Simple Summary
Viruses are pervasive components of aquatic ecosystems, and most of them are harmless to humans and animals; however, several aquatic viruses can infect animals, leading to diseases, especially when fish are confined, such as in aquaculture facilities. Traditional methods used to detect and study viruses have been widely applied to aquatic animals’ viruses, leading to the successful isolation, identification and understanding of several of them. However, they have limits, which can be overcome by molecular methods, such as polymerase chain reaction (PCR)-based assays, sequencing and in situ hybridisation. A standard PCR, followed by the sequencing of purified amplicons, is an effective method for both identifying well-known viruses and discovering new ones. In situ hybridisation, in which a labelled probe binds to a nucleic acid sequence in tissue, is able to correlate the presence of viruses to lesions. Novel molecular isothermal methods, such as loop-mediated isothermal amplification (LAMP), were also developed and applied to viral aquatic animal diseases, bringing molecular diagnosis into the field. This review considers the scientific literature dealing with the molecular methods employed hitherto to study the most relevant finfish and shellfish viral pathogens, stressing their advantages and disadvantages.
Abstract
Aquaculture is the fastest-growing food-producing sector, with a global production of 122.6 million tonnes in 2020. Nonetheless, aquatic animal production can be hampered by the occurrence of viral diseases. Furthermore, intensive farming conditions and an increasing number of reared fish species have boosted the number of aquatic animals’ pathogens that researchers have to deal with, requiring the quick development of new detection and study methods for novel unknown pathogens. In this respect, the molecular tools have significantly contributed to investigating thoroughly the structural constituents of fish viruses and providing efficient detection methods. For instance, next-generation sequencing has been crucial in reassignment to the correct taxonomic family, the sturgeon nucleo-cytoplasmic large DNA viruses, a group of viruses historically known, but mistakenly considered as iridoviruses. Further methods such as in situ hybridisation allowed objectifying the role played by the pathogen in the determinism of disease, as the cyprinid herpesvirus 2, ostreid herpesvirus 1 and betanodaviruses. Often, a combination of molecular techniques is crucial to understanding the viral role, especially when the virus is detected in a new aquatic animal species. With this paper, the authors would critically revise the scientific literature, dealing with the molecular techniques employed hitherto to study the most relevant finfish and shellfish viral pathogens.
1. Introduction
The advent of molecular methods has benefited all sectors of biology, expanding and deepening knowledge on the structures and operating mechanisms of all organisms. Molecular methods are powerful tools in the study of infectious diseases. They can provide valuable epidemiological insights, demonstrating, for example, relatedness between strains; furthermore, they have contributed to our understanding of pathogenesis and they are invaluable tools in the diagnosis of infectious diseases, expediting and improving pathogen detection and identifying the agents of diseases whose causes are unknown [1].
Polymerase chain reaction (PCR) is the most well-developed molecular technique and, since its invention by Kary Mullis in 1985, it has been used to create many microorganism detection methods and has been applied to all scientific sectors [2]. In PCR, a small amount of DNA can be copied in large quantities over a short period of time, achieving impressive sensitivity. PCR-based methods may overcome the limitations of traditional testing procedures. As a matter of fact, the virology sector profited handsomely from the molecular approach, especially for those viruses with no or few in vitro replication activities. Furthermore, the extensive use of molecular methods has now produced techniques that are also cost-effective when compared to traditional methods. All these features make the molecular approach ideal for the detection of viral aquatic animal diseases. The aquaculture sector is fast expanding and deals with multiple fish species, ranging across the animal kingdom and including freshwater and marine finfish, crustaceans and molluscs. In oceanic marine habitats, viruses can be found in concentrations of up to 108 viruses mL−1 [3] and some of them are well-known causes of fish diseases. However, the intensification of fish production and the movement of aquatic animals led to an increase in infectious diseases, requiring a continuous expansion of knowledge asset on fish viral diseases and a constant adjustment of diagnostic methods. In this respect, molecular methods provide the ideal tool to gain crucial knowledge about viral aquatic animal diseases.
The PCR approach has been expanded to detect multiple targets at the same time using multiplexing assays, which has found widespread application in the study and detection of viral aquatic animal diseases. Differential diagnosis, including clinical methods and gross pathology, is a milestone in the diagnosis process for infectious diseases; however, for fish, no pathognomonic signs are described and more than one aetiology needs to be investigated simultaneously to provide a correct and fast pathogen identification. In this respect, multiplex PCR can provide an efficient method to discriminate quickly among fish pathogens or genetic variants associated with similar clinical signs [4,5].
Real-time PCR is another widely applied PCR-based method able not only to detect the target DNA but also to quantify the number of DNA molecules initially present in the sample. For this reason, the method is also known as quantitative PCR (qPCR) [6]. In real-time PCR, the amplified product is monitored through the fluorescence of dyes or probes introduced into the reaction, eliminating post-PCR manipulations. For this reason, real-time PCR allows for processing a large number of samples with a lower level of contamination, making it more suitable for routine diagnostics compared to conventional PCR. On the other hand, a conventional PCR can be used to produce amplicons for sequencing and phylogenetic analyses [1]. In addition to diagnostics, real-time PCR can be very useful for virus detection and quantification in pathogenesis studies, especially for viruses with low or no cultivability [7].
More recently, digital PCR is taking hold thanks to the recent development of new instruments and chemistry, which have made it a much simpler and more practical technique [8]. This innovative approach has been recently applied for the diagnosis of carp edema virus (CEV), a pathogen that may lead to high mortality in both koi and common carp populations and for which no effective control methods have been developed. In this case, a prompt and efficient detection of the virus is the only approach for the prevention of further spread of the disease. In this regard, the developed digital PCR assay provides a robust diagnostic tool for the sensitive detection of CEV [9].
Despite their unparalleled contribution, PCR-based techniques also have some limits. In particular, they require expensive reagents and equipment. Isothermal amplification methods overcome this shortcoming as they are performed at one reaction temperature under simple conditions and thus do not rely on specialised equipment [10]. Among the several isothermal amplification techniques described so far, loop-mediated isothermal amplification (LAMP) is one of the most popular, with assays developed for the detection of several fish and shellfish viruses. More advanced and complex methods, such as the systematic evolution of ligands by exponential enrichment (SELEX), have been applied so far to a more limited number of fish viruses. This method was used to generate aptamers with high specificity and affinity for Singapore grouper iridovirus (SGIV)-infected cells. These aptamers can find several applications in both disease diagnosis and control; indeed, they can be used as molecular probes to set up rapid diagnostic assays and study the mechanism of virus infection as well as to develop antiviral treatments against SGIV infection in aquaculture [11]. In situ hybridisation (ISH) is another molecular method used for diagnostic purposes but above all to study the pathogenetic mechanisms of infectious diseases. ISH consists of a family of sensitive methods able to target specific nucleic acid sequences at the cellular level and potentially it can be used to correlate the presence of viruses to lesions [12,13]. The first in situ detection methods were characterised by the use of very sensitive radio labelled probes, but also by disadvantages as the difficulty in obtaining a precise localisation of the signals and the handling of radioisotopes. Hence, in the last few decades, several adjustments have been made in order to achieve greater sensitivity, practicability and safety. At present, non-radioactive methods such as chromogenic (CISH) and fluorescent (FISH) in situ hybridisation are used [13]. Moreover, if designed properly, these methods are also able to discriminate between the replicative and non-replicative status of a pathogen, by using specific sense or antisense probes [12,14]. Several ISH assays have successfully been developed in order to localise the presence of viruses in the tissues of aquatic animals. For instance, this approach is essential to distinguish true ostreid herpesvirus 1 (OsHV-1) active infection from passive contamination of the host, especially when the virus is detected in new host species and their role needs to be investigated [15,16].
The analysis of sequences has significantly contributed to gaining fundamental epidemiological information on viral diseases. The combination of the epidemiological approach and molecular methods has resulted in molecular epidemiology studies that help to understand disease distribution in populations and the relationship between viral strains and disease outbreaks. Sequence analysis has furthered the capacity to investigate viral evolutionary history through phylogenetic studies [17]. The genetic characterisation of novel North American marine viral haemorrhagic septicaemia virus (VHSV) strains and their comparison with traditional European freshwater strains have provided a remarkable insight into the origins of viral haemorrhagic septicaemia (VHS) in aquaculture. VHS was traditionally thought to be a disease exclusive to freshwater rainbow trout farming. North American and European strains belong to different genotypes that were indicated to have become separated a long time before fish farming was established on both continents. This finding has led to several new studies, which in the end proved the existence of long-standing natural marine reservoirs of the VHS virus [17].
At last, the advent of next generation sequencing (NGS) has greatly contributed to the discovery of emerging pathogens in the fish farming industry. The reduction of sequencing costs and the availability of efficient computational resources have led to a wider use of large-scale DNA/RNA sequencing. In particular, the research on unculturable and emerging viruses benefited above all by metagenomics and other methods that are not dependent on laboratory propagation [18]. The NGS approach unveils novel and often surprising findings for all viruses investigated, including the reassignment to the correct taxonomic family of an entire group of viruses such as the sturgeon nucleocytoplasmic large DNA viruses (NCLDVs), a group of viruses historically known, but mistakenly considered, as iridoviruses [19].
This review aims to illustrate how molecular methods contribute to gaining crucial knowledge about viral aquatic animal diseases reporting typical applications to some of the most challenging fish viruses. In particular, two groups of large dsDNA viruses, such as the nucleocytoplasmic large DNA viruses and the herpesvirales group, were chosen. Molecular methods have constituted a powerful tool to gain crucial knowledge on the large and complex genomes that characterise these viruses, overcoming limitations due to poor in vitro cultivability, strict species-specificity and different evolution dynamics. On the other hand, an example of a group of small RNA viruses is also reported: the nodaviruses. The research on this fast-evolving group of viruses greatly benefited from the application of molecular methods as well, providing successful diagnostic methods and allowing the exploration of the relationship between genes and biological characteristics.
2. Nucleocytoplasmic Large DNA Virus
The nucleocytoplasmic large DNA viruses (NCLDVs) are double-stranded DNA viruses of eukaryotes with a monophyletic origin [20]. This group includes several virus families, such as Poxviridae, Asfarviridae, Iridoviridae, Phycodnaviridae, Marseilleviridae, Ascoviridae and Mimiviridae. The sequencing of several viral genomes in the last two decades has substantially advanced knowledge of the evolutionary history of these viruses, and now all these families are officially joined in the phylum Nucleocytoviricota within the virus kingdom Bamfordvirae of the realm Varidnaviria by the International Committee on Taxonomy of Viruses (ICTV) (https://ictv.global/taxonomy accessed on 2 February 2023).
2.1. Iridoviridae
The family Iridoviridae comprises several viruses infecting fish, included in the subfamily Alphairidovirinae and grouped in three genera: Lymphocystivirus, Megalocytivirus and Ranavirus [21]. Lymphocystivirus is a group of viruses historically known and associated to a cutaneous proliferative benign disease of demersal fish since its first isolation in cell culture in 1962 [22]. Megalocytiviruses and ranaviruses have been reported long after in the 1990s and are responsible for systemic infections leading to high mortality outbreaks in cold-blooded vertebrates [23,24].
Iridoviruses possess large, linear, double-stranded DNA genomes that are circularly permuted and terminally redundant [25]. Virions have an icosahedral symmetry and infectious particles may be either “naked” or enveloped depending on their release by cell lysis or by budding from the plasma membrane, respectively [24].
PCR-based methods and phylogenetic analysis have been applied to the study of all vertebrate iridoviruses implementing diagnostic capacity, epidemiology knowledge including geographical distribution, host range and their taxonomy. Moreover, whole genome sequencing has permitted further exploration of their genome characteristics, bringing to attention interesting data on their genomic organisation and differences.
2.1.1. Lymphocystivirus
Lymphocystiviruses are responsible for lymphocystis, a self-limiting disease of teleosts known since the 19th century. Lymphocystis typically presents with cutaneous nodular masses consisting of hypertrophied fibroblasts.
On the basis of typical nodular masses on the skin and fins, lymphocystis has been described in more than 140 wild freshwater and marine water fish across the globe [26]; however, the actual identification of the virus has been conducted in a more limited number of cases. Electron microscopy can confirm the presence of icosahedral iridovirus-like virions inside hypertrophied fibroblasts, and this method has been used for a long time to identify the virus in pathological tissue [26]. However, this approach limited the identification, at most, to the family level, resulting in a considerable degree of uncertainty. When partial/complete genetic characterisation was conducted, considerable differences were reported among lymphocystiviruses, despite the consistent clinical presentation described across the affected species.
