A New High-Throughput Screening Method to Detect Antimicrobial Volatiles from Metagenomic Clone Libraries
Abstract
1. Introduction
2. Results
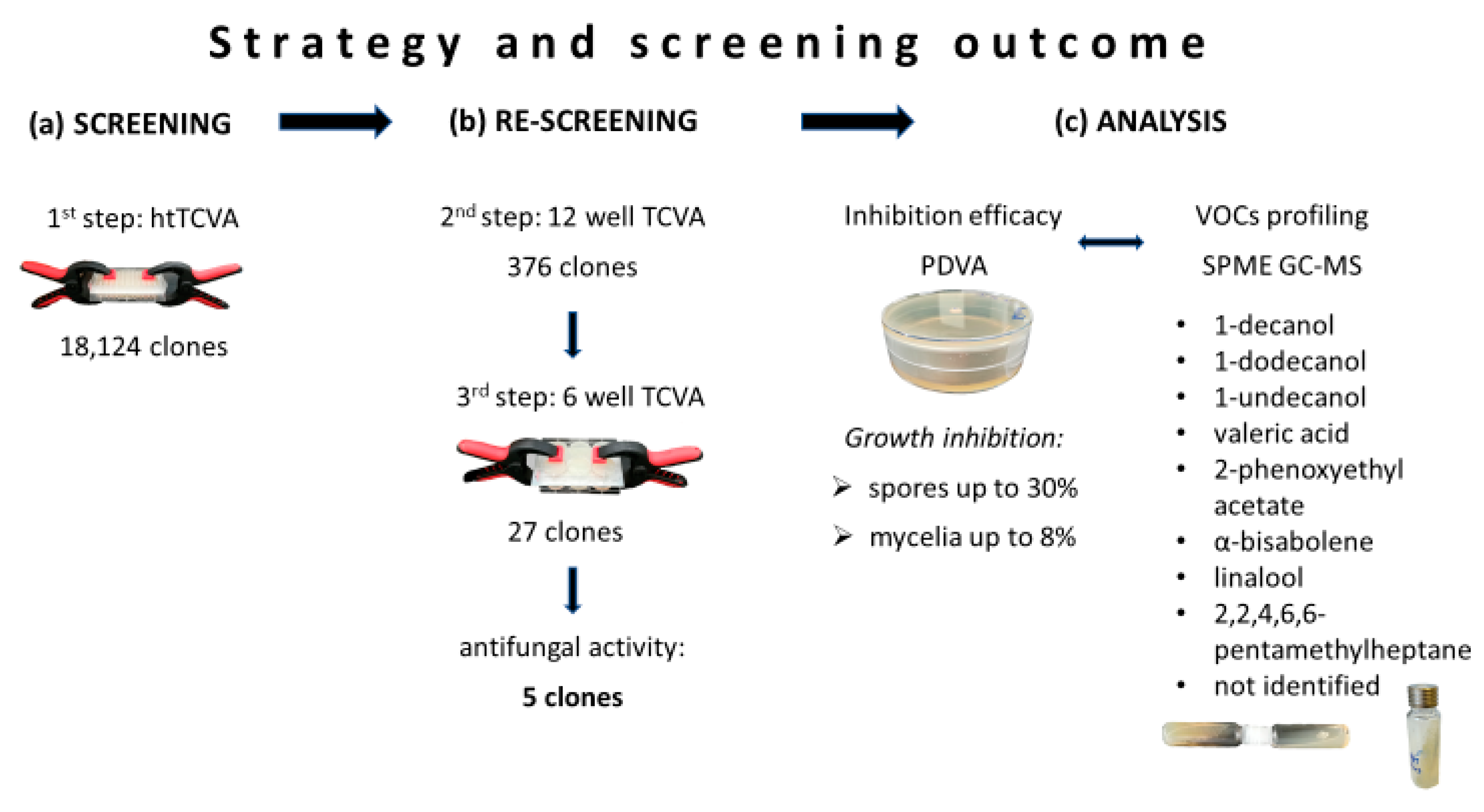
2.1. Screening of Metagenomic Libraries for Antifungal VOCs
2.2. PDVA: Determination of Growth Inhibition Rates
2.3. VOCs Profiling Through SPME GC-MS
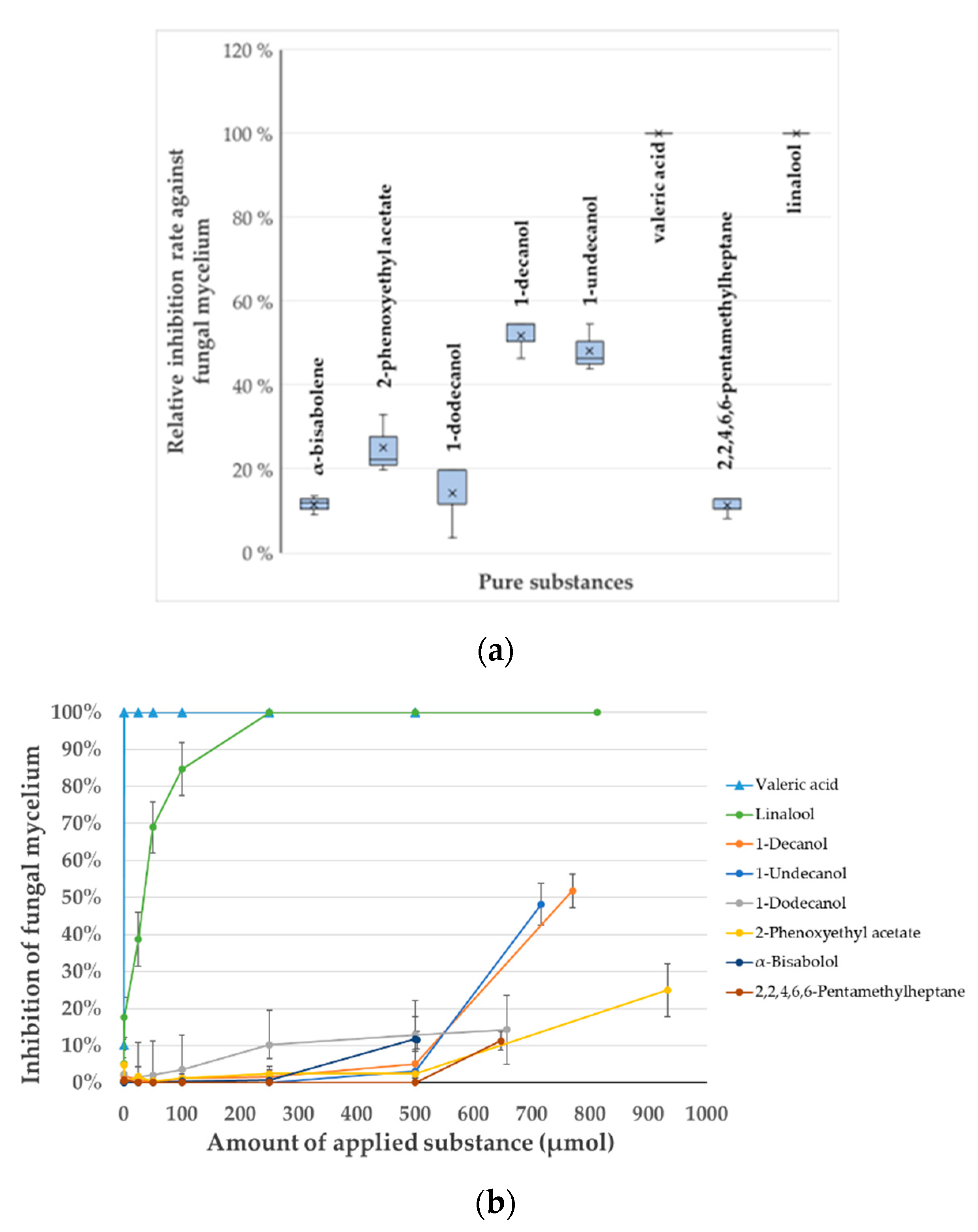
2.4. In Vitro Verification of the Identified VOCs and Determination of Their Minimal Inhibitory Concentration
2.5. Growth Inhibition of Different Fungi by 130 F2 and 131 E3
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Screening for Antimicrobial VOCs
4.3. Quantification and Validation of the Antagonistic Effect
4.4. Qualitative Detection of VOCs by SPME GC-MS
4.5. Data Accessibility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, 68–80. [Google Scholar] [CrossRef]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Review on Antimicrobial Resistance: London, UK, 2014; pp. 1–20. [Google Scholar]
- Schulz, S.; Dickschat, J.S. Bacterial volatiles: The smell of small organisms. Nat. Prod. Rep. 2007, 24, 814–842. [Google Scholar] [CrossRef] [PubMed]
- Kai, M.; Haustein, M.; Molina, F.; Petri, A.; Scholz, B.; Piechulla, B. Bacterial volatiles and their action potential. Appl. Microbiol. Biotechnol. 2009, 81, 1001–1012. [Google Scholar] [CrossRef]
- Berg, G.; Köberl, M.; Rybakova, D.; Müller, H.; Grosch, R.; Smalla, K. Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 2017, 93, 5. [Google Scholar] [CrossRef]
- Thomas, S.; Izard, J.; Walsh, E.; Batich, K.; Chongsathidkiet, P.; Clarke, G.; Sela, D.A.; Muller, A.J.; Mullin, J.M.; Albert, K.; et al. The host microbiome regulates and maintains human health: A primer and perspective for non-microbiologists. Cancer Res. 2017, 77, 1783–1812. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.S.; Mo, M.H.; Gu, Y.Q.; Zhou, J.P.; Zhang, K.Q. Possible contributions of volatile-producing bacteria to soil fungistasis. Soil Biol. Biochem. 2007, 39, 2371–2379. [Google Scholar] [CrossRef]
