Abstract
There is a scarcity of information on antibiotic resistance in goats. To understand shedding of resistant Escherichia coli in pastured goats, we collected fecal samples from a mixed age cohort over a one-year period. No antibiotic had been used on the study animals one year prior to and during the study period. Resistant isolates were detected in all age groups and prevalence in goat kids was significantly higher than adults; 43–48% vs. 8–25% respectively. The proportion of resistant isolates was higher when animals were congregated near handling facility than on pasture. Most isolates were resistant to tetracycline (51%) and streptomycin (30%), but also to antibiotics that had never been used on the farm; ampicillin (19%). TetB, bla-TEM, (aadA and strpA/strpB) genes were detected in 70%, 43%, (44% and 24%) of tetracycline, ampicillin, and streptomycin resistant isolates respectively. Resistant isolates also harbored virulent genes and some belonged to D and B2 phylogenetic groups. Thus, pastured goats, despite minimal exposure to antibiotics, are reservoirs of resistant E. coli that may contaminate the environment and food chain and spread resistant genes to pathogenic bacteria and some that are potential animal and human pathogens. Environmental sources may play a role in acquisition of resistant bacteria in pastured goats.
1. Introduction
The emergence of antibiotic resistant pathogens and commensal bacteria is a phenomenon important to both human and animal health globally [1]. While the origins and reservoirs of antimicrobial resistance in the environment is a complex topic which continues to be a subject of intensive research [2], extensive use of antibiotics in animal agriculture is considered one of the major causes of emergence and spread of antibiotic resistance organism in the environment [3]. Recent studies, however, have reported antibiotic resistant isolates from animals who have never been exposed to antibiotic use [4] and in humans living in remote parts far away from any agricultural or close human activities [5]. These findings indicate that other factors may have a role in acquisition and dissemination of antimicrobial resistance genes in the environment [6].
Many studies on antimicrobial resistance of commensals and clinical isolates have evaluated resistance in food animals including poultry, swine, and cattle [7,8,9,10,11,12,13,14,15,16,17,18,19,20]. Many of these studies report a high prevalence of antimicrobial resistance in both commensal bacteria and pathogenic bacteria to a variety of clinically relevant antibiotics [21]. On the other hand, little has been published on status of antimicrobial resistance in small ruminants and little surveillance on the status of antimicrobial resistance in these species is reported globally [22]. While pastured small ruminants are generally reared under system of low antibiotic use, these animals interact freely with the environment under grazing systems. Furthermore, antibiotic resistant Escherichia coli (E. coli) have been detected in wild animals that have no known prior exposure to antibiotics [2,4]. Thus, it is potentially possible pastured small ruminants may acquire or spread antibiotic resistant bacteria and genes to the broader environment or ultimately get to the public and food chain through many pathways [5,23,24,25,26,27]. Due to the abundance in the animal gut, ease of acquisition of antimicrobial resistance genes, and ease of cultivation in the laboratory, E. coli is often used as a sentinel to understand status and for surveillance of antimicrobial resistance [28]. Furthermore, small ruminant’s gastrointestinal tract is host to E. coli strains that are of public health significance, including those that belong to the O157, O26, and O145 serotypes [28,29,30]. The characteristics of gut E. coli strains in goats has not been fully characterized, despite their importance as a meat and milk source globally. In this study, we evaluated the temporal shedding patterns of antibiotic resistant E. coli in a cohort of young goat kids, nursing does, and other adult goats in the cohort over a one-year period. We further characterized the resistant isolates genes responsible for the antimicrobial resistance phenotypes, virulence genes, and phylogenetic grouping of the various resistant isolates to further understand the public health significance of the isolates.
2. Materials and Methods
2.1. Study Site Description and Livestock Management
The study cohort was a herd of Spanish and Myotonic goats which are part of Virginia State University (VSU), USA, research flock that are maintained on forty-five acres of land divided into fenced grazing paddocks and surrounded by wooded area. Animals are rotated between pasture lots year-round. A housing facility is available on site with small paddocks where animals routinely congregate in winter and for group procedures including hoof trimming, foot dips and during the weaning period. While grazing at the facility, animals are supplemented with baled hay. The breeding animals are routinely all bred in October/November and kidded on pasture around April/early May. During the experimental period, the animals were supplemented with hay as needed and a balanced corn-based concentrate. The animals are monitored daily during supplemental feeding for signs of ill health by the herd manager who communicated any concerns to the primary author and on all sampling days by the primary author. Interaction of the research animals and farm workers occur during supplemental feeding and during routine hoof trimming and deworming procedures. No antibiotics had been used on the participating animals at least one year before the initiation of the study and on both adults and kids in the study during the study period. Animals were cared for according to an approved Virginia State University Institutional Animal Care and Use Protocol (VSU 2017-02).
2.2. Study Groups and Sampling Protocol
Beginning May 2017, a cohort of 25 kids and their dams (15) were recruited in the study to understand and compare patterns of shedding of antibiotic resistant E. coli between nursing goat kids and their respective dams. The cohort was part of a larger flock and were all maintained on pastures and managed the same way throughout the study period. Animals in the initial cohort were first sampled at three weeks of age and thereafter monthly until weaning at about 3 months. In total, this cohort was sampled 4 times up to the weaning day. At 7 days post weaning, only kids in the initial cohort were sampled again to further understand shedding patterns around weaning time. To increase the sample size during subsequent samplings, the initial cohort of 25 kids and other goat kids born at the same period with the initial cohort and shared the same ecosystem and management were recruited into the study group. This latter group was sampled at six months and at one year of age.
For detection of shedding patterns based on location at the farm, 49 adult goats in the same flock under the same management were also included in the study. This group included animals sampled near the facility in March 2017 and on pasture in October 2017.
2.3. Fecal Sample Collection
At each sampling, individual fecal samples were collected from the rectum of kids and adults and transported in ice to laboratory for microbial isolation. The samples were processed the same day.100mg of fecal samples was mixed with 1ml of phosphate buffered saline and vortexed until the fecal pellet was well mixed with the saline. Samples were centrifuged at 700 rpm for three (3) minutes to sediment the fecal material and dilutions of supernatant were prepared and plated on MacConkey agar and incubated at 37 °C overnight. Two E. coli like colonies were picked from the plates with well separated colonies and transferred to Luria broth. Further transfer of isolates to Eosin Methylene Blue (EMB) agar was done for confirmation of E. coli. Colonies with metallic sheen growth on EMB media were assumed to be E. coli. Further confirmation utilized PCR amplification of 16S ribosomal gene amplification and visualization in agarose gel under UV light. Isolates were stored in 20% glycerol at −20 °C until processing for antimicrobial resistance.