Thanks to whole genome sequencing and phylogenetic analysis, four species of LCDV were recognised and named LCDV1-4. LCDV1 is reported in flatfishes (family Pleuronectidae), LCDV2 is isolated from olive flounder (Paralichthys olivaceus), LCDV3 is found in gilthead sea bream (Sparus aurata), and LCDV4 is detected in the whitemouth croaker (Micropogonias furnieri) [21,27]. Significant discrepancies have been pointed out in the genome size, whole genomic structure and organisation of different LCDV species [28]. In particular, LCDV1 differs significantly from other LCDVs with a genome of about half the size (1000 Kbp) compared to other LCDV species (180–203 Kbp). Comparison between the genomes of different LCDV showed a low percentage of nucleotide identities, indicating the almost complete genomic dissimilarities and non-relatedness. In particular, only some LCDV1 genomic regions were found in LCDV2-4, but the spatial arrangement of these regions was different [28]. On the other hand, viruses within a species showed a similar genome size and similar conserved genomic regions as demonstrated for viruses belonging to LCDV1 [28], at least in the case where more than one virus per species was fully sequenced.
The family Iridoviridae belongs to the NCLDVs, whose genome is characterised by a small number of widely shared genes and unique or species-specific operating reading frames (ORFs). These distinctive genes are possibly acquired by gene duplications, deletions, lateral gene transfers from their hosts and the de novo creation of protein-coding genes; more strikingly, it is increasingly understood that these mechanisms are driven by environmental factors [29]. Even if not fully demonstrated, differences in LCDV genomes may be due to these mechanisms of rearrangement induced by the environment in which they evolved; in fact, all LCDV have a common lifestyle, having been detected all from demersal fish [28].
The use of the major capsid protein (MCP) gene, a more limited area of the genome, allowed us to include in the analysis a broader number of LCDV detected in a wide range of fish host species. The MCP gene, which is characterised by extensive conservation, is employed in iridovirus comparative phylogenetic research. Sequence comparison of the MCP genes of several lymphocystiviruses clusters LCDVs into nine genotypes [26]. LCDV4 was recently added to them [27]. Full genome sequencing clearly distinguishes LCDV4 as a separate viral species; accordingly, analysis of the MCP gene segregates it separately from all other known genotypes, despite this being supported by a quite low bootstrap value (Figure 1). The size of the sequence and the genes used in the phylogenetic analysis can have a significant impact on the output and its robustness.
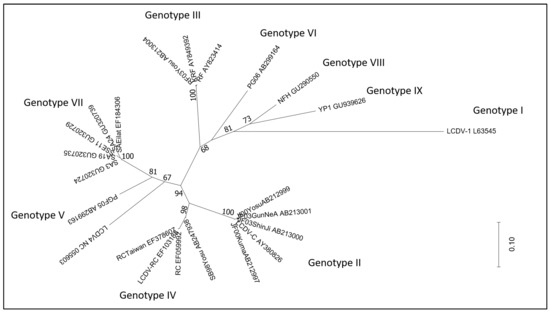
Figure 1.
Maximum likelihood phylogenetic tree (MEGA 6 software) constructed with the major capsid protein (MCP) gene of LCDVs detected in several fish species. Strains are reported with the isolate name and GenBank accession number. Bootstrap values (1000 replicates) above 65% are shown. Substitutions per site are reflected by branch lengths (scale at bottom right).
A phylogenetic analysis of a number of MCP gene nucleotide sequences showed that most of the LCDV were grouped by similar hosts [28].
Through experimental trials, species-specificity has been confirmed for some genotypes. Indeed, olive flounder and rockfish (Sebastes schlegeli) isolates were experimentally infected by their respective homologous isolates but not by the heterologous isolates [30]. However, MCP gene sequencing revealed that the majority of the genotypes are species-specific or have a limited range of host while some exceptions were identified. Gilthead sea bream and Senegalese sole (Solea senegalensis) isolates showed very high MCP nucleotide similarity and clustered in the same genogroup by phylogenetic analysis [31].
Benkaroun and colleagues [28] suggest that the virus speciation could be possibly due to different geographic locations. The observation that the LCDV sequences characterised from three different wild fish species, with similar behaviours and habitats, and that are commonly found in the North Sea, grouped together support this.
Nevertheless, even in this case, some exceptions have been described. High similarity has been observed among sequences of LCDV found in different geographical areas. In some cases, it can be easily explained due to the commercial movement of fish, whereas in other circumstances it is more difficult to explain, such as in the case of an LCDV isolate from a grey gurnard (Eutrigla gurnardus) from the North Sea that clustered with an LCDV isolate from a tropical ornamental fish species from South Korea [28].
Moreover, the sequencing and phylogenetic analysis of a 306-bp fragment of the MCP gene of an isolate detected in juvenile gilthead seabream, imported from a Mediterranean hatchery to a farm in Egypt, demonstrated the presence of LCDV1, originally associated with lymphocystis disease in Northern European countries, expanding de facto both the host and the geographical range of this virus species [32].
Molecular methods also contribute to advancing the knowledge on lymphocystis pathogenesis. The LCDVs show tropism for the dermal tissues infecting fibroblast cells that typically appear enlarged with inclusions and a central nucleus. PCR-based investigations also showed the presence of the virus in many other tissues, including internal organs such as the brain and spleen, pointing out a systemic infection in several species affected by lymphocystis [7,33,34,35]. A further study, applying the ISH technique along with immunohistochemistry (ISH) to experimentally infected gilthead seabream, showed the involvement of hepatocytes and macrophages as target cells for virus replication in addition to the fibroblasts [36]. The implementation of real-time PCR methods, specific for a fragment of the LCDV MCP gene, further improved the diagnostic and study abilities. These methods, in fact, showed a high level of sensitivity and were successfully applied to detect LCDV directly from asymptomatic fish tissues. For this reason, this method can be applied to screen carrier fish [7,37]. The ability of this method not only to detect but also to quantify LCDV in both diseases and asymptomatic fish showed its suitability for pathogenesis studies [7].
To further make the LCDV diagnosis faster and easier, LAMP assays were developed to detect two different lymphocystiviruses. LAMP requires a set of several specific primers that recognise different sequences on the target DNA, achieving highly selective nucleic acid amplification. Due to its high specificity, two different assays were developed, one for the LCDV2, infecting the olive flounder [38] and one for the LCDV3 (also known as genotype VII), which is widespread in gilthead seabream populations [39]. Both the assays provide a fast, reliable, sensitive and specific diagnostic method that can be used for routine diagnosis and surveillance programs in well-equipped laboratories as well as under field conditions.
2.1.2. Megalocytivirus
The implication of iridoviruses other than lymphocystivirus in fish pathology has further stressed the importance of using reliable and detailed (in-depth) identification methods able to distinguish different viruses that can show a similar morphology (irido-like) and cause similar effects on tissues, such as cell enlargement.
Viruses within the genus Megalocytivirus are causative agents of severe disease accompanied by high mortality in multiple species of marine and freshwater fish. Hitherto, megalocytivirus infection has been reported in more than 50 species of fish, resulting in epidemics in ornamental fish, fish farmed for food and also wild fish [1,23].
Sequence analysis clearly segregates megalocytivirus as a separate cluster of viruses with respect to other fish iridoviruses [23]. Adenosine triphosphatase (ATPase) and MCP are key viral genes for megalocytivirus phylogenetic analysis. In fact, most megalocytiviruses show >94% sequence identity within these genes, whereas sequence identity with ranaviruses and lymphocystiviruses is <50% [21]. Furthermore, phylogenetic analyses of these genes cluster megalocytiviruses into three groups: the red seabream iridovirus (RSIV), the infectious spleen and kidney necrosis (ISKNV) and the turbot reddish body iridovirus (TRBIV).
The red seabream iridovirus (RSIV) subgroup comprises megalocytivirus typically isolated from red seabream (Pagrus major) and other marine fish species; the infectious spleen and kidney necrosis (ISKNV) subgroup includes most of the megalocytivirus detected in ornamental fish species, primarily from freshwater fish and the turbot reddish body iridovirus (TRBIV) has been detected mainly from flatfishes, such as flounder and turbot, although some isolates have also been reported from barred knifejaw (Oplegnathus fasciatus) and from ornamental fish species [23,40].
Moreover, further viruses clustering with Megalocytivirus but forming separate branches within this genus have been reported. The threespine stickleback iridovirus (TSIV) is the first megalocytivirus isolated from a wild temperate North American fish, the threespine stickleback (Gasterosteus aculeatus) [41]. The scale drop disease virus (SDDV) was isolated from Asian seabass (Lates calcarifer) affected by the scale drop syndrome, a severe illness with, until then, an unknown aetiology causing significant economic losses in Asian seabass since its first description in 1992 [42]. The European chub iridovirus (ECI) was another divergent megalocytivirus isolated from moribund European chub (Squalius cephalus) [43]. Sequences obtained from traditional Sanger sequencing as well as from next-generation sequencing have been fundamental in the characterisation of these viruses and in their correct taxonomic placement. Particularly, the application of the virus discovery cDNA-AFLP (VIDISCA) approach played a fundamental role in the discovery of the scale drop disease virus [42]. This method consists of a library preparation method that has been successfully used to identify several novel viruses [44,45,46]. The next generation sequencing of the VIDISCA library of sera from fish affected by scale drop syndrome pointed out the sequences of an unknown virus, which was named scale drop disease virus. Starting from the sequences obtained by the VIDISCA approach, a real time qPCR was developed and the near complete genome sequence of the novel virus was obtained via genome walking. The correct identification of the aetiological agent was the starting point to develop efficient diagnostic and control methods [42].
The application of PCR-based methods, genetic and phylogenetic analyses was also very useful to investigate the distribution of already-known megalocytiviruses.
The first outbreak associated with a megalocytivirus was recorded in cultured red sea bream in Japan in 1990 and the virus was designated red sea bream iridovirus disease (RSIVD) [47]. A genetic investigation on archival ornamental fish samples, on the other hand, dated the megalocytivirus index case back to 1986 [40]. Furthermore, this study shows how the application of molecular techniques to archival formalin-fixed and paraffin-embedded (FFPE) materials can provide valuable taxonomic information relating to historical accounts of megalocytivirus-like infections in ornamental fish [40]. The sequence analysis of megalocytivirus cases in ornamental fish observed over an extended period of time showed an interesting distribution of different megalocytivirus species in this group of fish. The TRBIV genotype prevailed among megalocytiviruses affecting ornamental fish since the late 1980s and until the early 1990s, whereas after this date the ISKNV subgroup became the megalocytiviruses most frequently detected in ornamental fish [40].
Megalocytivirus infections are associated with severe systemic diseases. Histologically, enlarged, basophilic, inclusion body-bearing cells (IBC) can be observed within infected organs. IBC are made up of hypertrophied cells with large foamy or granular basophilic inclusions that enlarge the cytoplasm and displace the nucleus (Figure 2).
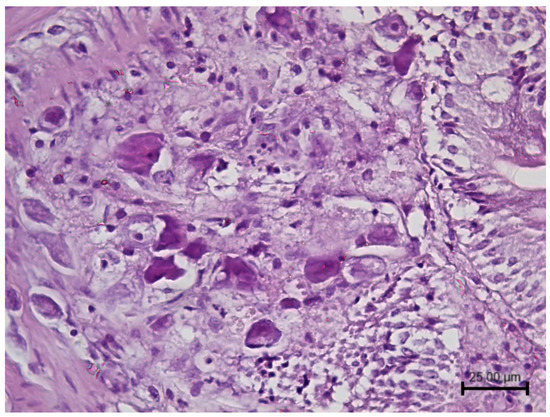
Figure 2.
Histopathology of intestine from a megalocytivirus-infected prickly leather-jacket (Chaetodermis penicilligerus) showing multiple hypertrophic cells containing granular basophilic viral inclusions. Haematoxylin and eosin (H&E) stain. (Courtesy Prof. Gian Enrico Magi University of Camerino, Italy).
IBC cells are considered pathognomonic for infection with megalocytiviruses; however, histology is not conclusive. IBC has been misinterpreted as amoebae, and other non-iridoviral viruses have been associated with some cases [40,48]. Furthermore, the presence of more than one viral species and several genotypes requires diagnostic methods able to distinguish among them. Being the RSIVD listed by the World Organisation for Animal Health (WOAH), it is critical to identify the correct megalocytivirus species to monitor and report it correctly [49]. For this purpose, the WOAH describes diagnostic PCRs that enable the differentiation of RSIV from ISKNV by sequencing or by an RSIV-specific PCR [50]. Moreover, universal and subgroup-specific PCR assays have been set up to amplify all megalocityviruses or only those belonging to a subgroup (RSIV, ISKNV and TRBIV), respectively [23]. In addition to a conventional PCR, a real-time PCR approach has also been applied to megalocytivirus to set up a rapid, specific and sensitive detection method to facilitate surveys of apparently healthy ornamental fish, especially for those subjected to movement, representing a risk of virus spreading across countries [51].
