Simple Summary
Lasiodiplodia species are plurivorous plant pathogens found worldwide, especially in tropical and subtropical regions, that result in fruit and root rot, die-back of branches and stem canker, etc. During the exploration of the fungal diversity of blighted stem samples collected in northern China, two new Lasiodiplodia species, L. acerina G.H. Qiao & W.T. Qin and L. cotini G.H. Qiao & W.T. Qin, were discovered based on integrated studies of phenotypic features, culture characteristics and molecular analyses. They were described and illustrated in detail. This work provided a better understanding of the biodiversity, phylogeny and established concepts of the genus Lasiodiplodia.
Abstract
The Lasiodiplodia are major pathogens or endophytes living on a wide range of plant hosts in tropical and subtropical regions, which can cause stem canker, shoot blight, and rotting of fruits and roots. During an exploration of the stem diseases on Acer truncatum and Cotinus coggygria in northern China, two novel species of Lasiodiplodia, L. acerina G.H. Qiao & W.T. Qin and L. cotini G.H. Qiao & W.T. Qin, were discovered based on integrated studies of the morphological characteristics and phylogenetic analyses of the internal transcribed spacer region (ITS), translation elongation factor 1-α (TEF1-α), beta-tubulin (TUB2) and RNA polymerase II subunit b genes (RPB2). Lasiodiplodia acerina is a sister taxon of L. henannica and distinguishable by smaller paraphysis and larger conidiomata. Lasiodiplodia cotini is closely related to L. citricola but differs in the sequence data and the size of paraphyses. Distinctions between the two novel species and their close relatives were compared and discussed in details. This study updates the knowledge of species diversity of the genus Lasiodiplodia. Furthermore, this is the first report of Lasiodiplodia associated with blighted stems of A. truncatum and C. coggygria in China.
1. Introduction
Lasiodiplodia, established in 1896, is a member of the family Botryosphaeriaceae [1]. Species in the genus Lasiodiplodia have been associated with different plant diseases including fruit and root rots, die-back of branches and stem cankers. The type species of Lasiodiplodia (L. theobromae) was regarded as one of the cosmopolitan, plurivorous pathogens mainly inhabiting tropical and subtropical regions [2,3].
The main morphological characteristics of Lasiodiplodia include hyaline, smooth, cylindrical to conical conidioenous cells, which produce subovoid to ellipsoid-ovoid conidia and the conidia are hyaline without septa or dark-brown with single septae [4]. Species in the genus Lasiodiplodia were mostly differentiated based on the characteristics of the conidia and paraphyses [5]. Some other morphological characteristics, such as annelations of conidiogenous cells, the dimensions and papillate nature of conidiomata, septate and pigmented conidia as well as the pycnidial paraphyses have been gradually used to recognize the Lasiodiplodia species, but to what extent these characteristics are phylogenetically significant warrants further investigation [6].
The Genealogical Concordance Phylogenetic Species Recognition (GCPSR) concept is widely used to delineate different fungal species. This approach relies on determining the concordance between multiple gene genealogies and delimiting species where the branches of multiple trees display congruence [7]. The widespread application of phylogenies based on ITS, TEF1-α, TUB2 and RPB2 genes promotes the accurate identification of species in the genus Lasiodiplodia, and more and more species have been successively introduced over the years; at present, more than 70 Lasiodiplodia species have been identified [8,9,10]. Among them, some species have been introduced almost entirely on the basis of DNA sequence phylogenies. Although the phylogenies were derived from the analysis of multiple loci, some species were introduced only on the basis of minor differences in only one locus, and some species cannot be clearly separated phylogenetically [11,12,13]. Several accepted Lasiodiplodia species (L. brasiliense, L. laeliocattleyae, L. missouriana, L. viticola) may be hybrids based on a detailed phylogenetic analyses of five loci from 19 Lasiodiplodia species [14].
To provide a better understanding of Lasiodiplodia species diversity in China, recent collections of the genus on Acer truncatum and Cotinus coggygria were examined. Two previously unrecognized Lasiodiplodia species were discovered based on integrated studies of phenotypic features, culture characteristics and phylogenetic analyses of the combined sequences of ITS, TEF1-α, TUB2 and RPB2. Detailed comparisons were made between the new taxa and their close relatives.