- Chaurasia, B.; Pandey, A.; Palni, L.M.S.; Trivedi, P.; Kumar, B.; Colvin, N. Diffusible and volatile compounds produced by an antagonistic Bacillus subtilis strain cause structural deformations in pathogenic fungi in vitro. Microbiol. Res. 2005, 160, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Cernava, T.; Aschenbrenner, I.A.; Grube, M.; Liebminger, S.; Berg, G. A novel assay for the detection of bioactive volatiles evaluated by screening of lichen-associated bacteria. Front. Microbiol. 2015, 6, 398. [Google Scholar] [CrossRef]
- Mülner, P.; Bergna, A.; Wagner, P.; Sarajlić, D.; Gstöttenmayr, B.; Dietel, K.; Grosch, R.; Cernava, T.; Berg, G. Microbiota associated with sclerotia of soilborne fungal pathogens—A novel source of biocontrol agents producing bioactive volatiles. Phytobiomes J. 2019, 3, 125–136. [Google Scholar] [CrossRef]
- Kai, M.; Effmert, U.; Berg, G.; Piechulla, B. Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol. 2007, 187, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Mulero-Aparicio, A.; Cernava, T.; Turrà, D.; Schaefer, A.; Di Pietro, A.; López-Escudero, F.J.; Trapero, A.; Berg, G. The role of volatile organic compounds and rhizosphere competence in mode of action of the non-pathogenic Fusarium oxysporum FO12 toward Verticillium wilt. Front. Microbiol. 2019, 10, 1808. [Google Scholar] [CrossRef]
- Stewart, E.J. Growing unculturable bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, 245–249. [Google Scholar] [CrossRef]
- Bragina, A.; Berg, C.; Cardinale, M.; Shcherbakov, A.; Chebotar, V.; Berg, G. Sphagnum mosses harbour highly specific bacterial diversity during their whole lifecycle. ISME J. 2012, 6, 802–813. [Google Scholar] [CrossRef]
- Opelt, K.; Berg, C.; Berg, G. The bryophyte genus Sphagnum is a reservoir for powerful and extraordinary antagonists and potentially facultative human pathogens. FEMS Microbiol. Ecol. 2007, 61, 38–53. [Google Scholar] [CrossRef]
- Opelt, K.; Berg, C.; Schönmann, S.; Eberl, L.; Berg, G. High specificity but contrasting biodiversity of Sphagnum-associated bacterial and plant communities in bog ecosystems independent of the geographical region. ISME J. 2007, 1, 502–516. [Google Scholar] [CrossRef]
- Opelt, K.; Chobot, V.; Hadacek, F.; Schönmann, S.; Eberl, L.; Berg, G. Investigations of the structure and function of bacterial communities associated with Sphagnum mosses. Environ. Microbiol. 2007, 9, 2795–2809. [Google Scholar] [CrossRef]