In addition to several PCR assays, the isothermal amplification approach has also been used to develop efficient diagnostic methods for megalocytivirus. A LAMP assay has been developed for each megalocytivirus subgroup: RSIV, ISKN and TRBIV. All three assays showed a sensitivity greater than a conventional PCR and the ability to detect the virus without the use of gel electrophoresis, making these assays suitable for rapid field diagnosis [52,53,54]. A similar approach has been used to set up a rapid field diagnostic method to detect the SDDV. This assay combined the clustered regularly interspaced short palindromic repeats-associated protein 12a (CRISPR-Cas12a)-based nucleic acid detection technology with recombinase polymerase amplification (RPA), reaching high sensitivity and accuracy and the ability to rapidly detect the virus in asymptomatic fish [55].
On the whole, the contribution of the molecular methods has been and continues to be invaluable in the diagnosis and study of megalocytiviruses and throughout the Iridoviridae. However, proper data handling and interpretation are required to avoid misunderstandings.
As above mentioned, MCP and ATPase are key genes in megalocytivirus phylogenesis and their analysis generally provides comparable results. However, genetic analysis also showed potential exceptions to this concordance, such as the megalocytivirus detected in African lampeyes (Lacustricola centralis). African lampeye iridovirus (ALIV) clusters with the red seabream-like megalocytivirus on the basis of the sequence of the ATPase gene, whereas phylogenetic analysis of the MCP gene sequence places this virus firmly as a member of the ISKNV subgroup. This result could be due to the presence of both ISKNV and RSIV-like viruses in African lampeye samples; alternatively, ALIV could be a chimeric virus displaying the MCP of ISKNV and the ATPase of RSIV, so further research is required to resolve this question [23,56].
2.2. Sturgeon NCLDVs
NCLDVs have also been detected in sturgeons; however, so far, sturgeon NCLDVs have not been officially classified as belonging to any of the virus families in this group. Sturgeon NCLDVs are increasingly widespread in sturgeon farming in North America and Europe, causing severe losses to the sector and therefore eliciting more and more interest. However, they have never been isolated in cell culture and this has significantly limited their study and detection. The identification of this viral infection in sturgeons has been historically made through histology and electron microscopy to visualise typical microscopic changes, consisting in prominent enlargement of the infected cells containing an abundant, homogeneous, amphophilic to basophilic cytoplasm and viral particles, respectively [57]. On the basis of the morphology of the virions, these viruses have long been reported as irido-like; however, sequence analyses unveiled a different scenario.
Due to their uncultivability, molecular methods have been the only essential but challenging tool to study these viruses. NCLDVs have been reported in nine sturgeon species of genera Acipenser, Scaphirhynchus and Huso, with significant sequence differences among the viral strains detected in different host species leading to the failure of PCR protocols developed on a different strain [58]. However, the design of degenerate primers on a vast alignment of MCP sequences, including several different strains, led to a PCR protocol able to detect several different sturgeon NCLDVs, including both European and American strains [59].
The phylogenesis of the NCLDVs is particularly difficult due to the fact that their genome evolution involved the lineage-specific expansion of some genes but also the acquisition of numerous genes via horizontal gene transfer from the eukaryotic hosts, other viruses and bacteria [20]. Therefore, the choice of the correct gene/genes for phylogenetic study is crucial. A reliable method for the taxonomic classification of NCLDVs to the species, genus and family levels is the phylogenetic analysis of MCP gene sequences [25].
Phylogenetic analyses of NCLDVs, including North American sturgeon viruses and members of the families Mimiviridae, Phycodnaviridae, Iridoviridae, Marseilleviridae, Ascoviridae and Asfarviridae, using the MCP gene, showed that sturgeon NCLDVs form a cohesive taxonomic group with a distinct evolutionary lineage within the class Megaviricetes to which all these viruses belong. Furthermore, this study shows that, on the basis of phylogenetic analyses using the MCP gene, the sturgeon NCLDVs could be identified to the species or possibly sub-species level. Sturgeon NCLDVs collected from different river basins in North America form distinct taxonomic units located at the tips of the MCP-based phylogenetic tree, defining different North American genotypes. Regarding host specificity, the virus MCP genotypes were not exclusively associated with host species, but MCP analysis suggests that they may be host family- or genus-specific [60]. Analysis of European NCLDVs based on the same gene showed that they form a homogeneous cluster that is closely but distinctively related to North American NCLDVs [59,61].
More recently, a phylogenetic study was conducted using nucleocytoplasmic virus orthologous genes (NCVOGs). NCVOGs are a set of genes that can be mapped onto the genomes of extant NCLDVs, with five NCVOGs shared by all NCLDVs. Going down to the family level, the number of common genes increases, with some genes proposed as hallmark features for a specific family that, in the case of Mimiviridae, have been named MimiCOGs. The application of next-generation sequencing to tissues collected from lake sturgeons associated with Namo virus (NV) mortality outbreaks led to the determination of a novel 306,448 bp long genome sequence, including orthologous genes. Nine orthologous protein sequences were specifically found by analysing this larger portion of the NV genome, one of the North American sturgeon NCLDVs, which enabled the expansion of the phylogenetic investigation of NV beyond the MCP. These nine proteins are regarded as appropriate phylogenetic markers since they are present in all or almost all of the seven virus families of the NCLDV. Despite not being conclusive, this study suggests that NCLDVs represent a new virus lineage in the family Mimiviridae [19]. The Mimiviridae family gathers together several peculiarities, most of which were discovered owing to the use of molecular methods. Mimiviridae includes some of the largest known DNA viruses, both in terms of particle size and genome complexity [62]. The Tupanvirus deep ocean genome consists of 1.51 Mbp, encoding 1425 predicted proteins, surpassing the number of genes encoded by many bacteria and by the smallest parasitic eukaryotic microorganisms [63,64]. However, Mimiviridae is a very large and diverse family of eukaryotic viruses, with the majority is isolated from aquatic environments, including fresh, brackish, and seawater environments. This family is known to have a vastly variable host range, infecting eukaryotic unicellular organisms from at least five major phyla, including Amoebozoa, Haptophyta, Chlorophyta, Excavata and Heterokonta. If NV is confirmed to be a member of the Mimiviridae, this family will have the broadest host range of any known aquatic virus, ranging from algae to fish [64].
4. Nodaviridae
The family Nodaviridae belongs to the order Nodamuvirales and it is currently divided into two genera: Alphanodavirus and Betanodavirus. Nodaviruses are small (29–32 nm in diameter), non-enveloped, icosahedral viruses whose genome is composed of two segments of positive strand single-stranded RNA named RNA1 and RNA2. The RNA1 encodes the viral RNA-dependent RNA polymerase (RdRp), whereas the RNA2 encodes the capsid protein. Alphanodaviruses primarily infect insects. They include Nodamura virus (NoV), Flock house virus (FHV), Black beetle virus (BBV), Boolarra virus (BoV) and Pariacoto virus (PaV) [137]. Betanodaviruses can infect over 50 fish species worldwide and they are the causative agents of viral nervous necrosis (VNN), also known as viral encephalopathy and retinopathy (VER). VNN outbreaks are characterised by clinical signs such as abnormal swimming such as erratic, frenzied or whirling swimming, loss of appetite, hyperinflation of the swim bladder and lightening or darkening of the coloration [138]. Histologically, the brain of the infected animals typically presents multiple intracytoplasmic vacuolations in the grey matter. Moreover, pyknosis, karyorrhexis, neuronal degeneration, inflammatory infiltration, congestion of the blood vessels and sometimes haemorrhages were also described in the nervous tissues of infected fish [139].
Before the 1990s, all known nodaviruses infected insects and were intensely studied because, due to their simple structure, they were considered a paradigm for understanding chemical biology [140]. However, the interest in developing efficient methods to study and detect these viruses further increased after the worldwide occurrence of several fish mortality outbreaks were associated with nodaviruses. Sequence analysis showed clearly that insect and fish viruses clustered separately according to significant differences in their genomes. In particular, a first study focused on the RNA2 segment showed the presence of a region conserved in all fish nodaviruses but absent in insect nodaviruses and the presence of a region more variable within the group of fish nodaviruses [141].
Betanodaviruses were, at first, identified by the name of the host species followed by “nervous necrosis virus” (NNV); however, the analysis of the viral genome showed that betanodaviruses infecting different species clustered together, forming a few genotypes. Phylogenetic analysis of the variable region within the RNA2 segment clusters betanodaviruses into four main genotypes [142]. Later research using the other viral genome fragment, the RNA1, confirmed the clustering of Betanodavirus into four main genotypes [143]. Currently, ICTV recognised the following official species: Striped jack nervous necrosis virus (SJNNV), Tiger puffer nervous necrosis virus (TPNNV), Barfin flounder nervous necrosis virus (BFNNV) and Redspotted grouper nervous necrosis virus (RGNNV) (https://ictv.global/taxonomy accessed on 2 February 2023).
More recently, further nodaviruses from invertebrate hosts other than insects have been detected and associated mainly with crustacean diseases and mortality, leading to a reconsideration of the taxonomy of the family Nodaviridae and the proposal of a new genus: Gammanodavirus. Indeed, the analysis of the genomes of these viruses shows they are clearly distinct from both alpha- and betanodaviruses. In particular, arthropod-infecting viruses have been included in this new proposed genus, such as Macrobrachium rosenbergii nodavirus (MrNV), Penaeus vannamei nodavirus (PvNV), Covert mortality nodavirus (CMNV) and Farfantepenaeus duorarum nodavirus (FdNV) [144,145,146].
Betanodavirus
Betanodavirus was described for the first time from infected larval stripped jack (Pseudocaranx dentex). The name ‘striped jack nervous necrosis virus’ (SJNNV) was consequently adopted [147]. Subsequently, other agents of VNN were isolated from diseased fish species [148]. The present classification of the four species (SJNNV, RGNNV, TPNNV and BFNNV) was proposed for the first time by Nishizawa and colleagues in 1997 [142]. The first studies on host-specificity determinants in betanodaviruses discovered that SJNNV and TPNNV cause diseases in striped jack and tiger puffer (Takifugu rubripes), respectively; BFNNV infects cold-water species, including barfin flounder (Verasper moseri), turbot (Scophthalmus maximus) and Atlantic halibut (Hippoglossus hippoglossus). Conversely, RGNNV has a broad host range and has been isolated from a wide variety of warm-water fish species, especially groupers and sea bass. In order to determine the mechanisms underlying the host specificity, Iwamoto and colleagues [149] developed in vitro reassortants of the RGNNV and SJNNV species. Interestingly, they have found out that the host specificity of SJNNV and RGNNV is determined by the RNA2 segment coding the capsid protein. Moreover, host shifts of betanodaviruses have been attributed to RNA reassortments. In fact, Toffolo and colleagues in 2007 [143] discovered for the first time natural evidence of reassortant strains harbouring the RNA1-SJNNV and RNA2-RGNNV types.
Since betanodavirus classification has been accepted by the scientific community and the ICTV, several betanodavirus strains have been reported. Most of them cluster within one of the well-known genotypes, whereas a few diverge. Turbot nodavirus (TNV) was detected in turbot larvae showing classical signs of VER with abnormal swimming behaviour and high mortality levels. Despite the fact that the virus could not be isolated in cell culture and it resulted negative for an RT-PCR using primers designed to detect the Atlantic halibut nodavirus, a viral genome fragment could be amplified and sequenced with primers complementary to a more conserved region of RNA2. RNA2 analysis demonstrated that TNV differs from previously described fish nodaviruses and that it was not included in any of the four already known betanodavirus genotypes, so it was proposed as a fifth genotype within this genus [150]. On the other hand, betanodaviruses isolated from Atlantic cod (Gadus morrhua), haddock (Melanogrammus aeglefinus) and winter flounder (Pseudopleuronectes americanus) from eastern North America form a monophyletic group, named ‘Atlantic cod nervous necrosis virus’ (ACNNV), within the BFNNV genotype [151]. RNA2 analysis showed that all eastern North American betanodaviruses formed a distinct cluster from the European isolates. Furthermore, these betanodavirus isolates clustered more according to their geographical origin than their host species [151]. More recently, a new genotype has been proposed. A new type of betanodavirus was identified in shellfish and called Korean shellfish nervous necrosis virus (KSNNV) [152]. Phylogenetic analysis shows that KSNNVs form a distinct group only distantly related to other betanodavirus clusters, suggesting that KSNNV is an ancient member of the Betanodavirus genus [152].