2. Materials and Methods
2.1. Isolates and Specimens
Cultures were isolated from the blighted stems of Cotinus coggygria and Acer Truncatum collected from Beijing, China, from 2018 to 2019. Stem segments (0.5 cm × 0.5 cm × 0.2 cm) were cut from the boundary of the lesion or dead tissues, surface sterilized subsequently and incubated on potato dextrose agar (PDA, peeled potatoes 200 g, glucose 20 g, agar 18 g, add water to 1 L) at 25 °C for fungal isolation [15]. Specimens, purified cultures and the ex-type strains were deposited in the culture collection of Institute of Plant Protection, Beijing Academy of Agriculture and Forestry Sciences.
2.2. Morphology and Growth Characterization
Morphological characterization of colonies, such as colony appearance, color and spore production were observed and recorded following the method of previous studies [5,11,16] on three media (PDA, malt extract agar (MEA, malt extract 20 g, agar 18 g, add water to 1 L) and synthetic nutrient-poor agar (SNA, monopotassium phosphate 1 g, potassium nitrate 1 g, Magnesium sulfate heptahydrate 0.5 g, potassium chloride 0.5 g, glucose 0.2 g, saccharose 0.2 g, agar 20 g, add water to 1 L)) with each isolate three replicates. Microscopic characteristics were recorded based on 20 paraphyses, 20 conidiogenous cells and 50 conidia on PDA at 25 °C in darkness. Photographs were taken from material mounted in lactic acid with Axiocam 506 color microscope (Carl Zeiss, Aalen, Germany) using Zeiss Imager Z2 software. The new species were established based on the guidelines outlined by Jeewon and Hyde [17].
2.3. DNA Extraction, PCR Amplification and Sequencing
Purified cultures were incubated on PDA with cellophane for 5 days at 25 °C in darkness. Genomic DNA was extracted using the TsingKe Plant Genomic DNA Extraction Kit® following the manufacturer’s protocol (Beijing, China). The ITS, TEF1-α, TUB2 and RPB2 gene sequences were amplified and sequenced using primer pairs ITS1/4 [18], EF1-728F/986R [19], Bt2a/2b [20] and RPB2-LasF/R [14], respectively. Each PCR reaction (25 μL) consisted of 1 μL 5–10 ng DNA, 22 μL TsingKe Golden Star T6 Super PCR Mix (1.1×) and 1 μL of each primer. PCR amplification followed the manufacturer’s protocol of TsingKe Golden Star T6 Super PCR Mix (Beijing, China), and products were sequenced by Beijing TsingKe Biotech Co. Ltd. (Beijing, China).
2.4. Sequence Alignment and Phylogenetic Analyses
Sequences of the investigated Lasiodiplodia species excluding those of our two new species for phylogenetic analyses were obtained from the NCBI using Tbtools v. 1.09876 [21] (Table 1). Sequences were assembled, aligned and manually adjusted with BioEdit v.7.2.5 [22]. To identify the phylogenetic positions of L. acerina and L. cotini, the combined sequences of ITS, TEF1-α, TUB2 and RPB2 for all strains were used for the phylogenetic analysis by methods of maximum parsimony (MP), maximum likelihood (ML) and MrBayes analyses (BI) with Diplodia mutila and D. seriata as outgroups. NEXUS files were generated with Clustal X 1.83 [23] in Phylosuit v.1.2.2 [24].

Table 1.
Details of Lasiodiplodia strains investigated in this study.
ML analyses with 1000 bootstrap replicates were conducted using raxmlGUI v. 2.06 [25]. The best-fit model of nucleotide substitution for each dataset was determined using ModelFinder [26]. Topological confidence of resulted trees was assessed by maximum likelihood bootstrap proportion (MLBP) with 1000 replicates.
MP trees were generated in PAUP v.4.0b [27], using the heuristic search function with tree bisection and reconstruction as branch swapping algorithms and 1000 random addition replicates. Gaps were treated as a fifth character and the characters were unordered and given equal weight. MAXTREES were set to 5000, branches of zero length were collapsed and all multiple, equally parsimonious trees were saved. Tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC) and homoplasy index (HI) were calculated. Topological confidence of resulting trees was tested by maximum parsimony bootstrap proportion (MPBP) with 1000 replications, each with 10 replicates of random addition of taxa.
BI analysis was conducted by MrBayes v. 3.2.6 [28] with Markov Chain Monte Carlo algorithm. Nucleotide substitution models were determined by ModelFinder and GTR + I+G + F was estimated as the best-fit model. Two MCMC chains were run from random trees for 2,000,000 generations and sampled every 100 generations. The first 2500 trees were discarded as the burn-in phase of the analyses, and Bayesian inference posterior probability (BIPP) was determined from the remaining trees. Trees were visualized in FigTree v1.4.4.