2.4. Antimicrobial Susceptibility Screening
The standard Kirby–Bauer agar disk diffusion method was used to screen for antimicrobial resistance following recommendations by the Clinical and Laboratory Standards Institute (2015) recommendations. A total of 12 antimicrobial discs were used based on importance in animal and human health and included: ampicillin, gentamicin, amoxicillin-clavulanic acid, streptomycin, tetracycline, tobramycin, amikacin, trimethoprim-sulphamethoxazole, nalidixic acid, meropenem, chloramphenicol, and ciprofloxacin. One confirmed E. coli isolate per animal was screened unless isolates from the same animal displayed different colony morphology on EMB (mucoid vs non mucoid) in which case both were screened. E. coli isolates were transferred to Muller Hinton broth and incubated at 37 °C to 0.5 McFarand standard turbidity. 100 µL of the broth was subsequently spread onto Muller Hinton agar 150mm plates. The twelve (12) antibiotic discs were placed using a Thermo Scientific™ Remel™ Antimicrobial Susceptibility 12-place 150 mm Disk Dispenser, incubated overnight, and zones of inhibition measured for each of the tested antibiotic. American Type Culture Collection (ATCC) E. coli 25922 was used as the quality control reference strain in all the screening tests. For each individual antibiotic tested, antimicrobial susceptibility description (“resistant”, “intermediate”, and “susceptible”) was based on the criteria outlined in the CLSI manual 2015 for Enterobacteriaceae. Isolates that were found to be resistant to any of the tested antimicrobials were further processed for determination of the commonly detected antimicrobial resistance genes/mutations, virulence genes, and also for phylogenetic grouping.
2.5. Characterization of Antibiotic Resistance Genes
Overnight Luria broth culture of each resistant isolate was used for DNA extraction following a simple boiling method previously described [31] with a few modifications. In brief, 2 mL of overnight Luria broth containing the isolates was centrifuged at 14,800 rpm (full speed) for 3 minutes at room temperature. The supernatant was poured out and the pellet further re-suspended in one milliliter (1 mL) of molecular grade water by vortexing. The bacteria suspension was further centrifuged at full speed for 3 minutes. The supernatant was poured out and the pellet re-suspended by vortexing in 200–500 µL molecular grade water. The suspension was boiled for 20 minutes at 100 °C using a table top heating block to lyse the bacteria. The suspension was centrifuged again at full speed for 4 minutes to pellet the bacteria lysate and 150–300 µL of the supernatant containing the DNA transferred to a new tube. DNA concentration was measured using a Nanodrop 2000c and samples stored at −20 °C until further processing. The presence of genes encoding TEM, OXA, and SHV β-lactamases was studied by PCR in all the ampicillin-and amoxicillin /clavulanic resistant isolates. The following genes were also studied by PCR: tet(A), tet(B),tet(C), tet(D), and tet(E) (in tetracycline resistant isolates), aadA and strA/B (in streptomycin and tobramycin resistant isolates), aac(3)-IV (in gentamicin, tobramycin and amikacin-resistant isolates), cmlA (in chloramphenicol-resistant isolates), and sul1, sul2, and sul3 (in trimethoprim–sulfamethoxazole-resistant isolates). The quinolone resistance-determining region (QRDR) of the gyrA gene was amplified in nalidixic acid resistant isolates [32] followed by sequencing using the same set of primers used for the PCR reactions. Sequences obtained were compared with those previously reported for gyrA (GenBank). The gene specific primers, their expected fragment sizes, and source references are listed in Table A1 (Appendix A) and were purchased from Thermofisher (Waltham, MA, USA). The gene products were visualized in ethidium bromide stained 1.5% agarose gel using UV light. A subset of PCR amplified fragment of the resistant genes were purified directly from the PCR reaction or after gel electrophoresis. The fragments were sequenced at MGH CCIB DNA Core (Cambridge, MA) and the resulting DNA sequence data were compared with corresponding genes in the GenBank database using the basic local alignment search tool (BLAST) of the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov) to confirm identity of antimicrobial resistance genes.
2.6. Detection of E. coli Virulence Genes and Phylogenetic Grouping
Resistant isolates were screened for four virulence genes commonly found in pathogenic E. coli; shiga toxins 1 and 2 (stx1 and stx2), hemolysin (hly), and also intimin (eae) by a polymerase chain reaction using gene specific primers listed in Table A1. The phylogenetic grouping of the isolates was also determined using primers targeting the chuA, yjaA, or tspE4.C2 genes as previously described [33].
2.7. Statistical Analysis
Proportions of resistant isolates were calculated for all the groups sampled. Differences in proportion of resistant isolates among sampling groups were tested using the MedCalc N-1 Chi-squared Comparison of proportions calculator 2018 [34]. A P value of <0.05 was considered significant for all comparisons. Results of the statistical analysis are shown in Table A2 (Appendix B).
3. Results
3.1. Antimicrobial Resistance Proportions and Resistance Phenotypes
A total of 408 isolates that included (196) from both nursing does and kids up to weaning day, twenty-two (22) from kids one week after weaning, fifty-four (54) from goat kids at six months, forty-three (43) from goat kids at one year old, and ninety-three (93) from other goats in the flock were screened for antimicrobial resistance. We detected antibiotic resistant E. coli isolates in all age groups sampled in the absence of use of antibiotics in the study animals during the previous one year prior to initiation of the study and during the study period. In total, 136 isolates (33%) were resistant to one or more antibiotic, while 272 isolates (67%) were sensitive to all tested antibiotics. Most of the isolates 105 (77%) showed resistance to a single antibiotic, while 27 (20%) showed resistance to two antibiotics, and three (2.2%) isolates to three antibiotic and only one isolate with resistance to four antibiotics (Table 1).

Table 1.
Number and percentages of antibiotic resistance phenotypes in 136 resistant E. coli isolates from pastured meat goats.
The highest resistance was reported for tetracycline (51%), followed by streptomycin (30%), ampicillin (19%), nalidixic acid (9%), amoxicillin/clavulanate (5%), and Chloramphenicol (5%), while for all the other antibiotics, resistance was less than 5% (Table 2).

Table 2.
Percentage of E. coli isolates from pastured meat goats showing each individual antibiotic resistance.
In the group of isolates showing resistance to two or more antibiotics (31), most showed a combination of tetracycline and streptomycin (12) (39%), followed by ampicillin/amoxicillin clavulanate (3) (10%), while other combinations were less than 10% (Table 1). Overall, isolates with resistance to two or more antibiotics tended to be detected in older goats (6 months and older) more frequently than in younger goats (less than 6 months) (Table 3).

Table 3.
Animal groups sampled and resistance phenotypes detected in 408 E. coli isolates from pastured meat goats.
3.2. Proportions of Resistant E. coli Isolates and Phenotypes Among Age Groups and at Different Farm Location
The proportion of antibiotic resistant E. coli isolates and resistance phenotypes detected differed among age groups. Resistant E. coli isolates in nursing goat kids ranged from 43% to 48% while that detected in the nursing does ranged from 8−25% up to 13 weeks of age (Figure 1).