The establishment of the striped snakehead cell line (SSN-1), derived from whole fry tissue of the striped snakehead (Ophicephalus striatus), allowed for the first effective isolation of a betanodavirus [153]. Even though NNV isolation in cell culture was a significant breakthrough and was considered the gold standard for detecting NNV infection for two decades, the procedure is time-consuming and labour-intensive, making it unsuitable for routine diagnosis, especially when processing large amounts of samples. For these reasons, molecular techniques have become increasingly relevant over the past few years. Indeed, several PCR-based techniques (RT-PCR, nested PCR and RT-qPCR), targeting RNA1, RNA2 or both genomic segments, have been reported [154]. Nowadays, the WOAH reference centre for viral encephalopathy and retinopathy suggests an RT-qPCR method for NNV diagnosis and surveillance [155] targeting the RNA1 genome segment, able to detect at least 50–100 copies of the target.
For a betanodavirus diagnosis, the isothermal amplification approach has also been explored. In particular, LAMP assays targeting the RNA2 segment of the RGNNV genotype have been developed in order to provide time- and cost-saving diagnostic tools. LAMP methods have the ability to amplify a target in one hour and do not require highly specialised laboratories. For these reasons, diagnostic methods using LAMP are also suitable for a company’s self-control procedures. However, the detection limit of the developed RT-LAMP applied for NNV detection was lower compared to qPCR assays [156,157,158].
Recently, evidence of natural reassortants between the RGNNV and SJNNV genotypes in southern Europe emerged from the sequencing of both genomic segments. In particular, both combinations of genomic segments, SJNNV/RGNNV and RGNNV/SJNNV (RNA1/RNA2), have been observed in viral isolates obtained from fish, although the second type has been detected more often [139,159,160,161]. The emergence of the reassortant strains and the large number of betanodavirus strains circulating at the same time in several geographical areas have promoted the application of molecular methods able to distinguish and identify the nature of the detected viruses. In this prospect, a multiplex RT-PCR assay able to distinguish among RGNNV, SJNNV and their reassortants has been developed, providing an easy, rapid and affordable molecular method to gain information about circulating NNV genotypes [5].
Moreover, the recent application of whole genome sequencing (WGS) to the study of aquatic viruses is gaining importance due to the large amount of information that it could generate. Accordingly, the WGS has permitted the identification of variable amino-acid residues probably correlated to the NNV virulence and pathogenicity thanks to the analysis of the NNV reassortant strains’ RdRp and capsid protein physico-chemical characteristics [162]. In particular, the amino acids (aa) 247 and 270 of capsid protein have been identified as putative viral receptor binding sites; accordingly, substitutions at these positions could affect the virus–host interaction. In addition, Biasini and colleagues [162] speculated on the potential existence of additional interesting residues that are all exposed on the outside of the structure of the trimeric P-domains. In particular, seven variable amino acids were identified and it was possible to speculate using protein-structure prediction that changes in these positions alter the hydrophobicity and the steric hindrance of the exposed surface region, changing the physiochemical properties of the capsid protein.
Molecular methods have also been successfully applied to identify specific areas of the central nervous system involved in NNV infection, providing a crucial understanding of the pathogenesis of this virus. Wang and colleagues [9], with a combined application of molecular-based methods such as real-time quantitative chain reaction (RT-qPCR) and FISH, were able to measure RGNNV content in different brain regions of an experimentally infected orange spotted grouper (Epinephelus coioides). The probe used in the FISH assay, targeting the capsid protein of RGNNV, revealed the presence of specific signals in the midbrain region. Furthermore, using quantitative (RT-qPCR) and qualitative (ISH) molecular methods, it is possible to detect the presence of the NNV genome as well as infectious particles in both nervous and non-nervous tissues and to study the viral uptake during the infection process. The application of ISH to an experimentally infected seven-band grouper (Hyporthodus septemfasciatus) was able to localise the RNA2 segment of the virus in the gill district during the first stages of NNV infection and in the blood–brain barrier, followed by the brain in the final stage of infection [163]. Lastly, the ISH has also been applied to investigate the NNV histopathology in other aquatic animal species, such as the big-belly seahorse (Hippocampus abdominalis). In particular, the application of different molecular methods (i.e., PCR detection, RT-qPCR and FISH) to the tissues of this aquatic vertebrate allowed the identification and localisation of a new NNV strain, closely related to the RGNNV genotype [164].
5. Discussion and Conclusions
With an average of 107 virus-like particles per mL−1, an abundance typically ranging from 104 to 108 mL–1 and an order of magnitude greater than bacteria, viruses are widespread elements of aquatic ecosystems [3]. The majority of them are harmless to humans and animals and are essential for global and small-scale biogeochemical cycles, community structure, gene transfer and the evolution of aquatic organisms. Furthermore, some of them are even beneficial to animals and ecosystems infecting and then controlling aquatic pathogenic bacteria or harmful algae [3]. However, several aquatic viruses can infect animals, leading to diseases, especially when they are confined, such as in aquaculture facilities.
Traditional methods, such as viral isolation in cell cultures and antigen/antibody detection assays, have been widely applied to detect and study aquatic animals’ viruses, leading to the successful isolation, identification and understanding of several of them. These methods offer several advantages and are still indispensable many times. However, they also present several limitations, especially in their application to aquatic animals’ virology. As seen from the many reported examples, several fish viruses are not cultivable in cell cultures; in fact, some of them can only hardly or very slowly replicate in vitro; furthermore, the use of virus isolation in cell cultures is barred from the study of shellfish diseases due to the lack of suitable cell cultures. As a matter of fact, despite decades of attempts, not even a single cell line of marine molluscs or crustaceans is available [165,166]. Cell cultures are systems with a sensitive balance that are often subjected to toxic samples. Nevertheless, isolation in cell culture is often time-consuming, making it not ideal for routine processing, especially for surveillance when a number of samples need to be tested.
Serology is another traditional approach used to detect and study viral infections, with a limited application to aquatic animals’ viral diseases. Obviously, it is not accessible for animals that do not produce antibodies; however, it is often considered an inappropriate or unreliable method for viral diagnosis in finfish too [167,168]. In fact, fish antibody production following infection is slow, especially at low water temperatures, resulting in the absence of antibodies or a lack of homogeneity in antibody production in infected populations.
Molecular methods have significantly contributed to overcoming the limitations of traditional ones. Furthermore, intensive farming conditions and an increasing number of farmed fish species across the animal kingdom, including different phyla, have boosted the number of pathogens that humans and animal production have to deal with, requiring the quick development of new detection and study methods for novel unknown pathogens.
Molecular methods provide undoubtedly a powerful tool to detect, identify and characterise both known and unknown pathogens. A standard PCR protocol followed by the sequencing of purified amplicons is an effective method for both identifying well-known viruses and discovering new ones. PCR-based methods are taking place in surveillance programmes, either alongside or, in some cases, replacing the traditional methods. The PCR method is quick, allows the simultaneous examination of numerous samples, and is highly sensitive, with some PCR assays even claiming to be able to detect just a single copy of the target DNA.
On the other hand, sequencing techniques, especially next generation sequencing have recently become irreplaceable tools in emerging infectious disease research. Moreover, the increasing availability of sequences in databases makes it easier to compare and ascribe new sequences to specific taxa. A wide range of sequences is the starting point of all epidemiological studies and the identification of viruses at the species or even sub-species level can further refine epidemiological studies.
However, molecular methods also have limitations that are good to know. PCR assays are able to detect nucleic acids regardless of virus integrity; obviously the persistence of nucleic acids outside virions, especially RNA, is limited; however, their occasional findings, especially in new host species or uncommon substrates, needs to be ascertained. Furthermore, PCR positivity cannot be univocally associated with active viral replication or active infection and needs to be confirmed with the additional analysis. This constraint has to be kept in mind, especially with animals able to passively accumulate viruses in their flesh, such as bivalve molluscs. In this regard, in addition to traditional methods, which are not always applicable, other molecular methods can be helpful. Transcript detection, for example, can be pointed out through an RT-PCR for DNA viruses or by ISH using complementary RNA probes, demonstrating an active replication status.
Another critical point is the optimisation and validation of protocols. PCR assays are developed through the design of primers, which need to be as specific as possible but, at the same time, have to match a conserved sequence to avoid mismatches in the case of sequence variations. As a result, not all PCR protocols can provide adequate specificity and sensitivity fit for purpose, so especially for surveillance, it is fundamental to use validated protocols.
Despite these constraints, in the last few years the study of viral pathogens has relied on the invaluable contribution of molecular techniques primarily based on PCR, sequencing and in situ hybridisation gaining crucial knowledge about viral aquatic animal diseases. Finally, the study of viral aquatic animal diseases should not be separated from the development of new innovative techniques or the application of already available molecular methods in order to further increase their in-depth understanding.
Author Contributions
Conceptualisation, S.C. and L.M.; writing—original draft preparation, S.C., F.E., L.M. and E.V.; writing—review and editing, S.C., F.E., L.M. and E.V.; visualisation, E.V., L.M. and S.C.; supervision, S.C. All authors have read and agreed to the published version of the manuscript.
Funding
The author E.V. was supported by the Project code CN_00000033, Concession Decree No. 1034 of 17 June 2022 adopted by the Italian Ministry of University and Research, CUP J33C22001190001, Project title “National Biodiversity Future Center—NBFC” under the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.4—Call for tender No. 3138 of 16 December 2021, rectified by Decree n.3175 of 18 December 2021 of Italian Ministry of University and Research funded by the European Union—NextGenerationEU.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fonseca, A.; Laguardia-Nascimento, M.; Scotá Ferreira, A.; Pinto, C.; Pereira Freitas, T.; Rivetti Júnior, A.; Ferreira Homem, V.; Camargos, M. Detection of Megalocytivirus in Oreochromis Niloticus and Pseudoplatystoma Corruscans in Brazil. Dis. Aquat. Organ. 2022, 149, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, S.; Afzal, M.; Sikandar, S.; Afzal, I. Polymerase Chain Reaction. In Genetic Engineering—A Glimpse of Techniques and Applications; Jamal, F., Ed.; IntechOpen: London, UK, 2020; ISBN 978-1-78985-179-3. [Google Scholar]
- Jacquet, S.; Miki, T.; Noble, R.; Peduzzi, P.; Wilhelm, S. Viruses in Aquatic Ecosystems: Important Advancements of the Last 20 Years and Prospects for the Future in the Field of Microbial Oceanography and Limnology. Adv. Oceanogr. Limnol. 2010, 1, 97–141. [Google Scholar] [CrossRef]
- Pinheiro, A.C.A.S.; Volpe, E.; Principi, D.; Prosperi, S.; Ciulli, S. Development of a Multiplex RT-PCR Assay for Simultaneous Detection of the Major Viruses That Affect Rainbow Trout (Oncorhynchus Mykiss). Aquac. Int. 2016, 24, 115–125. [Google Scholar] [CrossRef]
- Errani, F.; Volpe, E.; Riera-Ferrer, E.; Caffara, M.; Padrós, F.; Gustinelli, A.; Fioravanti, M.; Ciulli, S. Development and Diagnostic Validation of a One-Step Multiplex RT-PCR Assay as a Rapid Method to Detect and Identify Nervous Necrosis Virus (NNV) and Its Variants Circulating in the Mediterranean. PLoS ONE 2022, 17, e0273802. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The Real-Time Polymerase Chain Reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef]
- Ciulli, S.; Pinheiro, A.C.d.A.S.; Volpe, E.; Moscato, M.; Jung, T.S.; Galeotti, M.; Stellino, S.; Farneti, R.; Prosperi, S. Development and Application of a Real-Time PCR Assay for the Detection and Quantitation of Lymphocystis Disease Virus. J. Virol. Methods 2015, 213, 164–173. [Google Scholar] [CrossRef]
- Morley, A.A. Digital PCR: A Brief History. Biomol. Detect. Quantif. 2014, 1, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, Z.; Jing, H.; Zhang, M.; Wu, S.; Lin, X. Development of a Novel Droplet Digital PCR Assay for the Sensitive Detection of Carp Edema Virus. Aquaculture 2021, 545, 737162. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, M.-Z.; Xiao, H.-H.; Yi, Y.; Cheng, H.; Putra, D.-F.; Li, S.-Q.; Li, P.-F. Selection and Characterization of Aptamers for Specific Detection of Iridovirus Disease in Cultured Hybrid Grouper (Epinephelus Fuscoguttatus♀ × E. Lanceolatus♂). Chin. J. Anal. Chem. 2020, 48, 650–661. [Google Scholar] [CrossRef]
- Pfankuche, V.M.; Hahn, K.; Bodewes, R.; Hansmann, F.; Habierski, A.; Haverkamp, A.-K.; Pfaender, S.; Walter, S.; Baechlein, C.; Postel, A.; et al. Comparison of Different In Situ Hybridization Techniques for the Detection of Various RNA and DNA Viruses. Viruses 2018, 10, 384. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, A.; Jones, J. Developments in in Situ Hybridisation. Methods S. Diego Calif. 2014, 70, 39–45. [Google Scholar] [CrossRef]
- Jansen, G.J.; Wiersma, M.; van Wamel, W.J.B.; Wijnberg, I.D. Direct Detection of SARS-CoV-2 Antisense and Sense Genomic RNA in Human Saliva by Semi-Autonomous Fluorescence in Situ Hybridization: A Proxy for Contagiousness? PLoS ONE 2021, 16, e0256378. [Google Scholar] [CrossRef]
- Arzul, I.; Nicolas, J.-L.; Davison, A.J.; Renault, T. French Scallops: A New Host for Ostreid Herpesvirus-1. Virology 2001, 290, 342–349. [Google Scholar] [CrossRef] [PubMed]
- OIE CHAPTER 2.4.9 Infection with Ostreid Herpesvirus 1 Microvariant. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2013.