3. Results
3.1. Phylogenetic Analyses
The combined ITS, TEF1-α, TUB2 and RPB2 data set comprised 74 taxa with D. mutila and D. seriata as the outgroups. The MP dataset consisted of 1823 characters, of which 1358 characters were constant, 115 characters were parsimony informative and 366 variable characters were parsimony uninformative. A total of 284 most-parsimonious trees with the same topology were generated, one of them is shown in Figure 1 (tree length = 1075, CI = 0.5563, RI = 0.8692, RC = 0.4835, HI = 0.4437). In the ML analyses, GTRGAMMA was specified as the model. The best scoring RAxML tree with the final ML optimization likelihood value of −8913.786383 (ln) yielded. Estimated base frequencies were as follows: A = 0.224747, C = 0.283918, G = 0.271555, T = 0.219781; substitution rates AC = 0.836915, AG = 3.800207, AT = 1.307148, CG = 1.119223, CT = 6.358526, GT = 1.000000; gamma distribution shape parameter α = 0.220772. The ML, MP and BI methods for phylogenetic analyses resulted in trees with similar topologies.
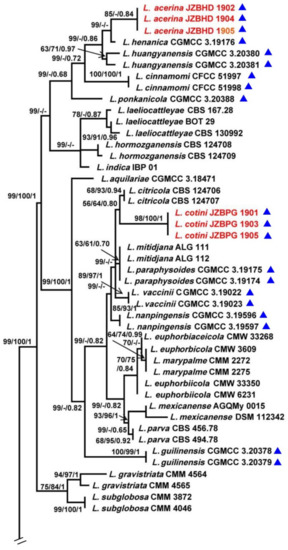
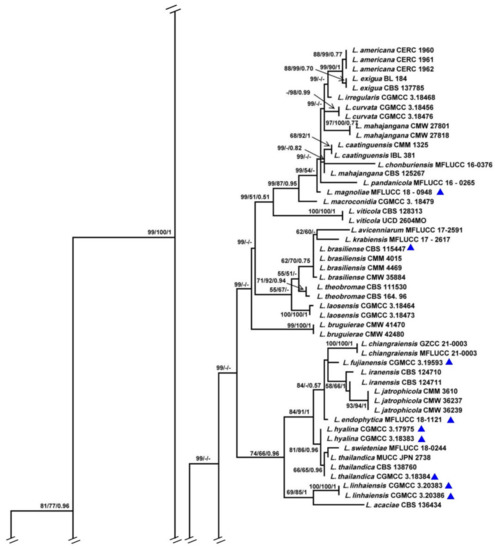
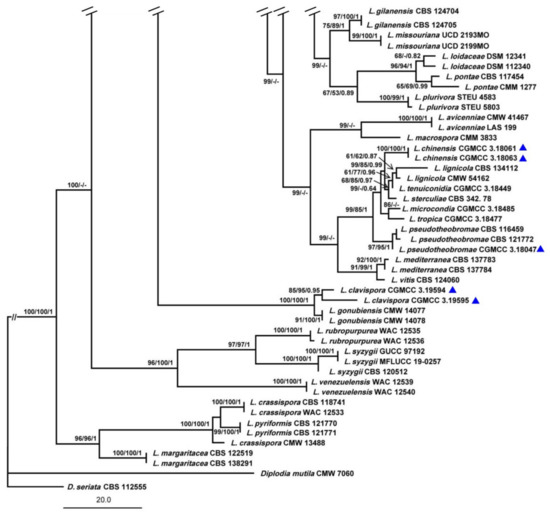
Figure 1.
Maximum parsimony phylogram reconstructed from the combined sequences of ITS, TEF1-α, TUB2 and RPB2 of Lasiodiplodia. MPBP above 50% (left), MLBP above 50% (middle), BIPP above 0.7 (right) are indicated at the nodes. New species proposed are indicated in red font. The tree is rooted to Diplodia mutila and D. seriata. The strains isolated from samples of China are marked in blue triangles.