- Khaneghah, A.M.; Martins, L.M.; von Hertwig, A.M.; Bertoldo, R.; Sant’Ana, A.S. Deoxynivalenol and its masked forms: Characteristics, incidence, control and fate during wheat and wheat based products processing—A review. Trends Food Sci. Technol. 2018, 71, 13–24. [Google Scholar] [CrossRef]
- Cordero, P.; Cavigliasso, A.; Príncipe, A.; Godino, A.; Jofré, E.; Mori, G.; Fischer, S. Genetic diversity and antifungal activity of native Pseudomonas isolated from maize plants grown in a central region of Argentina. Syst. Appl. Microbiol. 2012, 35, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, C.G.; Cassone, A.; Coda, R.; Gobbetti, M. Antifungal activity of sourdough fermented wheat germ used as an ingredient for bread making. Food Chem. 2011, 127, 952–959. [Google Scholar] [CrossRef]
- Xu, L.L.; Han, T.; Wu, J.Z.; Zhang, Q.Y.; Zhang, H.; Huang, B.K.; Rahman, K.; Qin, L.P. Comparative research of chemical constituents, antifungal and antitumor properties of ether extracts of Panax ginseng and its endophytic fungus. Phytomedicine 2009, 16, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, S.; Pandey, A.K. In vitro susceptibility of opportunistic Fusarium spp. to essential oils. Mycoses 1999, 42, 97–101.
- Raza, W.; Yuan, J.; Ling, N.; Huang, Q.; Shen, Q. Production of volatile organic compounds by an antagonistic strain Paenibacillus polymyxa WR-2 in the presence of root exudates and organic fertilizer and their antifungal activity against Fusarium oxysporum f. sp. niveum. Biol. Control 2015, 80, 89–95. [Google Scholar] [CrossRef]
- Kubo, I.; Muroi, H.; Kubo, A. Structural functions of antimicrobial long-chain alcohols and phenols. Bioorg. Med. Chem. 1995, 3, 873–880. [Google Scholar] [CrossRef]
- Msaada, K.; Salem, N.; Bachrouch, O.; Bousselmi, S.; Tammar, S.; Alfaify, A.; AlSane, K.; Ben Ammar, W.; Sana, A.; Brahim, A.H.; et al. Chemical composition and antioxidant and antimicrobial activities of wormwood (Artemisia absinthium L.) essential oils and phenolics. J. Chem. 2015, 2015, 12. [Google Scholar] [CrossRef]