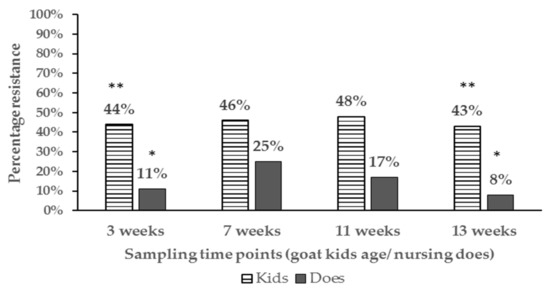
Figure 1.
Bar chart illustrating percentage of resistant E. coli isolates in pastured goat kids and the respective nursing does up to weaning. Proportions at the same sampling points with different number of asterisks ** vs. * are significantly different (P < 0.05).
In general, the proportion of antibiotic resistant E. coli tended to be higher in goat kids compared to the does irrespective of location at the farm and significant differences (P < 0.05) were detected at 3 weeks and 13 weeks of age (Appendix A for table of statistical analysis results). Additionally, the percentage of resistant isolates in goat kids decreased with age, with significant differences being detected between goat kids at 3 and 7 weeks compared to one year of age (46% vs. 12%) respectively (Figure 2).
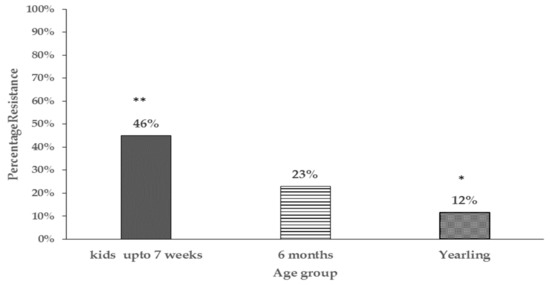
Figure 2.
Bar chart comparing percentage of resistant E. coli isolates in different age groups of pastured goat kids (only goat kids sampled at pasture are represented). Proportions with different number of asterisks ** vs. * are significantly different (P < 0.05).
We also observed unique patterns in resistance phenotype detected in E. coli over the growing period. We noted the resistance detected in young goat kids at 3 weeks of age was mostly of intermediate type (PR). Isolates with intermediate resistance however decreased over time and by the time the kids were 13 weeks and beyond, resistance detected tended to be full resistance (FR) (Figure 3).
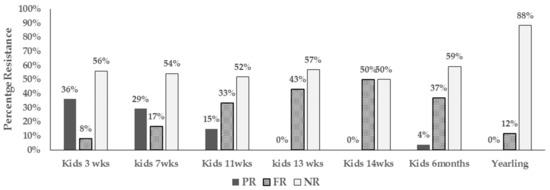
Figure 3.
Bar chart illustrating types of resistant (intermediate vs full resistance) in E. coli from pastured goat kids from 3 weeks of age to one year (PR-Intermediate resistance, FR-Full resistance, and NR-non-resistance).
This study also found that the resistance phenotype in nursing goat kids was unrelated to that detected in isolates from the respective nursing does despite the close interaction and shared ecosystem throughout the pre-weaning period. Resistance to streptomycin was the most prevalent during the first 14 weeks of age and was equally detected in both kids and does. However as mentioned above, resistance detected in young goat kids was mostly intermediate while that detected in nursing does was full resistance. Repeated fecal samplings also did not reveal persistence colonization by resistant isolates in any of the sampled animals over the study period. Instead, different resistance phenotype was detected in subsequent samplings and sometimes no resistant isolates was recovered at all in the same animal.
The proportion of E. coli isolates with antibiotic resistance phenotypes and also the type of resistance phenotype differed depending on the location of the animals (pasture vs paddocks around housing facility). A significantly higher percentage (P < 0.05) of antibiotic resistant isolates was detected in animals sampled while grazing at the paddocks surrounding the housing facility compared to animals sampled at pasture (Table 4). Additionally, tetracycline resistance was the predominant phenotype (31%) detected in isolates from animals at the facility compared to those located at pasture (2.5%). On the other hand, a significantly higher percentage of ampicillin resistant E. coli was detected in isolates from animals at pasture (11%) compared to those grazing around the housing facility (2%). The percentage of isolates resistant to streptomycin, nalidixic acid, chloramphenicol amoxicillin/clavulanate, and tobramycin did not differ significantly between E. coli isolates from animals at pasture or near the facility.

Table 4.
Resistance phenotype and the respective percentages of E. coli isolates detected in pastured goats at different locations at the research farm.
3.3. Antibiotic Resistance Mechanisms
To elucidate on possible mechanisms of antibiotic resistance in the E. coli from the goats in the study, all resistant isolates were screened for commonly encountered resistant genes for each antibiotic. The resistant genes detected in resistant isolates are shown in Table 5. Among 70 tetracycline resistant isolates, only tetB gene was detected in 69 isolates (97%) and no gene was detected in the other resistant isolate. In all ampicillin resistant isolates (28) the β- lactamases genes; blaTEM, blaOXA, and blaSHV were screened. The blaTEM gene was detected in twelve of the isolates (43%), while no gene was detected in the remaining isolates. One isolate harbored both the blaTEM and the blaSHV gene. The aminoglycoside modifying adenyltransferase enzyme gene aadA was detected in 18 of the 41 (44%) streptomycin resistant isolates. The strpA/strpB gene was also detected in ten (24%) of the streptomycin resistant isolates either alone or in combination with the aadA gene. Five streptomycin resistant isolates had both the aadA and strpA/B genes detected. The one isolate with resistance to sulfamethoxazole-trimethoprim harbored the sul2 gene while one isolate out of two with tobramycin resistance had the aminoglycoside acetyltransferase enzyme gene aac3 (iv) detected. In the four isolates with amikacin resistance and seven with chloramphenicol resistance, no antimicrobial resistance gene was detected using the primers used in this study. Mechanism of resistance to nalidixic acid was evaluated by sequencing of the amplified fragment of E. coli gyrase gene. All resistant isolates showed a substitution mutation of both the Serine 83 leucine and asparagine 87 aspartate amino acids.

Table 5.
Resistance genes detected among antibiotic resistant E. coli isolates of pastured goats origin.
3.4. Virulence Genes and Phylogenetic Grouping of Resistant Isolates
Four genes commonly associated with E. coli virulence in both animals and human were detected in resistant isolates in varying proportions. In one hundred and four (104) resistant isolates screened, shiga toxin 1(stx1), shiga toxin 2 (stx2), intimin (eae), and hemolysin (hly) genes were detected either individually or in combination in 84 (81%) of the resistant isolates (Figure 4). The most common virulence genes detected in the resistant isolates was for shiga toxin 1 (63%), followed by hemolysin (50%), and finally intimin (25%). Shiga toxin 2 (stx2) gene was only detected in two isolates. Resistant E. coli isolates were assigned to the four different phylogenetic groups based on the presence of the chuA, yjaA or tspE4.C2 genes in different combinations. Sixty percent of the isolates (60%) belonged to the D group, 28% belonged to B1, 9% to B2, and 3% to the A phylogenetic groups (Figure 5).