- Snow, M. The Contribution of Molecular Epidemiology to the Understanding and Control of Viral Diseases of Salmonid Aquaculture. Vet. Res. 2011, 42, 56. [Google Scholar] [CrossRef] [PubMed]
- Tengs, T.; Rimstad, E. Emerging Pathogens in the Fish Farming Industry and Sequencing-Based Pathogen Discovery. Dev. Comp. Immunol. 2017, 75, 109–119. [Google Scholar] [CrossRef]
- Clouthier, S.; Anderson, E.; Kurath, G.; Breyta, R. Molecular Systematics of Sturgeon Nucleocytoplasmic Large DNA Viruses. Mol. Phylogenet. Evol. 2018, 128, 26–37. [Google Scholar] [CrossRef]
- Iyer, L.M.; Balaji, S.; Koonin, E.V.; Aravind, L. Evolutionary Genomics of Nucleo-Cytoplasmic Large DNA Viruses. Virus Res. 2006, 117, 156–184. [Google Scholar] [CrossRef]
- Chinchar, V.G.; Waltzek, T.B.; Subramaniam, K. Ranaviruses and Other Members of the Family Iridoviridae: Their Place in the Virosphere. Virology 2017, 511, 259–271. [Google Scholar] [CrossRef]
- Wolf, K.; Gravell, M.; Malsberger, R.G. Lymphocystis Virus: Isolation and Propagation in Centrarchid Fish Cell Lines. Science 1966, 151, 1004–1005. [Google Scholar] [CrossRef]
- Kurita, J.; Nakajima, K. Megalocytiviruses. Viruses 2012, 4, 521–538. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.; Miller, D.; Hoverman, J. Ecology and Pathology of Amphibian Ranaviruses. Dis. Aquat. Organ. 2009, 87, 243–266. [Google Scholar] [CrossRef] [PubMed]
- Jancovich, J.K.; Chinchar, V.G.; Hyatt, A.D.; Miyazaki, T.; Williams, T.; Zhang, Q.Y. Family Iridoviridae. In Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier Academic Press: San Diego, CA, USA, 2011; pp. 193–210. [Google Scholar]
- Volpatti, D.; Ciulli, S. Chapter 15 Lymphocystis Virus Disease. In Aquaculture Pathophysiology Volume I. Fish Diseases; Elsevier Academic Press: New York, NY, USA, 2022; pp. 201–216. [Google Scholar]
- Doszpoly, A.; Kaján, G.L.; Puentes, R.; Perretta, A. Complete Genome Sequence and Analysis of a Novel Lymphocystivirus Detected in Whitemouth Croaker (Micropogonias Furnieri): Lymphocystis Disease Virus 4. Arch. Virol. 2020, 165, 1215–1218. [Google Scholar] [CrossRef] [PubMed]
- Benkaroun, J.; Bergmann, S.M.; Römer-Oberdörfer, A.; Demircan, M.D.; Tamer, C.; Kachh, G.R.; Weidmann, M. New Insights into Lymphocystis Disease Virus Genome Diversity. Viruses 2022, 14, 2741. [Google Scholar] [CrossRef]
- Mönttinen, H.A.M.; Bicep, C.; Williams, T.A.; Hirt, R.P. The Genomes of Nucleocytoplasmic Large DNA Viruses: Viral Evolution Writ Large. Microb. Genom. 2021, 7, 000649. [Google Scholar] [CrossRef]
- Kitamura, S.-I.; Jung, S.-J.; Kim, W.-S.; Nishizawa, T.; Yoshimizu, M.; Oh, M.-J. A New Genotype of Lymphocystivirus, LCDV-RF, from Lymphocystis Diseased Rockfish. Arch. Virol. 2006, 151, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Cano, I.; Valverde, E.J.; Lopez-Jimena, B.; Alonso, M.C.; Garcia-Rosado, E.; Sarasquete, C.; Borrego, J.J.; Castro, D. A New Genotype of Lymphocystivirus Isolated from Cultured Gilthead Seabream, Sparus Aurata L., and Senegalese Sole, Solea Senegalensis (Kaup). J. Fish Dis. 2010, 33, 695–700. [Google Scholar] [CrossRef]
- Shawky, M.; Taha, E.; Ahmed, B.; Mahmoud, M.A.; Abdelaziz, M.; Faisal, M.; Yousif, A. Initial Evidence That Gilthead Seabream (Sparus Aurata L.) Is a Host for Lymphocystis Disease Virus Genotype I. Animals 2021, 11, 3032. [Google Scholar] [CrossRef]
- Kvitt, H.; Heinisch, G.; Diamant, A. Detection and Phylogeny of Lymphocystivirus in Sea Bream Sparus Aurata Based on the DNA Polymerase Gene and Major Capsid Protein Sequences. Aquaculture 2008, 275, 58–63. [Google Scholar] [CrossRef]
- Wu, R.-H.; Tang, X.-Q.; Sheng, X.-Z.; Zhan, W.-B. Tissue Distribution of the 27.8 KDa Receptor and Its Dynamic Expression in Response to Lymphocystis Disease Virus Infection in Flounder (Paralichthys Olivaceus). J. Comp. Pathol. 2015, 153, 324–332. [Google Scholar] [CrossRef]
- Volpe, E.; Farneti, R.; Moscato, M.; Prosperi, S.; Ciulli, S. Distribuzione Di Lymphocystivirus in Organi Target e Non Target Di Orate (Sparus Aurata) Naturalmente Infette. Ittiopatologia 2015, 12, 129–137. Available online: https://core.ac.uk/display/226695907 (accessed on 2 February 2023).
- Cano, I.; Ferro, P.; Alonso, M.C.; Sarasquete, C.; Garcia-Rosado, E.; Borrego, J.J.; Castro, D. Application of in Situ Detection Techniques to Determine the Systemic Condition of Lymphocystis Disease Virus Infection in Cultured Gilt-Head Seabream, Sparus Aurata L. J. Fish Dis. 2009, 32, 143–150. [Google Scholar] [CrossRef]
- Valverde, E.J.; Cano, I.; Labella, A.; Borrego, J.J.; Castro, D. Application of a New Real-Time Polymerase Chain Reaction Assay for Surveillance Studies of Lymphocystis Disease Virus in Farmed Gilthead Seabream. BMC Vet. Res. 2016, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yue, Z.; Liu, H.; Liang, C.; Zheng, X.; Zhao, Y.; Chen, X.; Xiao, X.; Chen, C. Development and Evaluation of a Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Lymphocystis Disease Virus. J. Virol. Methods 2010, 163, 378–384. [Google Scholar] [CrossRef]
- Valverde, E.J.; Borrego, J.J.; Sarasquete, M.C.; Ortiz-Delgado, J.B.; Castro, D. Target Organs for Lymphocystis Disease Virus Replication in Gilthead Seabream (Sparus Aurata). Vet. Res. 2017, 48, 21. [Google Scholar] [CrossRef] [PubMed]
- Go, J.; Waltzek, T.; Subramaniam, K.; Yun, S.; Groff, J.; Anderson, I.; Chong, R.; Shirley, I.; Schuh, J.; Handlinger, J.; et al. Detection of Infectious Spleen and Kidney Necrosis Virus (ISKNV) and Turbot Reddish Body Iridovirus (TRBIV) from Archival Ornamental Fish Samples. Dis. Aquat. Organ. 2016, 122, 105–123. [Google Scholar] [CrossRef] [PubMed]
- Waltzek, T.; Marty, G.; Alfaro, M.; Bennett, W.; Garver, K.; Haulena, M.; Weber ES, I.; Hedrick, R. Systemic Iridovirus from Threespine Stickleback Gasterosteus Aculeatus Represents a New Megalocytivirus Species (Family Iridoviridae). Dis. Aquat. Organ. 2012, 98, 41–56. [Google Scholar] [CrossRef]
- de Groof, A.; Guelen, L.; Deijs, M.; van der Wal, Y.; Miyata, M.; Ng, K.S.; van Grinsven, L.; Simmelink, B.; Biermann, Y.; Grisez, L.; et al. A Novel Virus Causes Scale Drop Disease in Lates Calcarifer. PLoS Pathog. 2015, 11, e1005074. [Google Scholar] [CrossRef]
- Halaly, M.A.; Subramaniam, K.; Koda, S.A.; Popov, V.L.; Stone, D.; Way, K.; Waltzek, T.B. Characterization of a Novel Megalocytivirus Isolated from European Chub (Squalius Cephalus). Viruses 2019, 11, 440. [Google Scholar] [CrossRef]
- Canuti, M.; Eis-Huebinger, A.M.; Deijs, M.; de Vries, M.; Drexler, J.F.; Oppong, S.K.; Müller, M.A.; Klose, S.M.; Wellinghausen, N.; Cottontail, V.M.; et al. Two Novel Parvoviruses in Frugivorous New and Old World Bats. PLoS ONE 2011, 6, e29140. [Google Scholar] [CrossRef]
- Tan, L.V.; Van Doorn, H.R.; Van der Hoek, L.; Minh Hien, V.; Jebbink, M.F.; Quang Ha, D.; Farrar, J.; Van Vinh Chau, N.; de Jong, M.D. Random PCR and Ultracentrifugation Increases Sensitivity and Throughput of VIDISCA for Screening of Pathogens in Clinical Specimens. J. Infect. Dev. Ctries. 2011, 5, 142–148. [Google Scholar] [CrossRef]
- van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.M.; Wolthers, K.C.; Wertheim-van Dillen, P.M.E.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a New Human Coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef]
- Inouye, K.; Yamano, K.; Maeno, Y.; Nakajima, K.; Matsuoka, M.; Wada, Y.; Sorimachi, M. Iridovirus Infection of Cultured Red Sea Bream, Pagrus major. Fish Pathol. 1992, 27, 19–27. [Google Scholar] [CrossRef]
- Stephens, F.J.; Jones, J.B.; Hillier, P. Ornamental Fish Testing Project: Final Report; Government of Western Australia Department of Fisheries and Fisheries Research Division WA Marine Research Laboratories: Perth, WA, Australia, 2009.
- OIE CHAPTER 2.3.7 Red Sea Bream Iridoviral Disease. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2021; pp. 332–343.