- Maddula, S.; Blank, L.M.; Schmid, A.; Baumbach, J.I. Detection of volatile metabolites of Escherichia coli by multi capillary column coupled ion mobility spectrometry. Anal. Bioanal. Chem. 2009, 394, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.A.; Oberauner-Wappis, L.; Peyman, A.; Amos, G.C.A.; Wellington, E.M.H.; Berg, G. Mining for NRPS and PKS genes revealed a high diversity in the Sphagnum bog metagenome. Appl. Environ. Microbiol. 2015, 81, 5064–5072. [Google Scholar] [CrossRef]
- Opelt, K.; Berg, G. Diversity and antagonistic potential of bacteria associated with bryophytes from nutrient-poor habitats of the Baltic Sea Coast. Appl. Environ. Microbiol. 2004, 70, 6569–6579. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Miyazaki, K. Functional metagenomics for enzyme discovery: Challenges to efficient screening. Curr. Opin. Biotechnol. 2009, 20, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Obermeier, M.M.; Müller Bogotá, C.A. Prospects for biotechnological exploitation of endophytes using functional metagenomics. In Endophyte Biotechnology: Potential for Agriculture and Pharmacology, 1st ed.; Shouten, A., Ed.; CABI Publishing: Wallingford, UK, 2019; Volume 1, pp. 164–179. [Google Scholar]
- Simon, C.; Herath, J.; Rockstroh, S.; Daniel, R. Rapid identification of genes encoding DNA polymerases by function-based screening of metagenomic libraries derived from glacial ice. Appl. Environ. Microbiol. 2009, 75, 2964–2968. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Angelov, A.; Mientus, M.; Liebl, S.; Liebl, W. A two-host fosmid system for functional screening of (meta)genomic libraries from extreme thermophiles. Syst. Appl. Microbiol. 2009, 32, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Leis, B.; Angelov, A.; Liebl, W. Screening and expression of genes from metagenomes. Adv. Appl. Microbiol. 2013, 83, 1–68. [Google Scholar]
- Gabor, E.M.; Alkema, W.B.L.; Janssen, D.B. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ. Microbiol. 2004, 6, 879–886. [Google Scholar] [CrossRef]
- Gao, H.; Li, P.; Xu, X.; Zeng, Q.; Guan, W. Research on volatile organic compounds from Bacillus subtilis CF-3: Biocontrol effects on fruit fungal pathogens and dynamic changes during fermentation. Front. Microbiol. 2018, 9, 456. [Google Scholar] [CrossRef]