Among all the strains, 141 represented 76 Lasiodiplodia spp. clustered together with high support (MPBP/MLBP/BIPP = 100%/100%/1). Three isolates (JZBHD 1902, 1904 and 1905) representing L. acerina and three isolates (JZBPG 1901, 1903 and 1905) representing L. cotini clustered as distinct lineages from other Lasiodiplodia spp., with the support values MPBP/BIPP = 85%/0.84 and MPBP/MLBP/BIPP = 98%/100%/1, respectively. They showed a close phylogenetic relationship, respectively, with L. henanica and L. citricola.
3.2. Taxonomy
Lasiodiplodia acerina G. H. Qiao & W.T. Qin, sp. nov. MB845417; Figure 2.
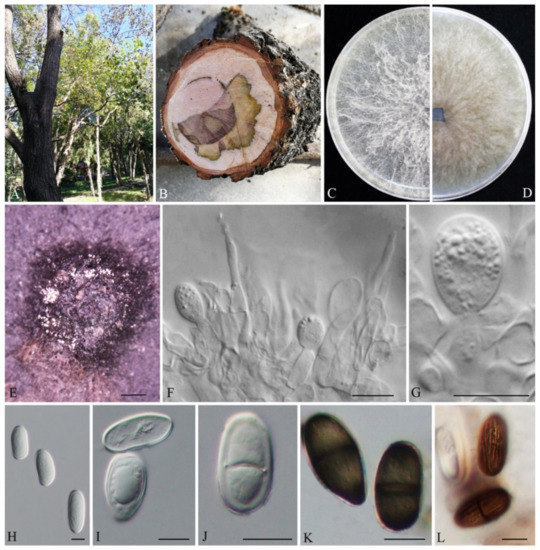
Figure 2.
Lasiodiplodia acerina (JZBHD 1904). (A) Disease tree in the field. (B) Cross-section of stem. (C,D) Culture grown on PDA. (E) Conidiomata developing on PDA. (F,G) Conidia developing on conidiogenous cells between paraphyses. (H–L) Conidia. Scale bars: E = 200 μm, F−L = 10 μm.
Etymology: The specific epithet is in reference to the host, Acer truncatum, from which the fungus was isolated.
Typification: China, Beijing, Haidian district, Summer Palace, Longevity Hill, from blighted stems of Acer truncatum, 18 September 2019, G. H. Qiao (Holotype: JZBHDT1904, ex-type isolate: JZBHD1904).
DNA barcodes: ITS = OP117391, TUB2 = OP141783, RPB2 = OP141788, TEF1-α = OP141777.
Conidiomata were semi-immersed or superficial stromatic on PDA within 14 d, and were solitary, smooth, globose, dark grey to black, covered by dark gray mycelia without conspicuous ostioles and up to 2525 µm in diameter. Paraphyses were filiform, cylindrical, aseptate, thin-walled, hyaline, apex rounded, occasionally swollen at the base and unbranched, arising from the conidiogenous layer, extending above the level of developing conidia, and were up to 39.4 µm long and 3.0 µm wide. Conidiophores were reduced to conidiogenous cells. Conidiogenous cells were hyaline, holoblastic, smooth, discrete, thin-walled, and were cylindrical to ampulliform. Conidia were initially hyaline, ovoid to cylindrical, with a 1-µm-thick wall, (21.64-)21.97–30.83(-30.96) × (10.61-)11.48–15.87(-16.72) µm (n = 50, av. = 26.9 µm × 13.5 µm, L/W ratio = 2.0, by range from 1.58 to 2.61. Mature conidia turned brown with a median septum and longitudinal striations and sometimes with one vacuole. The sexual stage and spermatia were not observed.
Culture characteristics: Colonies on PDA were initially white with thick aerial mycelia reaching the lid of the plate. After 7 d colonies were fluffy, grey to black, with reverse side of the colonies black. The colonies radius reached 32 mm on PDA after 24 h, and mycelia entirely covered the surface of the plate after 48 h in darkness at 25 °C. Aerial mycelia on MEA was moderately dense and reached the lid of the plate and became olive gray to black on the surface of the plate after 7 d. The colonies radius reached 30 mm after 24 h, and 76 mm after 48 h on MEA in darkness at 25 °C. Aerial mycelia on SNA were sparse, white. The colonies radius reached 22 mm after 24 h, and 58 mm after 48 h in darkness at 25 °C. Mycelia entirely covered the surface of the plate after 72 h on all the three culture media in darkness at 25 °C.
Additional strains examined: China, Beijing, Haidian district, Summer Palace, Longevity Hill, 39.91 °N 116.41 °E, from blighted stems of Acer truncatum, 18 September 2019, G. H. Qiao, HDyhy1902, JZBQHD1902; ibid., HDyhy1905, JZBQ1905.