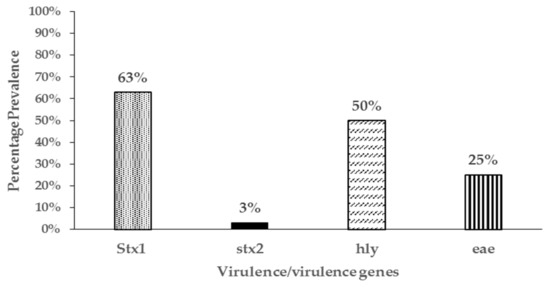
Figure 4.
Prevalence of virulence genes in 104 antibiotic resistant E. coli isolates from pastured meat goats.
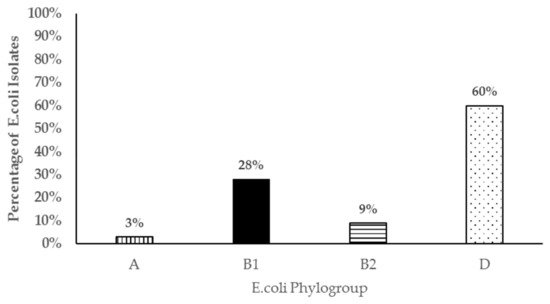
Figure 5.
Bar chart representing phylogenetic groups of 104 antibiotic resistant E. coli isolates from pastured meat goats.
4. Discussion
Surveillance of antibiotic resistance in food animals continues to be of importance worldwide due to the risk of spread of antibiotic resistance determinants to human pathogens, both zoonotic and non-zoonotic. Understanding the status of antibiotic resistance in animals is also important in veterinary medicine for disease control. Little is known on status or patterns of antibiotic resistance in small ruminants in North America, including the US [22], and little surveillance on the status of antimicrobial resistance in these species is reported globally.
Interestingly in this study, resistant E. coli isolates were detected in young goat kids on pasture as early as three weeks of age in the absence of use of any antibiotics in the goat kid cohort or the nursing does for the previous one year. While this was a unique finding not previously reported in goats, other studies have reported early colonization by resistant E. coli in young food animals of other species even before exposure to specific antibiotic or shortly after administration [35,36]. Furthermore, antibiotic resistant isolates were also reported in pristine ecosystems that have not been exposed to antibiotics before [2,36], which suggests that multiple factors may be responsible for antimicrobial resistance acquisition and spread in the environment. The role of horizontal transfer of resistant elements from environmental reservoirs in the soil or water or spread by wild animals has been thought to play a role [2] in some ecosystems, which may be the case in the current study, since the animals had not been treated with any antibiotics. In other studies, factors not associated with direct antimicrobial use, including geographic location, housing, animal age, and purpose of production, were also found to have an effect on the prevalence of E. coli resistant to antibiotics [7,9]. In this study, higher proportions of resistant E. coli isolates were detected in young goat kids than in nursing does in the same environment throughout the pre-weaning period. Prevalence of resistant isolates in the goat kids remained high but decreased post weaning, reaching significantly lower levels by one year of age. Our findings are similar to those reported by many other authors [9,35,37,38,39] who detected higher colonization by resistant E. coli in young calves and pigs that declined with age of the animals. Our findings also agree with Berge et al 2010 finding in cattle who reported a higher prevalence of antimicrobial resistance in younger animals than older animals and also early colonization by resistant isolates in the absence of antibiotic use. We also found that while most of the isolates found in the young animals at 3 weeks of age were of intermediate resistance, this pattern of colonization changed over time and by the time the animals were three months and beyond, most resistant E. coli were of the full resistance phenotype. It is unclear why this pattern would occur although our hypothesis is animal (more mature immune system) in combination with bacterial factors (lower fitness of strains with intermediate resistance compared to other gut bacteria) may play a role in reduced survival of isolates with intermediate resistance in older hosts. To our knowledge, this phenomenon has not been studied or reported in other studies. We also found that the resistant phenotypes found in goat kids were unrelated to those detected in the nursing does at all sampling points during the pre-weaning period despite sharing the same ecosystem. Furthermore, resistant phenotypes detected in individual animals during the same period varied between sampling time points. This pattern may imply that acquisition and colonization by resistant E. coli isolates was an independent dynamic event and persistence in individual animals did not occur under the study conditions.
Antibiotic resistance to a broad range and groups of antibiotics were detected in E. coli in goats involved in this study either singly or in combination. Detected resistance included to tetracycline, streptomycin, ampicillin, amoxicillin/clavulanic acid, nalidixic acid, tobramycin, gentamicin, and chloramphenicol. As reported in other studies in food animals, most resistance was against tetracycline, streptomycin, ampicillin, and amoxicillin/clavulanic acid, while resistance to other antibiotics was low [40]. Resistance to tetracycline and streptomycin are the most commonly detected antibiotics in food animals, which is as expected because they have been in the market and use in food animals for a long time. In the current study, there was a history of previous use of tetracycline for treatment on the farm albeit over a year before the study commenced. It is possible that bacteria isolates harboring tetracycline resistance existed or persisted in the soil or in the gut of some older animals and colonized the study animals during grazing. This is supported by studies that have shown that once bacteria develop resistance to antibiotics, this phenotype can be detected as long as four years after cessation of the antibiotic use [34,41]. Persistence of tetracycline genes for several months in the soil amended or in contact with animal manure has also been reported in some studies [11,42]. In this study, some older animals that had been on site for over one year harbored tetracycline resistant isolates. Resistance to β-lactams, especially ampicillin, was detected in a significant number of E. coli isolates in this study despite no previous use of this antibiotic in the farm. This indicates that this kind of resistance may have been acquired from other environmental sources unrelated to activities on the farm. Our finding are similar to other reported results of resistance to ampicillin and other antibiotics reported in wild animals unrelated to known exposure to these antibiotics [4].
Differences in proportion and phenotype of antibiotic resistant E. coli was reported depending on whether goats were located on pasture or on smaller paddocks near holding facility. Significantly higher proportions of antibiotic resistant E. coli was detected in animals located at or near the housing facility than at pasture. This resistance was mostly due to E. coli resistant to tetracycline which was also more frequently detected in fecal samples from animals grazing near or at the housing facility than animals on pasture. Although not tested, it is possible that there was a higher level of antibiotic resistant bacteria in the immediate environment (soil) near the housing facility as a result of frequent congregation of animals during treatment resulting in a high load of resistant microbes in the soil. Studies have shown that soils exposed to manure with antibiotic resistant bacteria are persistent sources of antibiotic resistant bacteria [42]. On the other hand, E. coli isolates resistant to ampicillin and other antibiotics were more frequently isolated from animals on pasture than those near the facility pointing to a distant environmental source than on farm use. The location of the pasture is frequently visited by migratory birds due to the presence of fish ponds but also deer from adjacent woodlands. Additionally, there are human settlements surrounding the research farm. Thus, it may be that resistance to these antibiotics may have been spread to the farm by these wild animals from other places or surface water run-off from distant places [43,44]. In a previous study involving small ruminants from the current study farm and wild birds captured in the farm vicinity, Salmonella and Campylobacter isolates were found to have antimicrobial resistance to a wide range of antibiotics [45].