- Kurita, J.; Nakajima, K.; Hirono, I.; Aoki, T. Polymerase Chain Reaction (PCR) Amplification of DNA of Red Sea Bream [Pagrus Major] Iridovirus (RSIV). Fish Pathol. Jpn. 1998, 33, 17–23. [Google Scholar] [CrossRef]
- Rimmer, A.E.; Becker, J.A.; Tweedie, A.; Whittington, R.J. Development of a Quantitative Polymerase Chain Reaction (QPCR) Assay for the Detection of Dwarf Gourami Iridovirus (DGIV) and Other Megalocytiviruses and Comparison with the Office International Des Epizooties (OIE) Reference PCR Protocol. Aquaculture 2012, 358–359, 155–163. [Google Scholar] [CrossRef]
- Caipang, C.M.A.; Haraguchi, I.; Ohira, T.; Hirono, I.; Aoki, T. Rapid Detection of a Fish Iridovirus Using Loop-Mediated Isothermal Amplification (LAMP). J. Virol. Methods 2004, 121, 155–161. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, C.; Huang, J.; Jia, K.; Chen, X.; Liu, H. Rapid Diagnosis of Turbot Reddish Body Iridovirus in Turbot Using the Loop-Mediated Isothermal Amplification Method. J. Virol. Methods 2009, 158, 18–23. [Google Scholar] [CrossRef]
- Ding, W.C.; Chen, J.; Shi, Y.H.; Lu, X.J.; Li, M.Y. Rapid and Sensitive Detection of Infectious Spleen and Kidney Necrosis Virus by Loop-Mediated Isothermal Amplification Combined with a Lateral Flow Dipstick. Arch. Virol. 2010, 155, 385–389. [Google Scholar] [CrossRef]
- Sukonta, T.; Senapin, S.; Meemetta, W.; Chaijarasphong, T. CRISPR-based Platform for Rapid, Sensitive and Field-deployable Detection of Scale Drop Disease Virus in Asian Sea Bass (Lates Calcarifer). J. Fish Dis. 2022, 45, 107–120. [Google Scholar] [CrossRef]
- Sudthongkong, C.; Miyata, M.; Miyazaki, T. Viral DNA Sequences of Genes Encoding the ATPase and the Major Capsid Protein of Tropical Iridovirus Isolates Which Are Pathogenic to Fishes in Japan, South China Sea and Southeast Asian Countries. Arch. Virol. 2002, 147, 2089–2109. [Google Scholar] [CrossRef]
- Adkinson, M.A.; Cambre, M.; Hedrick, R.P. Identification of an Iridovirus in Russian Sturgeon (Acipenser Guldenstadi) from Northern Europe. Bull. Eur. Assoc. Fish Pathol. UK 1998, 18, 29–32. [Google Scholar]
- Drennan, J.D.; Lapatra, S.E.; Samson, C.A.; Ireland, S.; Eversman, K.F.; Cain, K.D. Evaluation of Lethal and Non-Lethal Sampling Methods for the Detection of White Sturgeon Iridovirus Infection in White Sturgeon, Acipenser Transmontanus (Richardson). J. Fish Dis. 2007, 30, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Ciulli, S.; Volpe, E.; Sirri, R.; Passalacqua, P.L.; Cesa Bianchi, F.; Serratore, P.; Mandrioli, L. Outbreak of Mortality in Russian (Acipenser Gueldenstaedtii) and Siberian (Acipenser Baerii) Sturgeons Associated with Sturgeon Nucleo-Cytoplasmatic Large DNA Virus. Vet. Microbiol. 2016, 191, 27–34. [Google Scholar] [CrossRef]
- Clouthier, S.; VanWalleghem, E.; Anderson, E. Sturgeon Nucleo-Cytoplasmic Large DNA Virus Phylogeny and PCR Tests. Dis. Aquat. Organ. 2015, 117, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Bigarré, L.; Lesne, M.; Lautraite, A.; Chesneau, V.; Leroux, A.; Jamin, M.; Boitard, P.M.; Toffan, A.; Prearo, M.; Labrut, S.; et al. Molecular Identification of Iridoviruses Infecting Various Sturgeon Species in Europe. J. Fish Dis. 2017, 40, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; Audic, S.; Robert, C.; Abergel, C.; Renesto, P.; Ogata, H.; La Scola, B.; Suzan, M.; Claverie, J.-M. The 1.2-Megabase Genome Sequence of Mimivirus. Science 2004, 306, 1344–1350. [Google Scholar] [CrossRef]
- Abrahão, J.; Silva, L.; Silva, L.S.; Khalil, J.Y.B.; Rodrigues, R.; Arantes, T.; Assis, F.; Boratto, P.; Andrade, M.; Kroon, E.G.; et al. Tailed Giant Tupanvirus Possesses the Most Complete Translational Apparatus of the Known Virosphere. Nat. Commun. 2018, 9, 749. [Google Scholar] [CrossRef]
- Claverie, J.-M.; Abergel, C. Mimiviridae: An Expanding Family of Highly Diverse Large DsDNA Viruses Infecting a Wide Phylogenetic Range of Aquatic Eukaryotes. Viruses 2018, 10, 506. [Google Scholar] [CrossRef]
- Boutier, M.; Gao, Y.; Donohoe, O.; Vanderplasschen, A. Current Knowledge and Future Prospects of Vaccines against Cyprinid Herpesvirus 3 (CyHV-3). Fish Shellfish. Immunol. 2019, 93, 531–541. [Google Scholar] [CrossRef]
- Hanson, L.; Dishon, A.; Kotler, M. Herpesviruses That Infect Fish. Viruses 2011, 3, 2160–2191. [Google Scholar] [CrossRef]
- Poli, G.; Dall’Ara, P.; Martino, P.A.; Rosati, S. Microbiologia e Immunologia Veterinaria, 3rd ed.; Edra: Milan, Italy, 2017; ISBN 978-88-214-4227-8. [Google Scholar]
- Umene, K.; Sakaoka, H. Evolution of Herpes Simplex Virus Type 1 under Herpesviral Evolutionary Processes. Arch. Virol. 1999, 144, 637–656. [Google Scholar] [CrossRef]
- Sierra, E.; Fernández, A.; Fernández-Maldonado, C.; Sacchini, S.; Felipe-Jiménez, I.; Segura-Göthlin, S.; Colom-Rivero, A.; Câmara, N.; Puig-Lozano, R.; Rambaldi, A.M.; et al. Molecular Characterization of Herpesviral Encephalitis in Cetaceans: Correlation with Histopathological and Immunohistochemical Findings. Animals 2022, 12, 1149. [Google Scholar] [CrossRef]
- Lovy, J.; Friend, S.E. Cyprinid Herpesvirus-2 Causing Mass Mortality in Goldfish: Applying Electron Microscopy to Histological Samples for Diagnostic Virology. Dis. Aquat. Organ. 2014, 108, 1–9. [Google Scholar] [CrossRef]
- Doszpoly, A.; Benkő, M.; Bovo, G.; LaPatra, S.E.; Harrach, B. Comparative Analysis of a Conserved Gene Block from the Genome of the Members of the Genus Ictalurivirus. Intervirology 2011, 54, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Freitas, J.T.; Subramaniam, K.; Kelley, K.L.; Marcquenski, S.; Groff, J.; Waltzek, T.B. Genetic Characterization of Esocid Herpesvirus 1 (EsHV1). Dis. Aquat. Organ. 2016, 122, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sunarto, A.; McColl, K.A.; Crane, M.S.J.; Sumiati, T.; Hyatt, A.D.; Barnes, A.C.; Walker, P.J. Isolation and Characterization of Koi Herpesvirus (KHV) from Indonesia: Identification of a New Genetic Lineage: Indonesian KHV Characterization. J. Fish Dis. 2011, 34, 87–101. [Google Scholar] [CrossRef]
- Waltzek, T.B.; Kelley, G.O.; Alfaro, M.E.; Kurobe, T.; Davison, A.J.; Hedrick, R.P. Phylogenetic Relationships in the Family Alloherpesviridae. Dis. Aquat. Organ. 2009, 84, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Pellett, P.E.; Davison, A.J.; Ederle, R.; Ehlers, B.; Hayward, G.S.; Lacoste, V.; Minson, A.C.; Nicholas, J.; Roizman, B.; Studdert, M.J.; et al. Order Herpesvirales. In Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier Academic Press: San Diego, CA, USA, 2011; pp. 99–107. [Google Scholar]
- Gotesman, M.; Kattlun, J.; Bergmann, S.M.; El-Matbouli, M. CyHV-3: The Third Cyprinid Herpesvirus. Dis. Aquat. Organ. 2013, 105, 163–174. [Google Scholar] [CrossRef]
- McAllister, P.E.; Lidgerding, B.C.; Herman, R.L.; Hoyer, L.C.; Hankins, J. Viral Diseases of Fish: First Report of Carp Pox in Golden Ide (Leuciscus Idus) in North America. J. Wildl. Dis. 1985, 21, 199–204. [Google Scholar] [CrossRef]
- Hedrick, R.P.; Groff, J.M.; Okihiro, M.S.; McDowell, T.S. Herpesviruses Detected in Papillomatous Skin Growths of Koi Carp (Cyprinus Carpio). J. Wildl. Dis. 1990, 26, 578–581. [Google Scholar] [CrossRef]
- Sano, N.; Sano, M.; Sano, T.; Hondo, R. Herpesvirus Cyprini: Detection of the Viral Genome by in Situ Hybridization. J. Fish Dis. 1992, 15, 153–162. [Google Scholar] [CrossRef]
- Sano, N.; Moriwake, M.; Hondo, R.; Sano, T. Herpesvirus Cyprini: A Search for Viral Genome in Infected Fish by Infected Fish by in Situ Hybridization. J. Fish Dis. 1993, 16, 495–499. [Google Scholar] [CrossRef]
- Way, K.; Dixon, P.F. Koi Herpesvirus Disease. In Fish Viruses and Bacteria: Pathobiology and Protection; CABI Digital Library: Long Beach, CA, USA, 2017; pp. 115–127. ISBN 978-1-78064-778-4. [Google Scholar]
- Lievens, B.; Frans, I.; Heusdens, C.; Justé, A.; Jonstrup, S.P.; Lieffrig, F.; Willems, K.A. Rapid Detection and Identification of Viral and Bacterial Fish Pathogens Using a DNA Array-Based Multiplex Assay. J. Fish Dis. 2011, 34, 861–875. [Google Scholar] [CrossRef]
- Sirri, R.; Ciulli, S.; Barbé, T.; Volpe, E.; Lazzari, M.; Franceschini, V.; Errani, F.; Sarli, G.; Mandrioli, L. Detection of Cyprinid Herpesvirus 1 DNA in Cutaneous Squamous Cell Carcinoma of Koi Carp (Cyprinus Carpio). Vet. Dermatol. 2018, 29, 60-e24. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, K.; Kurita, J.; Kawana, M.; Kiryu, I.; Oseko, N.; Sano, M. Development of MRNA-Specific RT-PCR for the Detection of Koi Herpesvirus (KHV) Replication Stage. Dis. Aquat. Organ. 2012, 100, 11–18. [Google Scholar] [CrossRef]
- Xu, J.; Zeng, L.; Zhang, H.; Zhou, Y.; Ma, J.; Fan, Y. Cyprinid Herpesvirus 2 Infection Emerged in Cultured Gibel Carp, Carassius Auratus Gibelio in China. Vet. Microbiol. 2013, 166, 138–144. [Google Scholar] [CrossRef]
- Jiang, N.; Xu, J.; Ma, J.; Fan, Y.; Zhou, Y.; Liu, W.; Zeng, L. Histopathology and Ultrastructural Pathology of Cyprinid Herpesvirus II (CyHV-2) Infection in Gibel Carp, Carassius Auratus Gibelio. Wuhan Univ. J. Nat. Sci. 2015, 20, 413–420. [Google Scholar] [CrossRef]
- Fichi, G.; Susini, F.; Cocumelli, C.; Cersini, A.; Salvadori, M.; Guarducci, M.; Cardeti, G. New Detection of Cyprinid Herpesvirus 2 in Mass Mortality Event of Carassius Carassius (L.), in Italy. J. Fish Dis. 2016, 39, 1523–1527. [Google Scholar] [CrossRef]
- Tang, R.; Lu, L.; Wang, B.; Yu, J.; Wang, H. Identification of the Immediate-Early Genes of Cyprinid Herpesvirus 2. Viruses 2020, 12, 994. [Google Scholar] [CrossRef]
- Wang, H.; Xu, L.; Lu, L. Detection of Cyprinid Herpesvirus 2 in Peripheral Blood Cells of Silver Crucian Carp, Carassius Auratus Gibelio (Bloch), Suggests Its Potential in Viral Diagnosis. J. Fish Dis. 2016, 39, 155–162. [Google Scholar] [CrossRef]