- Yuan, J.; Raza, W.; Shen, Q.; Huang, Q. Antifungal activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. cubense. Appl. Environ. Microbiol. 2012, 78, 5942–5944. [Google Scholar] [CrossRef]
- Roujeinikova, A.; Simon, W.J.; Gilroy, J.; Rice, D.W.; Rafferty, J.B.; Slabas, A.R. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J. Mol. Biol. 2007, 365, 135–145. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2016, 1858, 936–946. [Google Scholar] [CrossRef]
- Croes, K.; Casteels, M.; Asselberghs, S.; Herdewijn, P.; Mannaerts, G.P.; Van Veldhoven, P.P. Formation of a 2-methyl-branched fatty aldehyde during peroxisomal α-oxidation. FEBS Lett. 1997, 412, 643–645. [Google Scholar] [CrossRef][Green Version]
- Zheng, Y.N.; Li, L.L.; Liu, Q.; Yang, J.M.; Wang, X.W.; Liu, W.; Xu, X.; Liu, H.; Zhao, G.; Xian, M. Optimization of fatty alcohol biosynthesis pathway for selectively enhanced production of C12/14 and C16/18 fatty alcohols in engineered Escherichia coli. Microb. Cell Fact. 2012, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.J.; Tantillo, D.J. Consequences of conformational preorganization in sesquiterpene biosynthesis: Theoretical studies on the formation of the bisabolene, curcumene, acoradiene, zizaene, cedrene, duprezianene, and sesquithuriferol sesquiterpenes. J. Am. Chem. Soc. 2009, 131, 7999–8015. [Google Scholar] [CrossRef] [PubMed]
- Jaroszuk-Scisel, J.; Kurek, E.; Slomka, A.; Janczarek, M.; Rodzik, B. Activities of cell wall degrading enzymes in autolyzing cultures of three Fusarium culmorum isolates: Growth-promoting, deleterious and pathogenic to rye (Secale cereale). Mycologia 2011, 103, 929–945. [Google Scholar] [CrossRef] [PubMed]
- Winkelhausen, E.; Pospiech, R.; Laufenberg, G. Antifungal activity of phenolic compounds extracted from dried olive pomace. Bull. Chem. Technol. Maced. 2005, 24, 41–46. [Google Scholar]
- Aakvik, T.; Degnes, K.F.; Dahlsrud, R.; Schmidt, F.; Dam, R.; Yu, L.; Völker, U.; Ellingsen, T.E.; Valla, S. A plasmid RK2-based broad-host-range cloning vector useful for transfer of metagenomic libraries to a variety of bacterial species. FEMS Microbiol. Lett. 2009, 296, 149–158. [Google Scholar] [CrossRef]