Notes: Phylogenetically, as a separated linage, three strains of L. acerina formed sister groups with L. henanica (MPBP = 99%) and L. huangyanensis (MPBP/BIPP = 99%/0.86). Compared with the sequences of TEF1-α for L. acerina, they shared low similarities with L. henanica (97.71%), L. huangyanensis CGMCC 3.20380 (96.08%) and L. huangyanensis CGMCC 3.20381 (96.41%) by 7, 12 and 11 bp divergent among 306 bp, respectively. Morphologically, mycelia of L. acerina on MEA grew faster than that of L. henanica (colony radius reached 26 mm on MEA after 24 h, and more than 65 mm after 48 h in darkness at 28 °C). The length of paraphysis were longer in L. henanica (105 μm) [6] and L. huangyanensis (82 μm) [9]. In addition, L. henanica had smaller conidiomata (520 µm) (Table 2), and vacuoles in the conidia, which were also different from L. acerina [6].

Table 2.
Morphological characteristic comparison between L. acerina, L. cotini and their close relatives.
Lasiodiplodia cotini G. H. Qiao & W.T. Qin, sp. nov. MB845418; Figure 3.
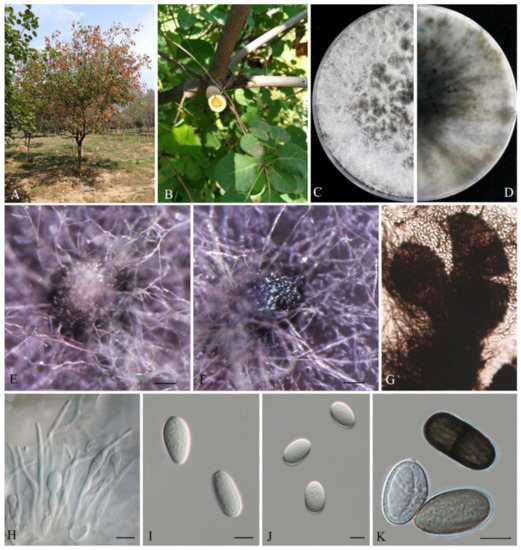
Figure 3.
Lasiodiplodia cotini (JZBPG 1905). (A) Diseased tree in the field. (B) Cross-section of the blighted stem. (C,D) Culture grown on PDA. (E,F) Conidiomata developing on PDA. (G) Crushed conidiomata with many conidia. (H) Conidia developing on conidiogenous cells between paraphyses. (I–K) Conidia. Scale bars: E − F = 100 μm, H − K = 10 μm.
Etymology: The specific epithet is in reference to the host, Cotinus coggygria, from which the fungus is isolated.
Typification: China, Beijing, Pinggu district, Huangsongyu Town, Dadonggou village, from blighted stems of Cotinus coggygria, 20 October 2018, G. H. Qiao (ex-type strain: JZBPG 1905).
DNA barcodes: ITS = OP117389, TUB2 = OP141781, RPB2 = OP141787, TEF1-α = OP141775.
Conidiomata were semi-immersed or superficial stromatic, produced on PDA within 14 d, solitary, smooth, globose, dark grey to black, covered by dark gray mycelia without a conspicuous ostiole, up to 415 µm in diameter. Paraphyses arise from the conidiogenous layer, filiform, extending above the level of developing conidia, up to 41.9 µm long and 2.6 µm wide, hyaline, cylindrical, aseptate, thin-walled, apex rounded, occasionally swollen at the base and unbranched. Conidiophores were reduced to conidiogenous cells. Conidiogenous cells were hyaline, cylindrical to ampulliform, holoblastic, discrete, thin-walled and smooth. Conidia were initially hyaline, ovoid to cylindrical, with a 1-µm-thick wall, mature conidia turned brown with a median septum and longitudinal striations and sometimes with one vacuole, (19.38-)20−27(-28.81) × (12.51-)13.61−16.55(-16.62) µm (n = 50, av. = 24.28 µm × 15.4 µm, L/W ratio = 1.58, by range from 1.40 to 1.69. The sexual stage and spermatia were not observed.