Most of the isolates in this cohort were resistant to either one or two antibiotics, while a few isolates had resistance to three antibiotics and one isolate to four antibiotics. This is a significant finding, given the fact there was no use of antibiotics in the cohort throughout the study period and these are animals predominantly reared on pasture. Resistant isolates found in these animals may reach far sites through surface water and runoff. Studies in poultry, swine, and cattle also report a high percentage of isolates with resistance to more than one antibiotic (reviewed in Reference [18]) in the US too. However, these animals are reared under intensive production systems in the US and in the past have been associated with relatively higher levels of use of antibiotics.
The antibiotic resistance genes detected in the E. coli isolates in this study are similar to those found in other studies. Most of the antibiotic resistant genes detected in the study are located in plasmids and transposons that can be transferred between bacteria indicating the potential of goats as reservoirs of antibiotic resistant genes in the environment. The finding could also offer insight on the potential source of the resistant bacteria in absence of antibiotic use in these animals; an environmental pool of resistant genes in plasmids. In particular, tetB gene, which codes for a tetracycline efflux protein [46] was the only gene detected in tetracycline resistant isolates in this study. TetB was also the most frequent gene detected in E. coli isolates from pigs, chicken, and turkey although tetA was also detected [16,17,47]. Similarly, in a study involving E. coli isolates from companion animals, tetB gene was the most frequently detected [48]. Resistance to ampicillin in this study, was found to be mainly due to the bla-TEM gene which codes for beta-lactamases. The significance of this form of resistant mechanism is the fact that bla-TEM genes are located in transposons found in plasmids which play a key role in their dispersal among bacteria [49]. Our findings are in agreement with other studies involving E. coli isolates from foods of animal origin, healthy animals, and human that found that this gene was the most frequently detected in over 80% of ampicillin resistant E. coli [50], a Danish study in food animals found the same gene in 91% of ampicillin resistant isolates [51] and a study in Portugal involving companion animals detected the gene in 70% of ampicillin resistant isolates [48]. In contrast Srinivasan reported a high frequency of the inducible Amp C (91%) gene in E. coli isolates from mastitic cows that were resistant to ampicillin, indicating other possible mechanisms of ampicillin resistance in E. coli. In isolates resistant to streptomycin, the adenyl transferase mechanism of aminoglycoside inactivation by the aadA gene product was the most frequently detected in this study. The strepA/strepB gene was also detected in a few streptomycin resistant isolates. Similar to our findings, aadA gene was also detected in streptomycin E. coli isolates in companion animals [48] and also human and food animals [3]. Additionally, Srinivasan et al [52] reported high frequency for aadA (51%) gene and strepA/strepB genes (21.6%) in E. coli isolates from cows with mastitis. Isolates with tobramycin resistance had the aminoglycoside modifying enzyme (acetyl transferase) aac(3)IV gene detected which is known to responsible for resistance to tobramycin and amikacin [43] and was also reported in apramycin resistant E. coli from farm workers and farm animals [53]. The Serine 83 leucine and aspartate 87 asparagine mutations detected in nalidixic acid resistant isolates were similar to those reported in isolates from dogs in previous studies [48] and also retail meats in the US [54].
The concurrent detection of virulent genes in isolates that were also resistant to antibiotics was another significant finding in this study as they may be potential animal or public health pathogens. While E. coli isolates possessing the stx1 and hly, eae, and hly virulent genes have been found in both healthy and diarrheic small ruminants, the full significance of these isolates has not been elucidated [53,55]. However, isolates possessing the eae and/or stx2 genes are known to be pathogenic in both animals and human. In this study, most resistant isolates belonged to the phylogenetic group D followed by group B1. However, we also detected resistant E. coli isolates that belonged to the B2 phylogenetic group in which most virulent extra-intestinal strains are known to belong. Those belonging to phylogenetic group B1 and D may also be potential intestinal pathogens further underscoring the significance of the resistant E. coli isolates detected in this study. Contrary to our study, Camilla et al 2010 [56] did not find any E. coli strains belonging to phylogenetic D or B2 in E. coli isolates from goats in their study. This may be due to the small number of isolates analyzed in Reference (16) compared to our study (104). To our knowledge, this is the first study reporting on the temporal dynamics, shedding patterns, and significance of antimicrobial resistant E. coli from pastured goats.
5. Conclusions
The study findings indicate that pastured goats, despite low exposure to antibiotics, are colonized by antibiotic resistant bacteria early in life. Additionally, it highlights shedding dynamics with higher proportions of fecal shedding of antibiotic resistant E. coli by younger goat kids compared to older animals. Higher proportions of resistant isolates were also detected in animals congregated in paddocks that have been under intensive animal use/treatment for a long time compared to animals spread out on pasture. The study also found colonization by bacteria resistant to antibiotics that had never been used on the farm, indicating a source from a broader environmental resistance gene pool. The genes detected in resistant isolates were similar to genes detected in other food and companion animals and human resistant E. coli isolates. A significant number of resistant E. coli isolates from the goats also belonged to phylogenetic groups of public health importance and harbored virulent genes further emphasizing the significance of the isolates. Findings underscore the fact that small ruminants are reservoirs of antibiotic resistant E. coli that are also potentially pathogenic and could spread antibiotic resistant genes to other gut pathogenic or commensal bacteria and also to the environment.
Author Contributions
Conceptualization E.N., methodology, E.N.; formal analysis, E.N. and H.A.; investigation, E.N., H.A., A.A.A., C.K.; resources, E.N., C.K.; data curation, E.N., H.A.; writing—original draft preparation, E.N., H.A. writing—review and editing, E.N., H.A., C.K., P.K; visualization, E.N. H.A.; supervision, E.N., C.K., P.K.; project administration, E.N.; funding acquisition, E.N.
Funding
This research was funded by USDA NIFA Evans Allen funds through Agricultural Research Station, Virginia State University, USA.
Acknowledgments
The authors wish to acknowledge the support given by Virginia State University herd manager Amanda Miller and her team for the assistance in animal management and handling.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Primers used in study.
Table A1.
