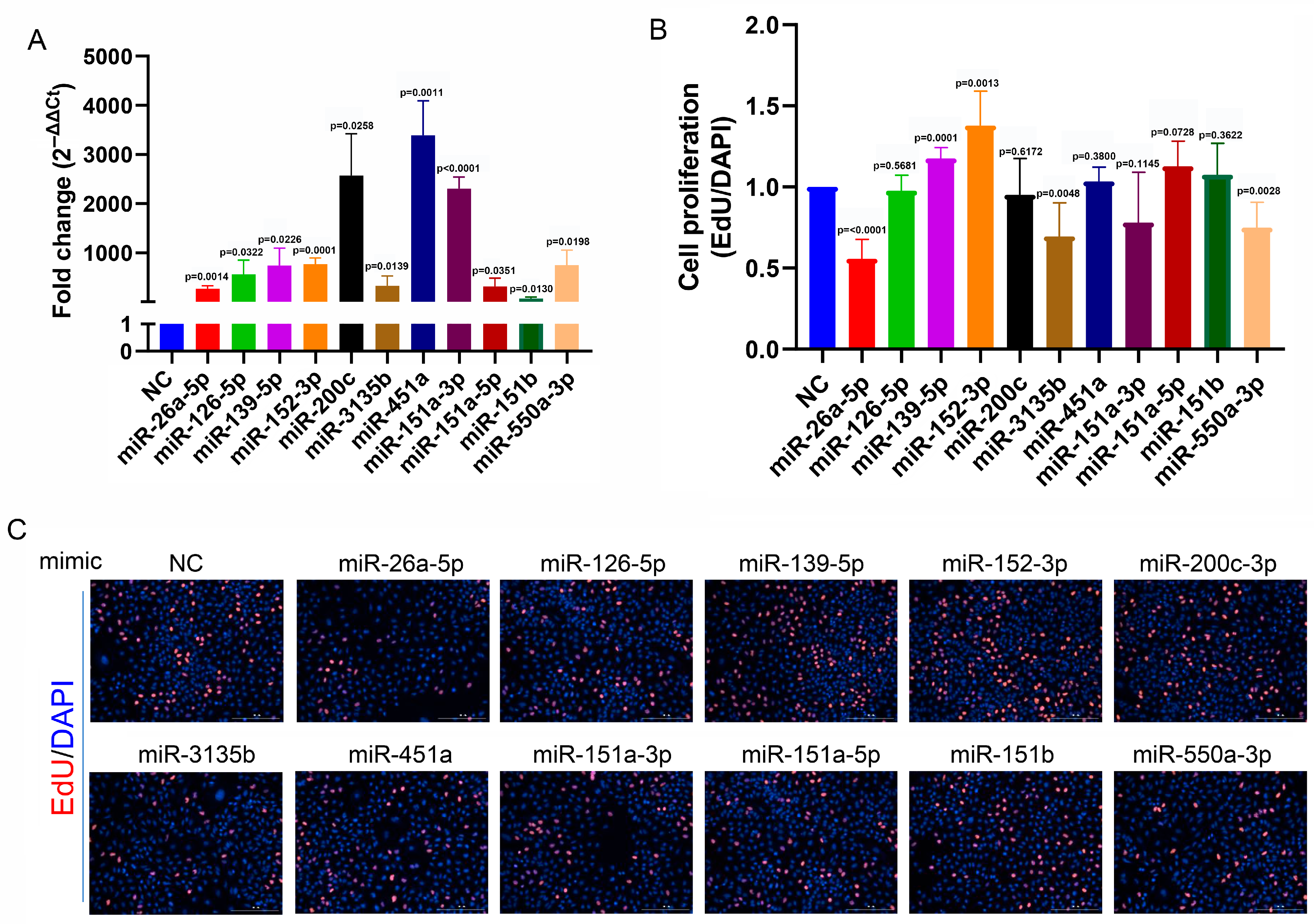
3.5. Functions of 11 miRNAs in A549 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| miRNA | microRNA |
| NSCLC | non-small-cell lung cancer |
| LUAD | lung adenocarcinoma |
| LUSC | lung squamous cell carcinoma |
| DE-miRNAs | differentially expressed miRNAs |
| ROC | receiver operating characteristic curve |
| GO | Gene Ontology |
| BP | biological process |
| CC | cellular component |
| MF | molecular function |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PPI | protein-protein interaction |
| TCGA | Cancer Genome Atlas |
| GEO | Gene Expression Omnibus |
| ROC | area under ROC curve |
| PCA | Principal component analysis |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022, GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K. Lung Cancer: Understanding Its Molecular Pathology and the 2015 WHO Classification. Front. Oncol. 2017, 7, 193. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J., Jr.; Wu, Y.-L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Kargutkar, N.; Hariharan, P.; Nadkarni, A. Dynamic interplay of microRNA in diseases and therapeutic. Clin. Genet. 2023, 103, 268–276. [Google Scholar] [CrossRef]
- Çakmak, H.A.; Demir, M. MicroRNA and Cardiovascular Diseases. Balkan. Med. J. 2020, 37, 60–71. [Google Scholar] [CrossRef]
- Pandey, M.; Mukhopadhyay, A.; Sharawat, S.K.; Kumar, S. Role of microRNAs in regulating cell proliferation, metastasis and chemoresistance and their applications as cancer biomarkers in small cell lung cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188552. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531, Erratum in Nat. Rev. Genet. 2004, 5, 631. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Wu, K.L.; Tsai, Y.M.; Lien, C.T.; Kuo, P.L.; Hung, A.J. The Roles of MicroRNA in Lung Cancer. Int. J. Mol. Sci. 2019, 20, 1611. [Google Scholar] [CrossRef]
- Qin, Q.; Wei, F.; Zhang, J.; Wang, X.; Li, B. miR-134 inhibits non-small cell lung cancer growth by targeting the epidermal growth factor receptor. J. Cell. Mol. Med. 2016, 20, 1974–1983. [Google Scholar] [CrossRef]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef]