Culture characteristics: Aerial mycelia on PDA were abundant, smoke-grey to olivaceous-grey with the colonies dark black on the reverse side of the plate after 7 d. The colonies radius reached 45 mm on PDA after 24 h, and mycelia entirely covered the surface of the plate after 48 h in darkness at 25 °C. The colonies radius reached 24 mm on MEA after 24 h in darkness at 25 °C, and 51 mm after 48 h. Aerial mycelium is moderately dense and grey. The colonies radius reached 14 mm on SNA after 24 h, and 43 mm after 48 h in darkness at 25 °C. Aerial mycelium on SNA is sparse and white. After 72 h mycelia entirely covered the surface of the plates of the three culture media.
Additional strains examined: China, Beijing, Pinggu district, Huangsongyu Town, Dadonggou village, 40.23 °N 117.29 °E from blighted stems of Cotinus coggygria, 20 October 2018, G. H. Qiao, PGhsy 1901, JZBPG1901; ibid., PGhsy 1903, JZBPG1903.
Notes: Phylogenetically, three strains of L. cotini clustered together (MPBP/MLBP/BIPP = 98%/100%/1) and are closely related to L. citricola (MPBP/MLBP/BIPP = 68%/93%/0.94). Comparison of the sequence data indicated that they shared 4 bp divergent among 259 bp for TEF1-α (98.46%). Morphologically, the colonies of L. citricola and L. cotini were not obviously different; however, L. cotini has smaller paraphyses than those of L. citricola (125 × 3–4 μm) [29] and L. cinnamomi (106 × 3–4 μm) [30]. In addition, larger conidia of L. cinnamomi (18.7–21.1 × 12.7–14.1 μm) also make it distinguishable from L. cotini (Table 2) [30].
4. Discussion
To explore the taxonomic positions of the genus Lasiodiplodia, the phylogenetic tree was constructed based on the combined sequences of ITS, TEF1-α, TUB2 and RPB2 with D. mutila and D. seriata used as outgroups. Two novel species, L. acerina and L. cotini, were found based on the integrated studies of phenotypic and molecular data. All investigated Lasiodiplodia species clustered together (Figure 1), which was basically congruent with the results of a previous study [6]. Lasiodiplodia acerina and L. cotini clustered as separated terminal branches at the top of the tree, and were closely related to L. henanica [6] and L. citricola [30], respectively, but they differed from each other in characters of conidiomata, conidia and paraphyses, etc. (Figure 2 and Figure 3; Table 2).
Although many species in Lasiodiplodia were differentiated on the basis of morphological characters, it is necessary to combine the morphology and molecular data for definitive identifications. The phylogenetic tree in this study was comprised of 76 Lasiodiplodia species represented by 141 strains. When our two new species joined, the tree topology was somewhat changed, including the relationships among species. Lasiodiplodia acerina and four newly reported species, L. henanica on blueberries [6], L. huangyanensis and L. ponkanicola on citrus [9], and L. cinnamomic on Cinnamomum camphora in China formed a separated terminal branch [29]. Lasiodiplodia citricola was reported as the sister group of L. paraphysoides and L. aquilariae [6,9]; however, in this study, four strains representing L. citricola were closely related to L. cotini represented by our three strains (MPBP/MLBP/BIPP = 56%/64%/0.8). Lasiodiplodia citricola were far away from L. paraphysoides, a novel species reported on blueberries [6] as a result of L. cotini in our study and L. mitidjana on citrus [30] joining in the phylogenetic tree.
Further analysis showed that the Lasiodiplodia species sampling from China tend to cluster together (Table 1 and Figure 1), which may be the result of the comprehensive action of fungal adaptive ability, regional climate and human-mediated factors. For example, five newly reported species sampling from China in recent years, L. acerina, L. cinnamomi, L. henanica, L. huangyanensis and L. ponkanicola formed a high-supported group (MPBP/BIPP = 99%/0.68). Geographically, species in the genus Lasiodiplodia tend to live in tropical or subtropical areas or in warm temperature areas associated with stem diseases of woody substrates [30,31]. In this study, two newly described species of Lasiodiplodia were also isolated from the blighted stem of A. truncatum and C. coggygria in Beijing, which are distributed in subtropical or warm temperate areas in China (Table 1).
Acer truncatum and Cotinus coggygria are two kinds of landscape trees that play important roles in urban greening construction. Botryosphaeria dothidea, Fusarium oxysporum, Neofusicoccum parvum and Pestalotiopsis microspora have been reported to be associated with diseased leaves and stems of Acer spp. [32,33,34,35], and Alternaria alternata, Botryosphaeria dothidea and Verticillum dahlia have been isolated from diseased leaves and stems of C. coggygria [36,37,38]; to our knowledge, this is the first report of Lasiodiplodia being associated with A. truncatum and C. coggygria.