Primers used in study.
| Primer Name | Sequence (5’ 3’) | Target Gene(s) or Region | Amplicon Size | Ref |
|---|---|---|---|---|
| TEM-F | TTCTTGAAGACGAAAGGGC | blaTEM | 1150 | [48] |
| TEM-R | ACGCTCAGTGGAACGAAAAC | |||
| SHV-SE | ACTCAAGGATGTATTGTG | blaSHV | 747 | [57] |
| SHV-AS | TTAGGGTTGCCAGTGCTCG | |||
| TEM-C | ATCAGCAATAAACCAGC | blaTEM | 516 | [58] |
| TEM-H | CCCCGAAGAACGTTTTC | |||
| HE605 | TTTCGTGTCGCCCTTATTCC | blaTEM | 690 | [59] |
| HE606 | CCGGCTCCAGATTTATCAGC | |||
| TEM-164.SE | TCGCCGCATACACTATTCTCAGAATGA | blaTEM | 447 | [57] |
| TEM-165.AS | ACGCTCACCGGCTCCAGATTTAT | |||
| Tet A-F | GTAATTCTGAGCACTGTCGC | tetA | 937 | [48] |
| Tet A-R | CTGTCCTGGACAACATTGCTT | |||
| Tet B-F | CTCAGTATTCCAAGCCTTTG | tetB | 416 | [48] |
| Tet B-R | CTAAGCACTTGTCTCCTGTT | |||
| Tet C-F | TCTAACAATGCGCTCATCGT | tetC | 570 | [48] |
| Tet C-R | GGTTGAAGGCTCTCAAGGGC | |||
| Tet D-F | ATTACACTGCTGGACGCGAT | tetD | 1104 | [48] |
| Tet D-R | CTGATCAGCAGACAGATTGC | |||
| Tet E-F | GTGATGATGGCACTGGTCAT | tetE | 1,179 | [48] |
| Tet E-R | CTCTGCTGTACATCGCTCTT | |||
| Str A-R | ATGGTGGACCCTAAAACTCT | streptA/B | 893 | [13] |
| Str B-F | CGTCTAGGATCGAGACAAAG | |||
| strA-F | CTTGGTGATAAGGCAATTC | strept A | 548 | [40] |
| strA-R | CCAATCGCAGATAGAAGGC | |||
| strA-F | GTCAAGGGATTGAAACC | streptA/B | 509 | [40] |
| strB-R | GGATCGTAGAACATATTGGC | |||
| AadA F | GTGGATGGCGGCCTGAAGCC | aadA | 525 | [40] |
| AadA R | AATGCCCAGTCGGCAGCG | |||
| Sul1-F | TGGTGACGGTGTTCGGCATTC | sul1 | 789 | [48] |
| Sul1-R | GCGAGGGTTTCCGAGAAGGCC | |||
| Sul2-F | CGGCATCAACATAACC | sul2 | 722 | [48] |
| Sul2-R | GTGTGCGGATGAAGTCAG | |||
| Sul3-F | GAGCAAGATTTTTGGAATCG | sul3 | 792 | [48] |
| Sul3-R. | CATCTGCAGCTAACCTAGGGCTTTGGA | |||
| GyrA-F | TACACCGGTCAACATTGAGG | gyr | 648 | [48] |
| Gyra-R. | TTAATGATTGCCGCCGTCGG | |||
| CmlA.F | TGTCATTTACGGCATACTCG | cml | 455 | [60] |
| CmlA.R | ATCAGGCATCCCATTCCCAT | |||
| aac(3)IV F | TGCTGGTCCACAGCTCCTTC | aac(3)IV | 653 | [61] |
| aac(3)IVR | CGGATGCAGGAAGATCAA | |||
| Stx1-a | TCTCAGTGGGCGTTCTTATG | stx1 | 338 | [62] |
| Stx1-b | TACCCCCTCAACTGCTAATA | |||
| Stx2-a | GCGGTTTTATTTGCATTAGC | stx2 | 115 | [62] |
| Stx2-b | TCCCGTCAACCTTCACTGTA | |||
| EAE-a | ATGCTTAGTGCTGGTTTAGG | eae | 248 | [62] |
| EAE-b | GCCTTCATCATTTCGCTTTC | |||
| HlyA-a | AGCTGCAAGTGCGGGTCTG | hly | 569 | [62] |
| HlyA-b | TACGGGTTATGCCTGCAAGTTCAC | |||
| ChuA.1 | GACGAACCA ACGGTCAGGAT | chuA | 279 | [33] |
| ChuA.2 | TGCCGCCAGTACC AAAGACA | |||
| YjaA.1 | TGAAGTGTCAGGAGACGCTG | yjaA | 211 | [33] |
| YjaA.2 | ATGGAGAATGCGTTCCTCAAC | |||
| TspE4C2.1 | GAGTAATGTCGGGGCATTCA | tspE4.C2 | 152 | [33] |
| TspE4C2.2 | CGCGCCAACAAAGTATTACG | |||
| EC16S-a | CCCCCTGGACGAAGACTGAC | E. coli 16s | 401 | [62] |
| EC16S-b | ACCGCTGGCAACAAAGGATA |
Appendix B

Table A2.
Statistical results of comparisons of proportions of antibiotic resistant isolates analyzed using MedCalc Software.
Table A2.
Statistical results of comparisons of proportions of antibiotic resistant isolates analyzed using MedCalc Software.
| Comparison Groups | Chi Square Value | P Value |
|---|---|---|
| Kids vs does 3 weeks | 5.282 | P = 0.0215 |
| Kids vs does 7 weeks | 2.03 | P = 0.1543 |
| Kids vs does 11 weeks | 3.287 | P = 0.0698 |
| Kids vs does 13 weeks | 5.151 | P = 0.0232 |
| Kids 7 weeks vs kids six months | 2.611 | P = 0.1061 |
| Kids 7 weeks vs kids one year | 9.734 | P = 0.0018 |
| Goats on pasture vs facility | 8.968 | P = 0.0027 |
| Tetracycline resistance on facility vs pasture | 57.654 | P < 0.0001 |
| Ampicillin resistance on pasture vs facility | 14.742 | P = 0.0001 |
References
- Bengtsson, B.; Greko, C. Antibiotic resistance—Consequences for animal health, welfare, and food production. Ups. J. Med. Sci. 2014, 119, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Schroeder, C.M.; Meng, J.; White, D.G.; McDermott, P.F.; Wagner, D.D.; Yang, H.; Simjee, S.; DebRoy, C.; Walker, R.D. Identification of antimicrobial resistance and class 1 integrons in Shiga toxin-producing Escherichia coli recovered from humans and food animals. J. Antimicrob. Chemother. 2005, 56, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Poeta, P.; Senz, Y.; Vinu, L.; Coelho, A.C.; Matos, M.; Rojo-Bezares, B.; Rodrigues, J.; Torres, C. Mechanisms of antibiotic resistance in Escherichia coli isolates recovered from wild animals. Microb. Drug. Resi. 2008, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Pallecchi, L.; Bartoloni, A.; Paradisi, F.; Rossolini, G.M. Antibiotic resistance in the absence of antimicrobial use: Mechanisms and implications. Expert Rev. Anti-Infect. Ther. 2008, 6, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Pallecchi, L.; Bartoloni, A.; Riccobono, E.; Fernandez, C.; Mantella, A.; Magnelli, D.; Mannini, D.; Strohmeyer, M.; Bartalesi, F.; Rodriguez, H. Quinolone resistance in absence of selective pressure: The experience of a very remote community in the Amazon forest. PLoS Negl. Trop. Dis. 2012, 6, e1790. [Google Scholar] [CrossRef] [PubMed]
- Berge, A.C.; Hancock, D.D.; Sischo, W.M.; Besser, T.E. Geographic, farm, and animal factors associated with multiple antimicrobial resistance in fecal Escherichia coli isolates from cattle in the western United States. J. Am. Vet. Med. Assoc. 2010, 236, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- De Verdier, K.; Nyman, A.; Greko, C.; Bengtsson, B. Antimicrobial resistance and virulence factors in Escherichia coli from Swedish dairy calves. Acta Vet. Scand. 2012, 54, 2. [Google Scholar] [CrossRef]
- Duse, A.; Waller, K.P.; Emanuelson, U.; Unnerstad, H.E.; Persson, Y.; Bengtsson, B. Risk factors for antimicrobial resistance in fecal Escherichia coli from preweaned dairy calves. J. Dairy Sci. 2015, 98, 500–516. [Google Scholar] [CrossRef]