- Chen, L.; Gibbons, D.L.; Goswami, S.; Cortez, M.A.; Ahn, Y.H.; Byers, L.A.; Zhang, X.; Yi, X.; Dwyer, D.; Lin, W.; et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014, 5, 5241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Jin, M.; Li, J.; Kong, X. Identifying circulating miRNA biomarkers for early diagnosis and monitoring of lung cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165847. [Google Scholar] [CrossRef] [PubMed]
- Andersen, G.B.; Tost, J. Circulating miRNAs as Biomarker in Cancer. Recent Results Cancer Res. 2020, 215, 277–298. [Google Scholar]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef]
- Stenvold, H.; Donnem, T.; Andersen, S.; Al-Saad, S.; Busund, L.T.; Bremnes, R.M. Stage and tissue-specific prognostic impact of miR-182 in NSCLC. BMC Cancer 2014, 14, 138. [Google Scholar] [CrossRef]
- Wu, L.-N.; Hu, B.; Zhao, B.-T.; Liu, Y.-N.; Yang, Y.; Zhang, L.-J.; Chen, J.-F. Circulating microRNA-422a is associated with lymphatic metastasis in lung cancer. Oncotarget 2017, 8, 42173–42188. [Google Scholar] [CrossRef]
- Szpechcinski, A.; Florczuk, M.; Duk, K.; Zdral, A.; Rudzinski, S.; Bryl, M.; Czyzewicz, G.; Rudzinski, P.; Kupis, W.; Wojda, E.; et al. The expression of circulating miR-504 in plasma is associated with EGFR mutation status in non-small-cell lung carcinoma patients. Cell. Mol. Life Sci. 2019, 76, 3641–3656. [Google Scholar] [CrossRef]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef]
- Turchinovich, A.; Samatov, T.R.; Tonevitsky, A.G.; Burwinkel, B. Circulating miRNAs: Cell-cell communication function? Front. Genet. 2013, 4, 119. [Google Scholar] [CrossRef] [PubMed]
- Igaz, I.; Igaz, P. Tumor surveillance by circulating microRNAs: A hypothesis. Cell Mol. Life Sci. 2014, 71, 4081–4087. [Google Scholar] [CrossRef]
- Niu, Y.; Su, M.; Wu, Y.; Fu, L.; Kang, K.; Li, Q.; Li, L.; Hui, G.; Li, F.; Gou, D. Circulating Plasma miRNAs as Potential Biomarkers of Non-Small Cell Lung Cancer Obtained by High-Throughput Real-Time PCR Profiling. Cancer Epidemiol. Biomark. Prev. 2019, 28, 327–336. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, 98–102. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, 216–221. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, 587–592. [Google Scholar] [CrossRef]
- Deng, L.; Chen, J.; Chen, B.; Wang, T.; Yang, L.; Liao, J.; Yi, J.; Chen, Y.; Wang, J.; Linneman, J.; et al. LncPTSR Triggers Vascular Remodeling in Pulmonary Hypertension by Regulating [Ca2+]i in Pulmonary Arterial Smooth Muscle Cells. Am. J. Respir. Cell Mol. Biol. 2022, 66, 524–538. [Google Scholar] [CrossRef]