Along with an increasing number of species recognized in the genus Lasiodiplodia, our understanding of the genus will become more sophisticated and intelligible through the integrated studies on morphology and phylogeny. Accumulations of our knowledge on Lasiodiplodia will provide useful information for establishing reasonable species concepts, and understand co-relations between morphology and sequence data in the future, which will lay further foundations for the scientific management of stem blight diseases and improvement in the landscape effect in the process of urban greening construction.
5. Conclusions
This study recognized two novel Lasiodiplodia species from blighted stems of A. truncatum and C. coggygria, which were the first reports of Lasiodiplodia associated with these two horticulture trees in China. The discovery provided a better understanding of the biodiversity and phylogeny of the genus Lasiodiplodia and is beneficial for future evaluation of the potential usages and functions of the new species.
Author Contributions
Conceptualization, W.Q. and G.Q.; methodology, J.Z.; software, G.Q.; validation, G.Q. and J.L.; writing—original draft preparation, G.Q.; writing—review and editing, W.Q. and J.Z.; visualization, G.Q. and X.T.; funding acquisition, W.Q. and G.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Innovation Foundation of Beijing Academy of Agriculture and Forestry Sciences (KJCX201910) and the Rural Revitalization Project of Beijing Municipal Bureau of Agriculture (BJXCZX20221229).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data analyzed in this study are curated from the public domain.
Acknowledgments
The authors would like to thank Xing-hong Li, Wei Zhang and Ying Zhou of the Institute of plant protection, Beijing Academy of Agriculture and Forestry Sciences for technical assistance in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Clendinin, I. Lasiodiplodia Ellis. and Everh. n. gen. Bot. Gaz. 1896, 21, 92–93. [Google Scholar] [CrossRef]
- De Silva, N.I.; Phillips, A.J.L.; Liu, J.-K.; Lumyong, S.; Hyde, K.D. Phylogeny and morphology of Lasiodiplodia species associated with Magnolia forest plants. Sci. Rep. 2019, 9, 14355. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.W.; Lima, N.B.; de Morais, M.A.; Barbosa, M.A.G.; Souza, B.O.; Michereff, S.J.; Phillips, A.J.L.; Câmara, M.P.S. Species of Lasiodiplodia associated with mango in Brazil. Fungal Divers. 2013, 61, 181–193. [Google Scholar] [CrossRef]
- Phillips, A.J.L.; Alves, A.; Abdollahzadeh, J.; Slippers, B.; Wingfield, M.J.; Groenewald, J.Z.; Crous, P.W. The Botryosphaeriaceae: Genera and species known from culture. Stud. Mycol. 2013, 76, 51–167. [Google Scholar] [CrossRef] [PubMed]
- Dou, Z.P.; He, W.; Zhang, Y. Does morphology matter in taxonomy of Lasiodiplodia? An answer from Lasiodiplodia hyalina sp. nov. Mycosphere 2017, 8, 1014–1027. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Bhoyroo, V.; Rampadarath, S.; Jeewon, R. Multigene phylogenetics and morphology reveal five novel Lasiodiplodia species associated with blueberries. Life 2021, 11, 657. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.W.; Jacobson, D.J.; Kroken, S.; Kasuga, T.; Geiser, D.M.; Hibbett, D.S.; Fisher, M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000, 31, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, A.; Phillips, A.; Li, X.; Hyde, K. Botryosphaeriaceae: Current status of genera and species. Mycosphere 2016, 7, 1001–1073. [Google Scholar] [CrossRef]
- Xiao, X.E.; Pu, Z.X.; Wang, W.; Zhu, Z.R.; Li, H.; Crous, P.W. Species of Botryosphaeriaceae associated with citrus branch diseases in China. Persoonia 2021, 47, 106–135. [Google Scholar] [CrossRef]
- Zhang, W.; Groenewald, J.Z.; Lombard, L.; Schumacher, R.K.; Phillips, A.J.L.; Crous, P.W. Evaluating species in Botryosphaeriales. Persoonia 2021, 46, 63–115. [Google Scholar] [CrossRef]