- Cox, P.A. The ethnobotanical approach to drug discovery: Strengths and limitations. Sci. Am. 2007, 270, 82–87. [Google Scholar] [CrossRef]


| Volatile Organic Compounds | Retention Time (min) | Library Clone | Reported Antifungal Activity |
|---|---|---|---|
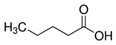 Valeric acid | 7.509 | 130 F2 | Fusarium oxysporum [23] |
 2,2,4,6,6-Pentamethylheptane (1) | 10.119 | 130 F2; 172 E4 | Candida albicans, Cryptococcus neoformans, Trichophyton rubrum, Aspergillus fumigatus [24] |
 Linalool (2) | 13.504 | 130 F2; 172 E4 | Fusarium spp. [25] |
 1-Decanol | 18.570 | 130 F2; 131 B5; 131 E3; 131 F2 | Fusarium oxysporum [26] |
 1-Undecanol | 19.990 | 131 B5; 131 E3; 131 F2 | Penicillium chrysogenum, Aspergillus niger, Mucor mucedo, Trichophyton mentagrophytes [27] |
 2-Phenoxyethyl acetate | 20.236 | 131 E3 | none |
 1-Dodecanol | 21.541 | 131 B5; 131 F2 | Fusarium oxysporum [26] |
 α-Bisabolene (2) | 22.533 | 131 E3 | Fusarium spp. [28] |
| Not identified (2) | 23.587 | 131 E3 | ---- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stocker, F.; Obermeier, M.M.; Resch, K.; Berg, G.; Müller Bogotá, C.A. A New High-Throughput Screening Method to Detect Antimicrobial Volatiles from Metagenomic Clone Libraries. Antibiotics 2020, 9, 726. https://doi.org/10.3390/antibiotics9110726
Stocker F, Obermeier MM, Resch K, Berg G, Müller Bogotá CA. A New High-Throughput Screening Method to Detect Antimicrobial Volatiles from Metagenomic Clone Libraries. Antibiotics. 2020; 9(11):726. https://doi.org/10.3390/antibiotics9110726
Chicago/Turabian StyleStocker, Franz, Melanie M. Obermeier, Katharina Resch, Gabriele Berg, and Christina A. Müller Bogotá. 2020. "A New High-Throughput Screening Method to Detect Antimicrobial Volatiles from Metagenomic Clone Libraries" Antibiotics 9, no. 11: 726. https://doi.org/10.3390/antibiotics9110726
APA StyleStocker, F., Obermeier, M. M., Resch, K., Berg, G., & Müller Bogotá, C. A. (2020). A New High-Throughput Screening Method to Detect Antimicrobial Volatiles from Metagenomic Clone Libraries. Antibiotics, 9(11), 726. https://doi.org/10.3390/antibiotics9110726