- Mounir, M.; Lucchetta, M.; Silva, T.C.; Olsen, C.; Bontempi, G.; Chen, X.; Noushmehr, H.; Colaprico, A.; Papaleo, E. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput. Biol. 2019, 15, e1006701. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Pirlog, R.; Chiroi, P.; Rusu, I.; Jurj, A.M.; Budisan, L.; Pop-Bica, C.; Braicu, C.; Crisan, D.; Sabourin, J.-C.; Berindan-Neagoe, I. Cellular and Molecular Profiling of Tumor Microenvironment and Early-Stage Lung Cancer. Int. J. Mol. Sci. 2022, 23, 5346. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Kumar, M.S.; Lu, J.; Mercer, K.L.; Golub, T.R.; Jacks, T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007, 39, 673–677. [Google Scholar] [CrossRef]
- Song, F.; Xuan, Z.; Yang, X.; Ye, X.; Pan, Z.; Fang, Q. Identification of key microRNAs and hub genes in non-small-cell lung cancer using integrative bioinformatics and functional analyses. J. Cell. Biochem. 2020, 121, 2690–2703. [Google Scholar] [CrossRef]
- Valihrach, L.; Androvic, P.; Kubista, M. Circulating miRNA analysis for cancer diagnostics and therapy. Mol. Asp. Med. 2020, 72, 100825. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, Y.; Fu, G.; Xia, S.; Li, M.; Qiu, L.; Wang, J.; Kang, K.; Gou, D. Three Circulating miRNAs Related to Non-Small-Cell Lung Cancer Progression: An Integrative Analysis of Their Biological Roles. Biology 2025, 14, 399. https://doi.org/10.3390/biology14040399
Niu Y, Fu G, Xia S, Li M, Qiu L, Wang J, Kang K, Gou D. Three Circulating miRNAs Related to Non-Small-Cell Lung Cancer Progression: An Integrative Analysis of Their Biological Roles. Biology. 2025; 14(4):399. https://doi.org/10.3390/biology14040399
Chicago/Turabian StyleNiu, Yanqin, Gaohui Fu, Sijian Xia, Menglong Li, Lin Qiu, Jun Wang, Kang Kang, and Deming Gou. 2025. "Three Circulating miRNAs Related to Non-Small-Cell Lung Cancer Progression: An Integrative Analysis of Their Biological Roles" Biology 14, no. 4: 399. https://doi.org/10.3390/biology14040399
APA StyleNiu, Y., Fu, G., Xia, S., Li, M., Qiu, L., Wang, J., Kang, K., & Gou, D. (2025). Three Circulating miRNAs Related to Non-Small-Cell Lung Cancer Progression: An Integrative Analysis of Their Biological Roles. Biology, 14(4), 399. https://doi.org/10.3390/biology14040399