- Ho, P.-L.; Wong, R.C.; Lo, S.W.; Chow, K.-H.; Wong, S.S.; Que, T.-L. Genetic identity of aminoglycoside-resistance genes in Escherichia coli isolates from human and animal sources. J. Med. Microbiol. 2010, 59, 702–707. [Google Scholar] [CrossRef]
- Hordijk, J.; Mevius, D.J.; Kant, A.; Bos, M.E.; Graveland, H.; Bosman, A.B.; Hartskeerl, C.M.; Heederik, D.J.; Wagenaar, J.A. Within-farm dynamics of ESBL/AmpC-producing Escherichia coli in veal calves: A longitudinal approach. J. Antimicrob. Chemother. 2013, 68, 2468–2476. [Google Scholar] [CrossRef] [PubMed]
- Kikuvi, G.; Schwarz, S.; Ombui, J.; Mitema, E.; Kehrenberg, C. Streptomycin and chloramphenicol resistance genes in Escherichia coli isolates from cattle, pigs, and chicken in Kenya. Microb. Drug Resi. 2007, 13, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Reid-Smith, R.J.; Jardine, C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 2009, 75, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 2003, 69, 6489–6494. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Silva, N.; Caniça, M.; Capelo-Martinez, J.L.; Brito, F.; Igrejas, G.; Poeta, P. High prevalence of antimicrobial-resistant Escherichia coli from animals at slaughter: A food safety risk. J. Sci. Food Agric. 2013, 93, 517–526. [Google Scholar] [CrossRef]
- Sengeløv, G.; Halling-Sørensen, B.; Aarestrup, F.M. Susceptibility of Escherichia coli and Enterococcus faecium isolated from pigs and broiler chickens to tetracycline degradation products and distribution of tetracycline resistance determinants in E. coli from food animals. Vet. Microbiol. 2003, 95, 91–101. [Google Scholar] [CrossRef]
- Smith, J.; Drum, D.; Dai, Y.; Kim, J.; Sanchez, S.; Maurer, J.; Hofacre, C.; Lee, M. Impact of antimicrobial usage on antimicrobial resistance in commensal Escherichia coli strains colonizing broiler chickens. Appl. Environ. Microbiol. 2007, 73, 1404–1414. [Google Scholar] [CrossRef]
- Tadesse, D.A.; Zhao, S.; Tong, E.; Ayers, S.; Singh, A.; Bartholomew, M.J.; McDermott, P.F. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg. Infect. Dis. 2012, 18, 741. [Google Scholar] [CrossRef]
- Witte, W. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents 2000, 16, 19–24. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Ding, L.J.; Fan, M.Z. Resistance patterns and detection of aac (3)-IV gene in apramycin-resistant Escherichia coli isolated from farm animals and farm workers in northeastern of China. Res. Vet. Sci. 2009, 87, 449–454. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.; Menzies, P. Antimicrobial resistance and small ruminant veterinary practice. Vet. Clin. North. Am. Food Anim. Pract. 2011, 27, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Motoi, Y.; Sato, M.; Maruyama, A.; Watanabe, H.; Fukumoto, Y.; Shimamoto, T. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl. Environ. Microbiol. 2007, 73, 6686–6690. [Google Scholar] [CrossRef] [PubMed]
- Roug, A.; Byrne, B.A.; Conrad, P.A.; Miller, W. Zoonotic fecal pathogens and antimicrobial resistance in county fair animals. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Sayah, R.S.; Kaneene, J.B.; Johnson, Y.; Miller, R. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic-and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 2005, 71, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. B: Biol. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef]
- Woolhouse, M.E.; Ward, M.J. Sources of antimicrobial resistance. Science 2013, 341, 1460–1461. [Google Scholar] [CrossRef]
- Espie, E.; Vaillant, V.; Mariani-Kurkdjian, P.; Grimont, F.; Martin-Schaller, R.; De Valk, H.; Vernozy-Rozand, C. Escherichia coli O157 outbreak associated with fresh unpasteurized goats’ cheese. Epidemiol. Infect. 2006, 134, 143–146. [Google Scholar] [CrossRef]
- Jacob, M.; Foster, D.; Rogers, A.; Balcomb, C.; Shi, X.; Nagaraja, T. Evidence of non-O157 Shiga toxin–Producing Escherichia coli in the feces of meat goats at a US slaughter plant. J. Food Prot. 2013, 76, 1626–1629. [Google Scholar] [CrossRef]
- Vu-Khac, H.; Cornick, N.A. Prevalence and genetic profiles of Shiga toxin-producing Escherichia coli strains isolated from buffaloes, cattle, and goats in central Vietnam. Vet. Microbiol. 2008, 126, 356–363. [Google Scholar] [CrossRef]
- Tobias, J.; Vutukuru, S.-R. Simple and rapid multiplex PCR for identification of the main human diarrheagenic Escherichia coli. Microbiol. Res. 2012, 167, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Sáenz, Y.; Zarazaga, M.; Briñas, L.; Ruiz-Larrea, F.; Torres, C. Mutations in gyrA and parC genes in nalidixic acid-resistant Escherichia coli strains from food products, humans and animals. J. Antimicrob. Chemother. 2003, 51, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Jernberg, C.; Löfmark, S.; Edlund, C.; Jansson, J.K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007, 1, 56. [Google Scholar] [CrossRef] [PubMed]
- Hoyle, D.V.; Shaw, D.J.; Knight, H.I.; Davison, H.C.; Pearce, M.C.; Low, J.C.; Gunn, G.J.; Woolhouse, M.E. Age-related decline in carriage of ampicillin-resistant Escherichia coli in young calves. Appl. Environ. Microbiol. 2004, 70, 6927–6930. [Google Scholar] [CrossRef] [PubMed]
- Pallecchi, L.; Lucchetti, C.; Bartoloni, A.; Bartalesi, F.; Mantella, A.; Gamboa, H.; Carattoli, A.; Paradisi, F.; Rossolini, G.M. Population structure and resistance genes in antibiotic-resistant bacteria from a remote community with minimal antibiotic exposure. Antimicrob. Agents Chemother. 2007, 51, 1179–1184. [Google Scholar] [CrossRef] [PubMed]
- Edrington, T.; Farrow, R.; Carter, B.; Islas, A.; Hagevoort, G.; Callaway, T.; Anderson, R.; Nisbet, D. Age and diet effects on fecal populations and antibiotic resistance of a multi-drug resistant Escherichia coli in dairy calves. Agric. Food Anal. Bacteriol. 2012, 2, 162–174. [Google Scholar]