- Ding, Z.; Xia, S.; Zhao, Z.; Xia, A.; Shen, M.; Tang, J.; Xue, H.; Geng, X.; Yuan, S. Histopathological Characterization and Fluorescence in Situ Hybridization of Cyprinid Herpesvirus 2 in Cultured Prussian Carp, Carassius Auratus Gibelio in China. J. Virol. Methods 2014, 206, 76–83. [Google Scholar] [CrossRef]
- Ma, J.; Jiang, N.; LaPatra, S.E.; Jin, L.; Xu, J.; Fan, Y.; Zhou, Y.; Zeng, L. Establishment of a Novel and Highly Permissive Cell Line for the Efficient Replication of Cyprinid Herpesvirus 2 (CyHV-2). Vet. Microbiol. 2015, 177, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Kurita, J.; Ozaki, A.; Sano, M.; Fukuda, H.; Ototake, M. Growth of Cyprinid Herpesvirus 2 (CyHV-2) in Cell Culture and Experimental Infection of Goldfish Carassius Auratus. Dis. Aquat. Organ. 2013, 105, 193–202. [Google Scholar] [CrossRef]
- Lu, J.; Xu, D.; Lu, L. A Novel Cell Line Established from Caudal Fin Tissue of Carassius Auratus Gibelio Is Susceptible to Cyprinid Herpesvirus 2 Infection with the Induction of Apoptosis. Virus Res. 2018, 258, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Engelsma, M.Y.; Way, K.; Dodge, M.J.; Voorbergen-Laarman, M.; Panzarin, V.; Abbadi, M.; El-Matbouli, M.; Frank Skall, H.; Kahns, S.; Stone, D.M. Detection of Novel Strains of Cyprinid Herpesvirus Closely Related to Koi Herpesvirus. Dis. Aquat. Organ. 2013, 107, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.; Merry, G.; Sadler, J. Detection of the Herpesviral Hematopoietic Necrosis Disease Agent (Cyprinid Herpesvirus 2) in Moribund and Healthy Goldfish: Validation of a Quantitative PCR Diagnostic Method. Dis. Aquat. Organ. 2006, 69, 137–143. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Shi, X.; Yu, L.; Zheng, X.; Lan, W.; Jia, P.; Wang, J.; Liu, H. Development and Evaluation of a Loop-Mediated Isothermal Amplification Assay for Diagnosis of Cyprinid Herpesvirus 2. J. Virol. Methods 2013, 194, 206–210. [Google Scholar] [CrossRef]
- Preena, P.G.; Kumar, T.V.A.; Johny, T.K.; Dharmaratnam, A.; Swaminathan, T.R. Quick Hassle-Free Detection of Cyprinid Herpesvirus 2 (CyHV-2) in Goldfish Using Recombinase Polymerase Amplification-Lateral Flow Dipstick (RPA-LFD) Assay. Aquac. Int. 2022, 30, 1211–1220. [Google Scholar] [CrossRef]
- Wang, H.; Sun, M.; Xu, D.; Podok, P.; Xie, J.; Jiang, Y.; Lu, L. Rapid Visual Detection of Cyprinid Herpesvirus 2 by Recombinase Polymerase Amplification Combined with a Lateral Flow Dipstick. J. Fish Dis. 2018, 41, 1201–1206. [Google Scholar] [CrossRef]
- Yang, J.; Wen, J.; Xiao, S.; Wei, C.; Yu, F.; Roengjit, P.; Lu, L.; Wang, H. Complete Genome and Molecular Characterization of a New Cyprinid Herpesvirus 2 (CyHV-2) SH-01 Strain Isolated from Cultured Crucian Carp. Viruses 2022, 14, 2068. [Google Scholar] [CrossRef]
- Chong, R. Chapter 14 Koi Herpesvirus Disease. In Aquaculture Pathophysiology Volume I. Fish Diseases; Elsevier Academic Press: New York, NY, USA, 2022; pp. 189–199. [Google Scholar]
- Hanson, L.; Doszpoly, A.; Van Beurden, S.J.; de Oliveira Viadanna, P.H.; Waltzek, T.B. Chapter 9 Alloherpesviruses of Fish. In Aquaculture Virology; Elsevier Academic Press: New York, NY, USA, 2016; pp. 153–172. [Google Scholar]
- Bercovier, H.; Fishman, Y.; Nahary, R.; Sinai, S.; Zlotkin, A.; Eyngor, M.; Gilad, O.; Eldar, A.; Hedrick, R.P. Cloning of the Koi Herpesvirus (KHV) Gene Encoding Thymidine Kinase and Its Use for a Highly Sensitive PCR Based Diagnosis. BMC Microbiol. 2005, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Gray, W.L.; Mullis, L.; LaPatra, S.E.; Groff, J.M.; Goodwin, A. Detection of Koi Herpesvirus DNA in Tissues of Infected Fish. J. Fish Dis. 2002, 25, 171–178. [Google Scholar] [CrossRef]
- Yuasa, K.; Sano, M.; Kurita, J.; Ito, T.; Iida, T. Improvement of a PCR Method with the Sph I-5 Primer Set for the Detection of Koi Herpesvirus (KHV). Fish Pathol. 2005, 40, 37–39. [Google Scholar] [CrossRef]
- Yuasa, K.; Sano, M. Koi Herpesvirus: Status of Outbreaks, Diagnosis, Surveillance, and Research. Isr. J. Aquac. Bamidgeh 2009, 61, 169–179. [Google Scholar] [CrossRef]
- OIE CHAPTER 2.3.6 Infection with Koi Herpesvirus. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2022.
- Yuasa, K.; Kawana, M.; Ito, T.; Kiryu, I.; Oseko, N.; Sano, M. Intra Vitam Assays for Detecting Fish Infected with Cyprinid Herpesvirus 3 (CyHV-3). Dis. Aquat. Organ. 2022, 149, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Soliman, H.; El-Matbouli, M. Rapid Detection and Differentiation of Carp Oedema Virus and Cyprinid Herpes Virus-3 in Koi and Common Carp. J. Fish Dis. 2018, 41, 761–772. [Google Scholar] [CrossRef]
- Boutier, M.; Ronsmans, M.; Rakus, K.; Jazowiecka-Rakus, J.; Vancsok, C.; Morvan, L.; Peñaranda, M.M.D.; Stone, D.M.; Way, K.; van Beurden, S.J.; et al. Cyprinid Herpesvirus 3: An Archetype of Fish Alloherpesviruses. Adv. Virus Res. 2015, 93, 161–256. [Google Scholar] [CrossRef] [PubMed]
- Cano, I.; Worswick, J.; Mulhearn, B.; Stone, D.; Wood, G.; Savage, J.; Paley, R. A Seasonal Study of Koi Herpesvirus and Koi Sleepy Disease Outbreaks in the United Kingdom in 2018 Using a Pond-Side Test. Animals 2021, 11, 459. [Google Scholar] [CrossRef]
- Thorstensen, M.J.; Vandervelde, C.A.; Bugg, W.S.; Michaleski, S.; Vo, L.; Mackey, T.E.; Lawrence, M.J.; Jeffries, K.M. Non-Lethal Sampling Supports Integrative Movement Research in Freshwater Fish. Front. Genet. 2022, 13, 795355. [Google Scholar] [CrossRef]
- Monaghan, S.J.; Thompson, K.D.; Adams, A.; Kempter, J.; Bergmann, S.M. Examination of the Early Infection Stages of Koi Herpesvirus (KHV) in Experimentally Infected Carp, Cyprinus Carpio L. Using in Situ Hybridization. J. Fish Dis. 2015, 38, 477–489. [Google Scholar] [CrossRef]
- Doszpoly, A.; Kovács, E.R.; Bovo, G.; LaPatra, S.E.; Harrach, B.; Benkő, M. Molecular Confirmation of a New Herpesvirus from Catfish (Ameiurus Melas) by Testing the Performance of a Novel PCR Method, Designed to Target the DNA Polymerase Gene of Alloherpesviruses. Arch. Virol. 2008, 153, 2123–2127. [Google Scholar] [CrossRef]
- Doszpoly, A.; Shchelkunov, I. Partial Genome Analysis of Siberian Sturgeon Alloherpesvirus Suggests Its Close Relation to AciHV-2—Short Communication. Acta Vet. Hung. 2010, 58, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Radosavljević, V.; Milićević, V.; Maksimović-Zorić, J.; Veljović, L.; Nešić, K.; Pavlović, M.; Pelić, D.L.; Marković, Z. Sturgeon Diseases in Aquaculture. Arch. Vet. Med. 2019, 12, 5–20. [Google Scholar] [CrossRef]
- Doszpoly, A.; Kalabekov, I.M.; Breyta, R.; Shchelkunov, I.S. Isolation and Characterization of an Atypical Siberian Sturgeon Herpesvirus Strain in Russia: Novel North American Acipenserid Herpesvirus 2 Strain in Europe? J. Fish Dis. 2017, 40, 1363–1372. [Google Scholar] [CrossRef]
- Kelley, G.O.; Waltzek, T.B.; McDowell, T.S.; Yun, S.C.; LaPatra, S.E.; Hedrick, R.P. Genetic Relationships among Herpes-Like Viruses Isolated from Sturgeon. J. Aquat. Anim. Health 2005, 17, 297–303. [Google Scholar] [CrossRef]
- Kurobe, T.; Kelley, G.O.; Waltzek, T.B.; Hedrick, R.P. Revised Phylogenetic Relationships among Herpesviruses Isolated from Sturgeons. J. Aquat. Anim. Health 2008, 20, 96–102. [Google Scholar] [CrossRef]
- Ciulli, S.; Volpe, E.; Sirri, R.; Tura, G.; Errani, F.; Zamperin, G.; Toffan, A.; Silvi, M.; Renzi, A.; Abbadi, M.; et al. Multifactorial Causes of Chronic Mortality in Juvenile Sturgeon (Huso Huso). Animals 2020, 10, 1866. [Google Scholar] [CrossRef]
- Hedrick, R.; McDowell, T.; Gilad, O.; Adkison, M.; Bovo, G. Systemic Herpes-like Virus in Catfish Ictalurus Melas (Italy) Differs from Ictalurid Herpesvirus 1 (North America). Dis. Aquat. Organ. 2003, 55, 85–92. [Google Scholar] [CrossRef]
- Alborali, L.; Paternello, P.; Lavazza, A.; Caggiano, M.; Lombardi, G. Descrizione Di Un Episodio Di Linfocisti Nel Sarago (Diplodus Puntazzo). Boll. Soc. Ital. Patol. Ittica 1996, 20, 2–7. [Google Scholar]
- Thompson, D.J.; Khoo, L.H.; Wise, D.J.; Hanson, L.A. Evaluation of Channel Catfish Virus Latency on Fingerling Production Farms in Mississippi. J. Aquat. Anim. Health 2005, 17, 211–215. [Google Scholar] [CrossRef]
- Gray, W.L.; Williams, R.J.; Jordan, R.L.; Griffin, B.R. Detection of Channel Catfish Virus DNA in Latently Infected Catfish. J. Gen. Virol. 1999, 80, 1817–1822. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.E.; Marecaux, E. Validation of a QPCR Assay for the Detection of Ictalurid Herpesvirus-2 (IcHV-2) in Fish Tissues and Cell Culture Supernatants. J. Fish Dis. 2010, 33, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Hao, K.; Liu, H.-Y.; Chen, X.-H.; Yuan, J.-F.; Li, L.-J.; Bian, W.-J.; Zhao, Z. Real-Time fluorescent loop mediated isothermal amplification for detection of channel catfish virus. Acta Hydrobiol. Sin. 2021, 45, 541–546. [Google Scholar] [CrossRef]
- Barbosa-Solomieu, V.; Miossec, L.; Vázquez-Juárez, R.; Ascencio-Valle, F.; Renault, T. Diagnosis of Ostreid Herpesvirus 1 in Fixed Paraffin-Embedded Archival Samples Using PCR and in Situ Hybridisation. J. Virol. Methods 2004, 119, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Batista, F.M.; Arzul, I.; Pepin, J.-F.; Ruano, F.; Friedman, C.S.; Boudry, P.; Renault, T. Detection of Ostreid Herpesvirus 1 DNA by PCR in Bivalve Molluscs: A Critical Review. J. Virol. Methods 2007, 139, 1–11. [Google Scholar] [CrossRef]
- Carter, M.J. Enterically Infecting Viruses: Pathogenicity, Transmission and Significance for Food and Waterborne Infection. J. Appl. Microbiol. 2005, 98, 1354–1380. [Google Scholar] [CrossRef]
- Errani, F.; Ponti, M.; Volpe, E.; Ciulli, S. Spatial and Seasonal Variability of Human and Fish Viruses in Mussels inside and Offshore of Ravenna’s Harbour (Adriatic Sea, Italy). J. Appl. Microbiol. 2021, 130, 994–1008. [Google Scholar] [CrossRef]
- Volpe, E.; Pagnini, N.; Serratore, P.; Ciulli, S. Fate of Redspotted Grouper Nervous Necrosis Virus (RGNNV) in Experimentally Challenged Manila Clam Ruditapes Philippinarum. Dis. Aquat. Organ. 2017, 125, 53–61. [Google Scholar] [CrossRef]