- Dou, Z.P.; He, W.; Zhang, Y. Lasiodiplodia chinensis, a new holomorphic species from China. Mycosphere 2017, 8, 521–532. [Google Scholar] [CrossRef]
- Burgess, T.I.; Barber, P.A.; Mohali, S.; Pegg, G.; de Beer, W.; Wingfield, M.J. Three new Lasiodiplodia spp. from the tropics, recognized based on DNA sequence comparisons and morphology. Mycologia 2006, 98, 423–435. [Google Scholar] [CrossRef]
- Liang, L.; Li, H.; Zhou, L.; Chen, F. Lasiodiplodia pseudotheobromae causes stem canker of Chinese hackberry in China. J. For. Res. 2019, 31, 2571–2580. [Google Scholar] [CrossRef]
- Cruywagen, E.M.; Slippers, B.; Roux, J.; Wingfield, M.J. Phylogenetic species recognition and hybridisation in Lasiodiplodia: A case study on species from baobabs. Fungal Biol. 2017, 121, 420–436. [Google Scholar] [CrossRef]
- Pavlic, D.; Slippers, B.; Coutinho, T.A.; Gryzenhout, M.; Wingfield, M.J. Lasiodiplodia gonubiensis sp. nov., a new Botryosphaeria anamorph from native Syzygium cordatum in South Africa. Stud. Mycol. 2004, 50, 313–322. [Google Scholar]
- Ismail, A.; Cirvilleri, G.; Polizzi, G.; Crous, P.; Groenewald, J.; Lombard, L. Lasiodiplodia species associated with dieback disease of mango (Mangifera indica) in Egypt. Australas. Plant Pathol. 2012, 41, 649–660. [Google Scholar] [CrossRef]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Oxford University Press: Oxford, UK, 1999; pp. 95–98. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2019, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.P. Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4.0; Sinauer Associates: Sunderland, UK, 2002. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Jiang, N.; Wang, X.-W.; Liang, Y.-M.; Tian, C.-M. Lasiodiplodia cinnamomi sp. nov. from Cinnamomum camphora in China. Mycotaxon 2018, 133, 249–259. [Google Scholar] [CrossRef]
- Abdollahzadeh, J.; Javadi, A.; Mohammadi Goltapeh, E.; Zare, R.; Phillips, A.J. Phylogeny and morphology of four new species of Lasiodiplodia from Iran. Persoonia 2010, 25, 1–10. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, S.; Zhao, L.; Sun, X.; He, W.; Zhang, Y.; Dai, Y.-C. Lasiodiplodia spp. associated with Aquilaria crassna in Laos. Mycol. Prog. 2019, 18, 683–701. [Google Scholar] [CrossRef]
- Zhao, X.; Li, H.; Zhou, L.; Chen, F.; Chen, F. Wilt of Acer negundo L. caused by Fusarium nirenbergiae in China. J. For. Res. 2020, 31, 2013–2022. [Google Scholar] [CrossRef]
- Cui, C.; Wang, Y.; Jiang, J.; Hui, O.; Qin, S.; Huang, T. Identification of the pathogen causing brown spot disease of ‘October Glory’. Sci. Silvae Sin. 2015, 51, 142–147. [Google Scholar] [CrossRef]
- Moricca, S.; Uccello, A.; Ginetti, B.; Ragazzi, A. First Report of Neofusicoccum parvum Associated with Bark Canker and Dieback of Acer pseudoplatanus and Quercus robur in Italy. Plant Dis. 2012, 96, 1699. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.X.; Dong, H.X.; Jia, X.Z.; Zhang, X.Y. First report of Botryosphaeria dothidea causing canker of Acer platanoides in China. Plant Dis. 2015, 99, 1857. [Google Scholar] [CrossRef]
- Zhang, S.; Liang, W.; Yang, Q. First report of Alternaria alternata causing leaf spot on Cotinus coggygria Scop. in China. Plant Dis. 2018, 102, 2644. [Google Scholar] [CrossRef]
- Xiong, D.G.; Wang, Y.L.; Ma, J.; Klosterman, S.J.; Xiao, S.; Tian, C. Deep mRNA sequencing reveals stage-specific transcriptome alterations during microsclerotia development in the smoke tree vascular wilt pathogen, Verticillium dahliae. BMC Genom. 2014, 15, 324. [Google Scholar] [CrossRef]
- Fan, S.S.; Huang, Y.J.; Zhang, X.J.; Chen, G.H.; Zhou, J.; Li, X.; Han, M.Z. First report of Botryosphaeria dothidea causing canker on Cotinus coggygria in China. Plant Dis. 2019, 103, 2678. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).