- Hoyle, D.; Davison, H.; Knight, H.; Yates, C.; Dobay, O.; Gunn, G.; Amyes, S.; Woolhouse, M. Molecular characterisation of bovine faecal Escherichia coli shows persistence of defined ampicillin resistant strains and the presence of class 1 integrons on an organic beef farm. Vet. Microbiol. 2006, 115, 250–257. [Google Scholar] [CrossRef]
- Langlois, B.E.; Dawson, K.A.; Leak, I.; Aaron, D.K. Effect of age and housing location on antibiotic resistance of fecal coliforms from pigs in a non-antibiotic-exposed herd. Appl. Environ. Microbiol. 1988, 54, 1341–1344. [Google Scholar]
- Srinivasan, V.; Nam, H.-M.; Sawant, A.A.; Headrick, S.I.; Nguyen, L.T.; Oliver, S.P. Distribution of tetracycline and streptomycin resistance genes and class 1 integrons in Enterobacteriaceae isolated from dairy and nondairy farm soils. Microb. Ecol. 2008, 55, 184–193. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Jernberg, C.; Andersson, A.F.; Sjölund-Karlsson, M.; Jansson, J.K.; Engstrand, L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 2010, 5, e9836. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; LaPara, T.M. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 2007, 1, 191. [Google Scholar] [CrossRef] [PubMed]
- Finley, R.L.; Collignon, P.; Larsson, D.J.; McEwen, S.A.; Li, X.-Z.; Gaze, W.H.; Reid-Smith, R.; Timinouni, M.; Graham, D.W.; Topp, E. The scourge of antibiotic resistance: The important role of the environment. Clin. Infect. Dis. 2013, 57, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Radhouani, H.; Silva, N.; Poeta, P.; Torres, C.; Correia, S.; Igrejas, G. Potential impact of antimicrobial resistance in wildlife, environment and human health. Front. Microbiol. 2014, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Kim, C. Prevalence of antimicrobial resistance (AMR) in bacteria isolated from farm animals, wildlife, and food samples in the eastern United States between 2007 and 2013. EC Nutrition 2017, 7, 264–274. [Google Scholar]
- Roberts, M.C. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Bryan, A.; Shapir, N.; Sadowsky, M.J. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 2004, 70, 2503–2507. [Google Scholar] [CrossRef]
- Costa, D.; Poeta, P.; Sáenz, Y.; Coelho, A.C.; Matos, M.; Vinué, L.; Rodrigues, J.; Torres, C. Prevalence of antimicrobial resistance and resistance genes in faecal Escherichia coli isolates recovered from healthy pets. Vet. Microbiol. 2008, 127, 97–105. [Google Scholar] [CrossRef]
- Bailey, J.K.; Pinyon, J.L.; Anantham, S.; Hall, R.M. Distribution of the bla TEM gene and bla TEM-containing transposons in commensal Escherichia coli. J. Antimicrob. Chemother. 2011, 66, 745–751. [Google Scholar] [CrossRef]
- Briñas, L.; Zarazaga, M.; Sáenz, Y.; Ruiz-Larrea, F.; Torres, C. β-Lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 2002, 46, 3156–3163. [Google Scholar] [CrossRef]
- Olesen, I.; Hasman, H.; Møller Aarestrup, F. Prevalence of β-lactamases among ampicillin-resistant Escherichia coli and Salmonella isolated from food animals in Denmark. Microb. Drug. Resi. 2004, 10, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.; Gillespie, B.E.; Lewis, M.J.; Nguyen, L.T.; Headrick, S.I.; Schukken, Y.H.; Oliver, S.P. Phenotypic and genotypic antimicrobial resistance patterns of Escherichia coli isolated from dairy cows with mastitis. Vet. Microbiol. 2007, 124, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Fuente, R.D.L.; García, S.; Orden, J.A.; Ruiz-Santa-Quiteria, J.A.; Díez, R.; Cid, D. Prevalence and characteristics of attaching and effacing strains of Escherichia coli isolated from diarrheic and healthy sheep and goats. J. Am. Vet. Med. Assoc. 2002, 63, 262–266. [Google Scholar]
- Zhao, S.; Blickenstaff, K.; Bodeis-Jones, S.; Gaines, S.; Tong, E.; McDermott, P. Comparison of the prevalences and antimicrobial resistances of Escherichia coli isolates from different retail meats in the United States, 2002 to 2008. Appl. Environ. Microbiol. 2012, 78, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Cortés, C.; De la Fuente, R.; Blanco, J.; Blanco, M.; Blanco, J.; Dhabi, G.; Mora, A.; Justel, P.; Contreras, A.; Sanchez, A. Serotypes, virulence genes and intimin types of verotoxin-producing Escherichia coli and enteropathogenic E. coli isolated from healthy dairy goats in Spain. Vet. Microbiol. 2005, 110, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Carlos, C.; Pires, M.M.; Stoppe, N.C.; Hachich, E.M.; Sato, M.I.; Gomes, T.A.; Amaral, L.A.; Ottoboni, L.M. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 2010, 10, 161. [Google Scholar] [CrossRef]
- Monstein, H.J.; Östholm-Balkhed, Å.; Nilsson, M.; Nilsson, M.; Dornbusch, K.; Nilsson, L. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 2007, 115, 1400–1408. [Google Scholar] [CrossRef]
- Colom, K.; Pérez, J.; Alonso, R.; Fernández-Aranguiz, A.; Lariño, E.; Cisterna, R. Simple and reliable multiplex PCR assay for detection of bla TEM, bla SHV and bla OXA–1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 2003, 223, 147–151. [Google Scholar] [CrossRef]
- Bailey, J.K.; Pinyon, J.L.; Anantham, S.; Hall, R.M. Commensal Escherichia coli of healthy humans: A reservoir for antibiotic-resistance determinants. J. Med. Microbiol. 2010, 59, 1331–1339. [Google Scholar] [CrossRef]
- Sáenz, Y.; Brinas, L.; Domínguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 2004, 48, 3996–4001. [Google Scholar] [CrossRef]
- Boerlin, P.; Travis, R.; Gyles, C.L.; Reid-Smith, R.; Lim, N.J.H.; Nicholson, V.; McEwen, S.A.; Friendship, R.; Archambault, M. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 2005, 71, 6753–6761. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Clark, C.G.; Rodgers, F.G. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157: H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 2002, 40, 3613–3619. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).