- Furhmann, M.; Patirana, E.; de Kantzou, M.; Hick, P. Chapter 63 Ostreid Herpesvirus Disease. In Aquaculture Pathophysiology Volume II. Crustacean and Molluscan Diseases; Elsevier Academic Press: New York, NY, USA, 2022; pp. 473–488. ISBN 978-0-323-95434-1. [Google Scholar]
- Evans, O.; Paul-Pont, I.; Whittington, R.J. Detection of Ostreid Herpesvirus 1 Microvariant DNA in Aquatic Invertebrate Species, Sediment and Other Samples Collected from the Georges River Estuary, New South Wales, Australia. Dis. Aquat. Organ. 2017, 122, 247–255. [Google Scholar] [CrossRef]
- Bueno, R.; Perrott, M.; Dunowska, M.; Brosnahan, C.; Johnston, C. In Situ Hybridization and Histopathological Observations during Ostreid Herpesvirus-1-Associated Mortalities in Pacific Oysters Crassostrea Gigas. Dis. Aquat. Organ. 2016, 122, 43–55. [Google Scholar] [CrossRef]
- Toldrà, A.; Furones, M.D.; O’Sullivan, C.K.; Campàs, M. Detection of Isothermally Amplified Ostreid Herpesvirus 1 DNA in Pacific Oyster (Crassostrea Gigas) Using a Miniaturised Electrochemical Biosensor. Talanta 2020, 207, 120308. [Google Scholar] [CrossRef]
- Dotto-Maurel, A.; Pelletier, C.; Morga, B.; Jacquot, M.; Faury, N.; Dégremont, L.; Bereszczynki, M.; Delmotte, J.; Escoubas, J.-M.; Chevignon, G. Evaluation of Tangential Flow Filtration Coupled to Long-Read Sequencing for Ostreid Herpesvirus Type 1 Genome Assembly. Microb. Genom. 2022, 8, mgen000895. [Google Scholar] [CrossRef]
- Trancart, S.; Tweedie, A.; Liu, O.; Paul-Pont, I.; Hick, P.; Houssin, M.; Whittington, R.J. Diversity and Molecular Epidemiology of Ostreid Herpesvirus 1 in Farmed Crassostrea Gigas in Australia: Geographic Clusters and Implications for “Microvariants” in Global Mortality Events. Virus Res. 2023, 323, 198994. [Google Scholar] [CrossRef] [PubMed]
- Thiéry, R.; Johnson, K.L.; Nakai, T.; Schneemann, A.; Bonami, J.R.; Lightner, D.V. Family Nodaviridae. In Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier Academic Press: San Diego, CA, USA, 2011; pp. 1061–1067. [Google Scholar]
- Bandín, I.; Souto, S. Betanodavirus and VER Disease: A 30-Year Research Review. Pathogens 2020, 9, 106. [Google Scholar] [CrossRef] [PubMed]
- Volpe, E.; Gustinelli, A.; Caffara, M.; Errani, F.; Quaglio, F.; Fioravanti, M.L.; Ciulli, S. Viral Nervous Necrosis Outbreaks Caused by the RGNNV/SJNNV Reassortant Betanodavirus in Gilthead Sea Bream (Sparus Aurata) and European Sea Bass (Dicentrarchus Labrax). Aquaculture 2020, 523, 735155. [Google Scholar] [CrossRef]
- Schneemann, A.; Reddy, V.; Johnson, J.E. The Structure and Function of Nodavirus Particles: A Paradigm for Understanding Chemical Biology. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 1998; Volume 50, pp. 381–446. ISBN 978-0-12-039850-8. [Google Scholar]
- Nishizawa, T.; Mori, K.-I.; Furuhashi, M.; Nakai, T.; Furusawa, I.; Muroga, K. Comparison of the Coat Protein Genes of Five Fish Nodaviruses, the Causative Agents of Viral Nervous Necrosis in Marine Fish. J. Gen. Virol. 1995, 76, 1563–1569. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, T.; Furuhashi, M.; Nagai, T.; Nakai, T.; Muroga, K. Genomic Classification of Fish Nodaviruses by Molecular Phylogenetic Analysis of the Coat Protein Gene. Appl. Environ. Microbiol. 1997, 63, 1633–1636. [Google Scholar] [CrossRef] [PubMed]
- Toffolo, V.; Negrisolo, E.; Maltese, C.; Bovo, G.; Belvedere, P.; Colombo, L.; Valle, L.D. Phylogeny of Betanodaviruses and Molecular Evolution of Their RNA Polymerase and Coat Proteins. Mol. Phylogenet. Evol. 2007, 43, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.L.; Gabrielsen, M.; Beh, P.L.; Kueh, C.L.; Thong, Q.X.; Streetley, J.; Tan, W.S.; Bhella, D. Structure of the Macrobrachium Rosenbergii Nodavirus: A New Genus within the Nodaviridae? PLoS Biol. 2018, 16, e3000038. [Google Scholar] [CrossRef]
- NaveenKumar, S.; Shekar, M.; Karunasagar, I.; Karunasagar, I. Genetic Analysis of RNA1 and RNA2 of Macrobrachium Rosenbergii Nodavirus (MrNV) Isolated from India. Virus Res. 2013, 173, 377–385. [Google Scholar] [CrossRef]
- Johnson, K.L.; Moore, J.S. Nodaviruses of Invertebrates and Fish (Nodaviridae). In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 819–826. ISBN 978-0-12-814516-6. [Google Scholar]
- Mori, K.; Nakai, T.; Muroga, K.; Arimoto, M.; Mushiake, K.; Furusawa, I. Properties of a New Virus Belonging to Nodaviridae Found in Larval Striped Jack (Pseudocaranx Dentex) with Nervous Necrosis. Virology 1992, 187, 368–371. [Google Scholar] [CrossRef] [PubMed]
- Munday, B.L.; Kwang, J.; Moody, N. Betanodavirus Infections of Teleost Fish: A Review. J. Fish Dis. 2002, 25, 127–142. [Google Scholar] [CrossRef]
- Iwamoto, T.; Okinaka, Y.; Mise, K.; Mori, K.-I.; Arimoto, M.; Okuno, T.; Nakai, T. Identification of Host-Specificity Determinants in Betanodaviruses by Using Reassortants between Striped Jack Nervous Necrosis Virus and Sevenband Grouper Nervous Necrosis Virus. J. Virol. 2004, 78, 1256–1262. [Google Scholar] [CrossRef]
- Johansen, R.; Sommerset, I.; Tørud, B.; Korsnes, K.; Hjortaas, M.J.; Nilsen, F.; Nerland, A.H.; Dannevig, B.H. Characterization of Nodavirus and Viral Encephalopathy and Retinopathy in Farmed Turbot, Scophthalmus Maximus (L.). J. Fish Dis. 2004, 27, 591–601. [Google Scholar] [CrossRef]
- Gagné, N.; Johnson, S.C.; Cook-Versloot, M.; MacKinnon, A.M.; Olivier, G. Molecular Detection and Characterization of Nodavirus in Several Marine Fish Species from the Northeastern Atlantic. Dis. Aquat. Organ. 2004, 62, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Kwon, W.J.; Min, J.G.; Kim, K.I.; Jeong, H.D. Complete Genome Sequence and Pathogenic Analysis of a New Betanodavirus Isolated from Shellfish. J. Fish Dis. 2019, 42, 519–531. [Google Scholar] [CrossRef]
- Frerichs, G.N.; Rodger, H.D.; Peric, Z. Cell Culture Isolation of Piscine Neuropathy Nodavirus from Juvenile Sea Bass, Dicentrarchus Labrax. J. Gen. Virol. 1996, 77 Pt 9, 2067–2071. [Google Scholar] [CrossRef]
- Zrnčić, S.; Padros, F.; Mladineo, I.; Fioravanti, M.-L.; Gustinelli, A.; Palenzuela, O.; Toffan, A.; Cuilli, S.; Fouz, B.; Breton, A.L.; et al. Bottlenecks in Diagnostics of Mediterranean Fish Diseases; European Association of Fish Pathologists: Aberdeen City, UK, 2022; Volume 40. [Google Scholar]
- Baud, M.; Cabon, J.; Salomoni, A.; Toffan, A.; Panzarin, V.; Bigarré, L. First Generic One Step Real-Time Taqman RT-PCR Targeting the RNA1 of Betanodaviruses. J. Virol. Methods 2015, 211, 1–7. [Google Scholar] [CrossRef]
- Sung, C.-H.; Lu, J.-K. Reverse Transcription Loop-Mediated Isothermal Amplification for Rapid and Sensitive Detection of Nervous Necrosis Virus in Groupers. J. Virol. Methods 2009, 159, 206–210. [Google Scholar] [CrossRef]
- Xu, H.-D.; Feng, J.; Guo, Z.-X.; Ou, Y.-J.; Wang, J.-Y. Detection of Red-Spotted Grouper Nervous Necrosis Virus by Loop-Mediated Isothermal Amplification. J. Virol. Methods 2010, 163, 123–128. [Google Scholar] [CrossRef]
- Hwang, J.; Suh, S.-S.; Park, M.; Oh, M.-J.; Kim, J.-O.; Lee, S.; Lee, T.-K. Detection of Coat Protein Gene of Nervous Necrosis Virus Using Loop-Mediated Isothermal Amplification. Asian Pac. J. Trop. Med. 2016, 9, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Toffan, A.; Pascoli, F.; Pretto, T.; Panzarin, V.; Abbadi, M.; Buratin, A.; Quartesan, R.; Gijón, D.; Padrós, F. Viral Nervous Necrosis in Gilthead Sea Bream (Sparus Aurata) Caused by Reassortant Betanodavirus RGNNV/SJNNV: An Emerging Threat for Mediterranean Aquaculture. Sci. Rep. 2017, 7, 46755. [Google Scholar] [CrossRef]
- Olveira, J.G.; Souto, S.; Dopazo, C.P.; Thiéry, R.; Barja, J.L.; Bandín, I. Comparative Analysis of Both Genomic Segments of Betanodaviruses Isolated from Epizootic Outbreaks in Farmed Fish Species Provides Evidence for Genetic Reassortment. J. Gen. Virol. 2009, 90, 2940–2951. [Google Scholar] [CrossRef]
- Panzarin, V.; Fusaro, A.; Monne, I.; Cappellozza, E.; Patarnello, P.; Bovo, G.; Capua, I.; Holmes, E.C.; Cattoli, G. Molecular Epidemiology and Evolutionary Dynamics of Betanodavirus in Southern Europe. Infect. Genet. Evol. 2012, 12, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Biasini, L.; Berto, P.; Abbadi, M.; Buratin, A.; Toson, M.; Marsella, A.; Toffan, A.; Pascoli, F. Pathogenicity of Different Betanodavirus RGNNV/SJNNV Reassortant Strains in European Sea Bass. Pathogens 2022, 11, 458. [Google Scholar] [CrossRef]
- Krishnan, R.; Kim, J.-O.; Qadiri, S.S.N.; Kim, J.-O.; Oh, M.-J. Early Viral Uptake and Host-Associated Immune Response in the Tissues of Seven-Band Grouper Following a Bath Challenge with Nervous Necrosis Virus. Fish Shellfish. Immunol. 2020, 103, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Qi, J.; He, L.; Luo, H.; Lin, J.; Qiu, F.; Wang, Q.; Zheng, L. Isolation and Identification of a New Strain of Nervous Necrosis Virus from the Big-Belly Seahorse Hippocampus Abdominalis. Virol. J. 2022, 19, 109. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, S.; Singh, I.S.B.; Puthumana, J. Status in Molluscan Cell Line Development in Last One Decade (2010–2020): Impediments and Way Forward. Cytotechnology 2022, 74, 433–457. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.E.J.; Bufalino, M.R.; Christie, A.E.; Frischer, M.E.; Soin, T.; Tsui, C.K.M.; Hanner, R.H.; Smagghe, G. Misidentification of OLGA-PH-J/92, Believed to Be the Only Crustacean Cell Line. In Vitro Cell. Dev. Biol. Anim. 2011, 47, 665–674. [Google Scholar] [CrossRef]
- OIE CHAPTER 2.3.8 Infection with Salmon Alphavirus. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2021; pp. 344–359.
- OIE CHAPTER 2.3.10 Infection with Viral Haemorrhagic Septicaemia Virus. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2021; pp. 377–401.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).